Newborn Screening for Fabry Disease in Northeastern Italy: Results of Five Years of Experience
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
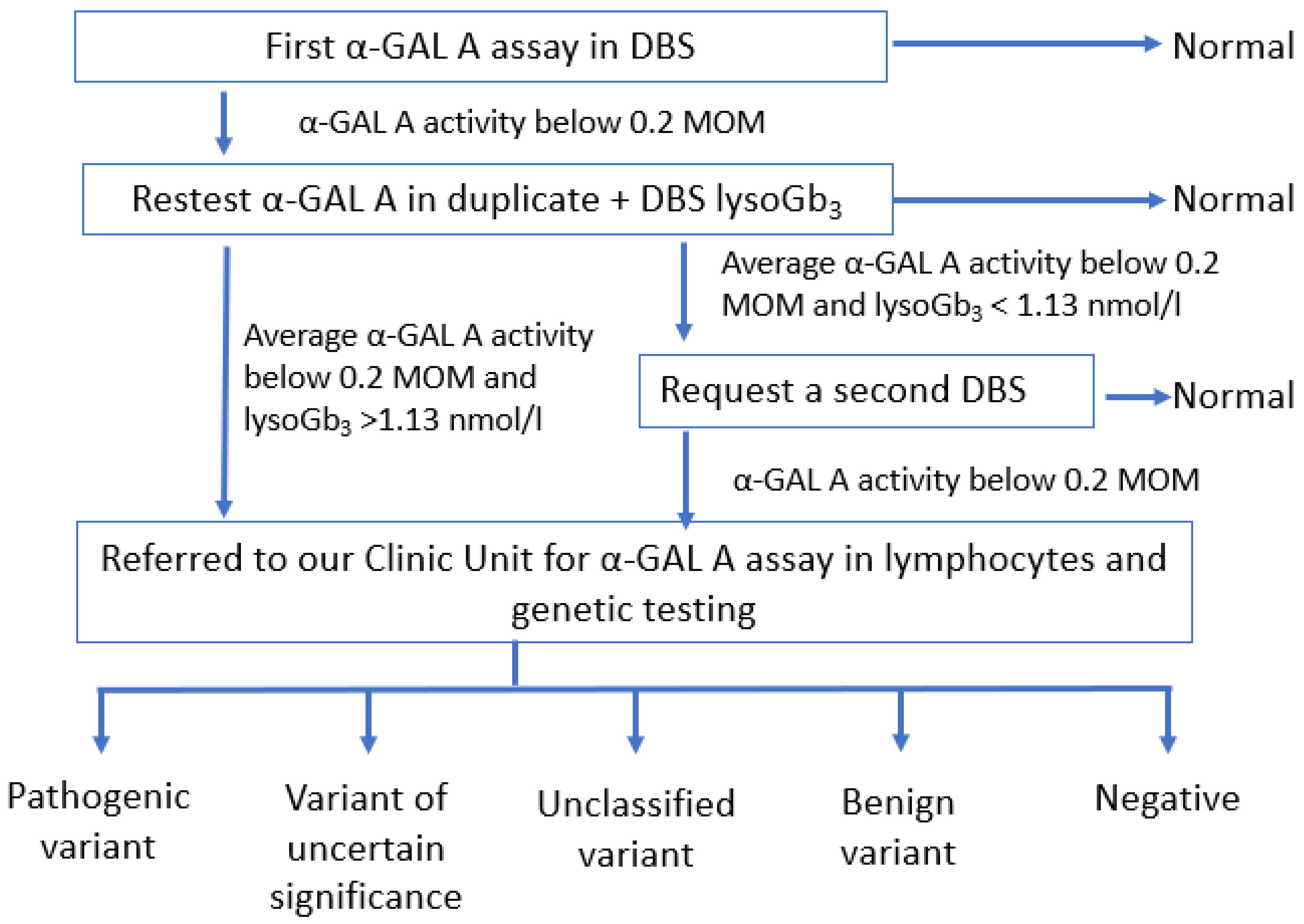
2.2. Methods
3. Results
3.1. Enzyme Activity
3.2. Genetic Testing
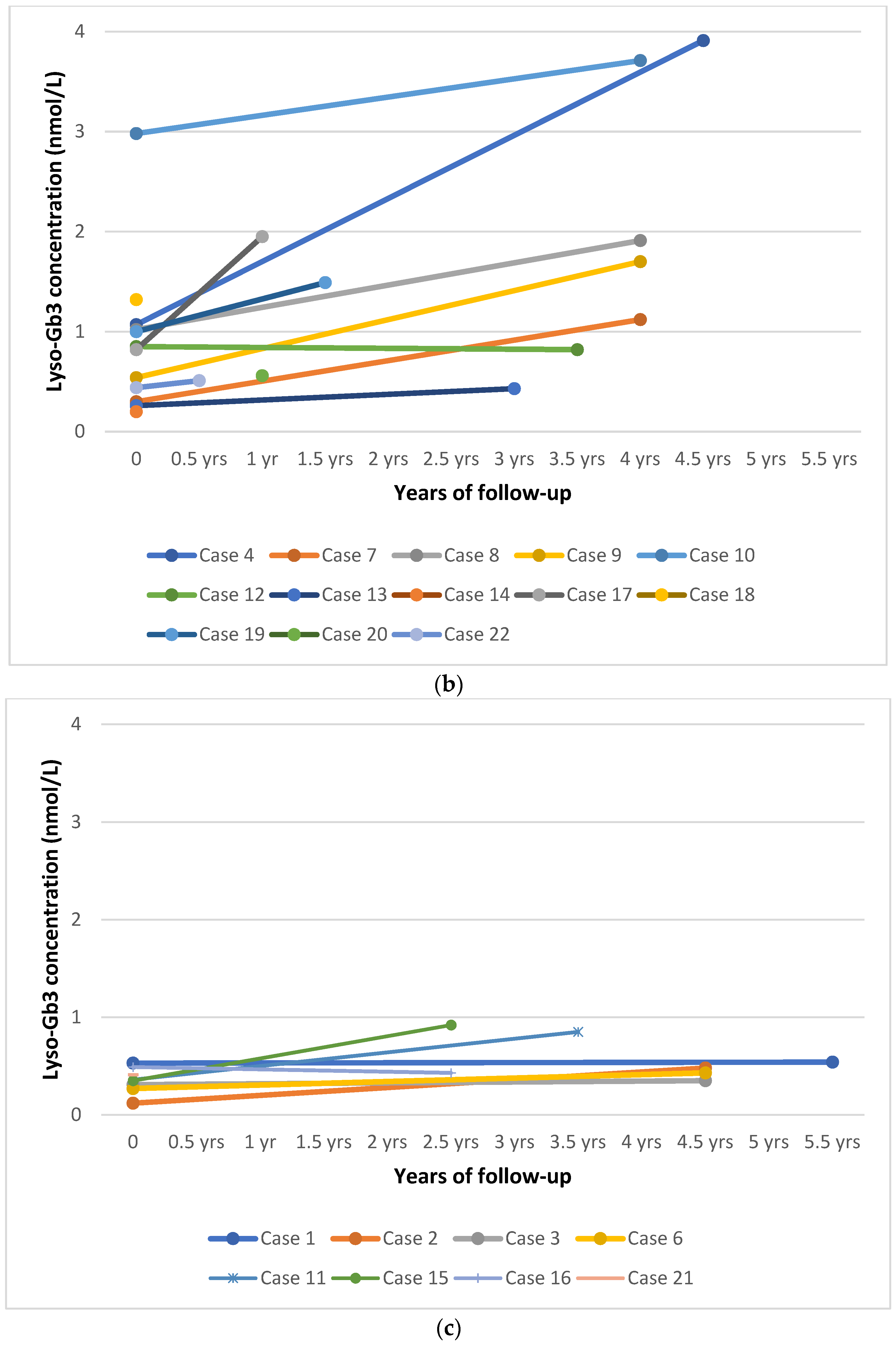
3.3. DBS Lyso-Gb3
3.4. Plasma Lyso-Gb3
3.5. Follow-Up
4. Discussion
4.1. Epidemiology
4.2. Interpretation of Genetic Variants
4.3. Clinical Follow-Up
4.4. Biomarkers and Biochemical
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Germain, D.P. Fabry disease. Orphan J. Rare Dis. 2010, 5, 30. [Google Scholar] [CrossRef]
- Kok, K.; Zwiers, K.C.; Boot, R.G.; Overkleeft, H.S. Fabry Disease: Molecular Basis, Pathophysiology, Diagnostics and Potential Therapeutic Directions. Biomolecules 2021, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Burlina, A.P.; Politei, J. Fabry disease. In Neurometabolic Hereditary Diseases of Adults; Burlina, A.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 67–98. [Google Scholar]
- Desnick, R.J.; Ioannou, Y.A.; Eng, C.M. A-Galactosidase A Deficiency: Fabry Disease; McGraw Hill: New York, NY, USA, 2021. [Google Scholar]
- Germain, D.P.; Brand, E.; Burlina, A.; Cecchi, F.; Garman, S.C.; Kempf, J.; Laney, D.A.; Linhart, A.; Maródi, L.; Nicholls, K.; et al. Phenotypic Characteristics of the p.Asn215Ser (p.N215S) GAL Mutation in Male and Female Patients with Fabry Disease: A Multicenter Fabry Registry Study. Mol. Genet. Genom. Med. 2018, 6, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Echevarria, L.; Benistan, K.; Toussaint, A.; Dubourg, O.; Hagege, A.A.; Eladari, D.; Jabbour, F.; Beldjord, C.; De Mazancourt, P.; Germain, D.P. X-chromosome inactivation in female patients with Fabry disease. Clin. Genet. 2016, 89, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Massaccesi, L.; Burlina, A.; Baquero, C.J.; Goi, G.; Burlina, A.P.; Tettamanti, G. Whole-Blood Alpha-D-Galactosidase A Activity for the Identification of Fabry’s Patients. Clin. Biochem. 2011, 44, 916–921. [Google Scholar] [CrossRef]
- Gal, A.; Beck, M.; Höppner, W.; Germain, D.P. Clinical utility gene card for Fabry disease—Update 2016. Eur. J. Hum. Genet. 2017, 25, e1–e3. [Google Scholar] [CrossRef]
- Rombach, S.M.; Dekker, N.; Bouwman, M.G.; Linthorst, G.E.; Zwinderman, A.H.; Wijburg, F.A.; Kuiper, S.; vd Bergh Weerman, M.A.; Groener, J.E.M.; Poorthuis, B.J.; et al. Plasma Globotriaosylsphingosine: Diagnostic Value and Relation to Clinical Manifestations of Fabry Disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2010, 1802, 741–748. [Google Scholar] [CrossRef]
- Polo, G.; Burlina, A.P.; Ranieri, E.; Colucci, F.; Rubert, L.; Pascarella, A.; Duro, G.; Tummolo, A.; Padoan, A.; Plebani, M.; et al. Plasma and Dried Blood Spot Lysosphingolipids for the Diagnosis of Different Sphingolipidoses: A Comparative Study. Clin. Chem. Lab. Med. 2019, 57, 1863–1874. [Google Scholar] [CrossRef]
- Effraimidis, G.; Feldt-Rasmussen, U.; Rasmussen, Å.K.; Lavoie, P.; Abaoui, M.; Boutin, M.; Auray-Blais, C. Globotriaosylsphingosine (Lyso-Gb3) and Analogues in Plasma and Urine of Patients with Fabry Disease and Correlations with Long-Term Treatment and Genotypes in a Nationwide Female Danish Cohort. J. Med. Genet. 2020. [Google Scholar] [CrossRef]
- Schiffmann, R.; Murray, G.J.; Treco, D.; Daniel, P.; Sellos-Moura, M.; Myers, M.; Quirk, J.M.; Zirzow, G.C.; Borowski, M.; Loveday, K.; et al. Infusion of alpha-Galactosidase A Reduces Tissue Globotriaosylceramide Storage in Patients with Fabry Disease. Proc. Natl. Acad. Sci. USA 2000, 97, 365–370. [Google Scholar] [CrossRef]
- Eng, C.M.; Guffon, N.; Wilcox, W.R.; Germain, D.P.; Lee, P.; Waldek, S.; Caplan, L.; Linthorst, G.E.; Desnick, R.J. For The International Collaborative Fabry Disease Study Group: Safety and efficacy of recombinant human alphagalactosidase A—replacement therapy in Fabry’s disease. N. Engl. J. Med. 2001, 345, 9–16. [Google Scholar] [CrossRef]
- Germain, D.P.; Arad, M.; Burlina, A.; Elliott, P.M.; Falissard, B.; Feldt-Rasmussen, U.; Hilz, M.J.; Hughes, D.A.; Ortiz, A.; Wanner, C.; et al. The effect of enzyme replacement therapy on clinical outcomes in female patients with Fabry disease—A systematic literature review by a European panel of experts. Mol. Genet. Metab. 2018, 126, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, A.; Germain, D.P.; Desnick, R.J.; Politei, J.; Mauer, M.; Burlina, A.; Eng, C.; Hopkin, R.J.; Laney, D.; Linhart, A.; et al. Fabry Disease Revisited: Management and Treatment Recommendations for Adult Patients. Mol. Genet. Metab. 2018, 123, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Germain, D.P.; Charrow, J.; Desnick, R.J.; Guffon, N.; Kempf, J.; Lachmann, R.H.; Lemay, R.; Linthorst, G.E.; Packman, S.; Scott, C.R.; et al. Ten-Year Outcome of Enzyme Replacement Therapy with Agalsidase Beta in Patients with Fabry Disease. J. Med. Genet. 2015, 52, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Chamoles, N.A.; Blanco, M.; Gaggioli, D. Fabry disease enzymatic diagnosis in dried blood spot on filter paper. Clinica Chimica Acta 2001, 308, 195–196. [Google Scholar] [CrossRef]
- Gelb, M.H.; Turecek, F.; Scott, C.R.; Chamoles, N.A. Direct Multiplex Assay of Enzymes in Dried Blood Spots by Tandem Mass Spectrometry for the Newborn Screening of Lysosomal Storage Disorders. J. Inherit. Metab Dis. 2006, 29, 397–404. [Google Scholar] [CrossRef]
- Zhang, X.K.; Elbin, C.S.; Chuang, W.-L.; Cooper, S.K.; Marashio, C.A.; Beauregard, C.; Keutzer, J.M. Multiplex Enzyme Assay Screening of Dried Blood Spots for Lysosomal Storage Disorders by Using Tandem Mass Spectrometry. Clin. Chem. 2008, 54, 1725–1728. [Google Scholar] [CrossRef]
- Sista, R.S.; Eckhardt, A.E.; Wang, T.; Graham, C.; Rouse, J.L.; Norton, S.M.; Srinivasan, V.; Pollack, M.G.; Tolun, A.A.; Bali, D.; et al. Digital Microfluidic Platform for Multiplexing Enzyme Assays: Implications for Lysosomal Storage Disease Screening in Newborns. Clin. Chem. 2011, 57, 1444–1451. [Google Scholar] [CrossRef]
- Mechtler, T.P.; Metz, T.F.; Müller, H.G.; Ostermann, K.; Ratschmann, R.; De Jesus, V.R.; Shushan, B.; Di Bussolo, J.M.; Herman, J.L.; Herkner, K.R.; et al. Short-Incubation Mass Spectrometry Assay for Lysosomal Storage Disorders in Newborn and High-Risk Population Screening. J. Chromatogr. B 2012, 908, 9–17. [Google Scholar] [CrossRef][Green Version]
- Scott, C.R.; Elliott, S.; Buroker, N.; Thomas, L.I.; Keutzer, J.; Glass, M.; Gelb, M.H.; Turecek, F. Identification of Infants at Risk for Developing Fabry, Pompe or Mucopolysaccharidosis-I from Newborn Blood Spots by Tandem Mass Spectrometry. J. Pediatr. 2013, 163, 498–503. [Google Scholar] [CrossRef]
- Hopkins, P.V.; Campbell, C.; Klug, T.; Rogers, S.; Raburn-Miller, J.; Kiesling, J. Lysosomal Storage Disorder Screening Implementation: Findings from the First Six Months of Full Population Pilot Testing in Missouri. J. Pediatr. 2015, 166, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Burton, B.K. Newborn Screening for Lysosomal Storage Disorders in Illinois: The Initial 15-Month Experience. J. Pediatr. 2017, 190, 130–135. [Google Scholar] [CrossRef]
- Wittmann, J.; Karg, E.; Turi, S.; Legnini, E.; Wittmann, G.; Giese, A.-K.; Lukas, J.; Gölnitz, U.; Klingenhäger, M.; Bodamer, O.; et al. Newborn Screening for Lysosomal Storage Disorders in Hungary. In JIMD Reports—Case and Research Reports, 2012/3; SSIEM, Ed.; JIMD Reports; Springer: Berlin/Heidelberg, Germany, 2012; Volume 6, pp. 117–125. [Google Scholar] [CrossRef]
- Mechtler, T.P. Neonatal Screening for Lysosomal Storage Disorders: Feasibility and Incidence from a Nationwide Study in Austria. Lancet 2012, 379, 335–341. [Google Scholar] [CrossRef]
- Colon, C.; Ortolano, S.; Melcon-Crespo, C.; Alvarez, J.V.; Lopez-Suarez, O.E.; Couce, M.L.; Fernández-Lorenzo, J.R. Newborn Screening for Fabry Disease in the North-West of Spain. Eur. J. Pediatr. 2017, 176, 1075–1081. [Google Scholar] [CrossRef]
- Burlina, A.B.; Polo, G.; Salviati, L.; Duro, G.; Zizzo, C.; Dardis, A.; Bembi, B.; Cazzorla, C.; Rubert, L.; Zordan, R.; et al. Newborn Screening for Lysosomal Storage Disorders by Tandem Mass Spectrometry in North East Italy. J. Inherit. Metab Dis. 2018, 41, 209–219. [Google Scholar] [CrossRef]
- Chien, Y.-H.; Lee, N.-C.; Chiang, S.-C.; Desnick, R.J.; Hwu, W.-L. Fabry Disease: Incidence of the Common Later-Onset α-Galactosidase A IVS4+919G→A Mutation in Taiwanese Newborns—Superiority of DNA-Based to Enzyme-Based Newborn Screening for Common Mutations. Mol. Med. 2012, 18, 780–784. [Google Scholar] [CrossRef]
- Inoue, T.; Hattori, K.; Ihara, K.; Ishii, A.; Nakamura, K.; Hirose, S. Newborn Screening for Fabry Disease in Japan: Prevalence and Genotypes of Fabry Disease in a Pilot Study. J. Hum. Genet. 2013, 58, 548–552. [Google Scholar] [CrossRef]
- Meikle, P.J. Prevalence of Lysosomal Storage Disorders. JAMA 1999, 281, 249. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hedge, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Spada, M.; Pagliardini, S.; Yasuda, M.; Tukel, T.; Thiagarajan, G.; Sakuraba, H.; Ponzone, A.; Desnick, R.J. High Incidence of Later-Onset Fabry Disease Revealed by Newborn Screening. Am. J. Hum. Genet. 2006, 79, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Hwu, W.-L.; Chien, Y.-H.; Lee, N.-C.; Chiang, S.-C.; Dobrovolny, R.; Huang, A.-C.; Yeh, H.-Y.; Chao, M.-C.; Lin, S.-J.; Kitagawa, T.; et al. Newborn Screening for Fabry Disease in Taiwan Reveals a High Incidence of the Later-Onset GLA Mutation c.936+919G>A (IVS4+919G>A). Hum. Mutat. 2009, 30, 1397–1405. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Chong, K.-W.; Hsu, J.-H.; Yu, H.-C.; Shih, C.-C.; Huang, C.-H.; Lin, S.-J.; Chen, C.-H.; Chiang, C.-C.; Ho, H.-J.; et al. High Incidence of the Cardiac Variant of Fabry Disease Revealed by Newborn Screening in the Taiwan Chinese Population. Circ. Cardiovasc. Genet. 2009, 2, 450–456. [Google Scholar] [CrossRef]
- Burton, B.; Charrow, J.; Angle, B.; Widera, S.; Waggoner, D. A pilot newborn screening program for lysosomal storage disease in Illinois. Mol. Genet. Metab. 2012, 105, S23–S24. [Google Scholar] [CrossRef]
- Paciotti, S.; Persichetti, E.; Pagliardini, S.; Deganuto, M.; Rosano, C.; Balducci, C.; Codini, M.; Filocamo, M.; Menghini, A.R.; Pagliardini, V.; et al. First Pilot Newborn Screening for Four Lysosomal Storage Diseases in an Italian Region: Identification and Analysis of a Putative Causative Mutation in the GBA Gene. Clin. Chim. Acta 2012, 5, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-C.; Chiang, C.-C.; Niu, D.-M.; Wang, C.-H.; Kao, S.-M.; Tsai, F.-J.; Huang, Y.-H.; Liu, H.-C.; Huang, C.-K.; Gao, H.-J.; et al. Detecting Multiple Lysosomal Storage Diseases by Tandem Mass Spectrometry—A National Newborn Screening Program in Taiwan. Clin. Chim. Acta 2014, 431, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Sanders, K.A.; Gavrilov, D.K.; Oglesbee, D.; Raymond, K.M.; Tortorelli, S.; Hopwood, J.J.; Lorey, F.; Majumdar, R.; Kroll, C.A.; McDonald, A.M.; et al. A Comparative Effectiveness Study of Newborn Screening Methods for Four Lysosomal Storage Disorders. IJNS 2020, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Chinen, Y.; Nakamura, S.; Yoshida, T.; Maruyama, H.; Nakamura, K. A New Mutation Found in Newborn Screening for Fabry Disease Evaluated by Plasma Globotriaosylsphingosine Levels. Hum. Genome Var. 2017, 4, 17002. [Google Scholar] [CrossRef]
- Liao, H.-C.; Hsu, T.-R.; Young, L.; Chiang, C.-C.; Huang, C.-K.; Liu, H.-C.; Niu, D.-M.; Chen, Y.-J. Functional and Biological Studies of α-Galactosidase A Variants with Uncertain Significance from Newborn Screening in Taiwan. Mol. Genet Metab. 2018, 123, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.-R. Later Onset Fabry Disease, Cardiac Damage Progress in Silence. J. Am. Coll. Cardiol. 2016, 68, 2554–2563. [Google Scholar] [CrossRef]
- Navarrete-Martínez, J.I.; Limón-Rojas, A.E.; Gaytán-García, M.d.J.; Reyna-Figueroa, J.; Wakida-Kusunoki, G.; Delgado-Calvillo, M.d.R.; Cantú-Reyna, C.; Cruz-Camino, H.; Cervantes-Barragán, D.E. Newborn Screening for Six Lysosomal Storage Disorders in a Cohort of Mexican Patients: Three-Year Findings from a Screening Program in a Closed Mexican Health System. Mol. Genet. Metab. 2017, 121, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.; Buroker, N.; Cournoyer, J.J.; Potier, A.M.; Trometer, J.D.; Elbin, C.; Schermer, M.J.; Kantola, J.; Boyce, A.; Turecek, F.; et al. Pilot Study of Newborn Screening for Six Lysosomal Storage Diseases Using Tandem Mass Spectrometry. Mol. Genet. Metab. 2016, 118, 304–309. [Google Scholar] [CrossRef]
- Camargo Neto, E.; Schulte, J.; Pereira, J.; Bravo, H.; Sampaio-Filho, C.; Giugliani, R. Neonatal Screening for Four Lysosomal Storage Diseases with a Digital Microfluidics Platform: Initial Results in Brazil. Genet. Mol. Biol. 2018, 41, 414–416. [Google Scholar] [CrossRef]
- Sawada, T.; Kido, J.; Yoshida, S.; Sugawara, K.; Momosaki, K.; Inoue, T.; Tajima, G.; Sawada, H.; Mastumoto, S.; Endo, F.; et al. Newborn Screening for Fabry Disease in the Western Region of Japan. Mol. Genet. Metab. Rep. 2020, 22, 100562. [Google Scholar] [CrossRef] [PubMed]
- Wasserstein, M.P.; Caggana, M.; Bailey, S.M.; Desnick, R.J.; Edelmann, L.; Estrella, L.; Holzman, I.; Kelly, N.R.; Kornreich, R.; Kupchik, S.G.; et al. The New York Pilot Newborn Screening Program for Lysosomal Storage Diseases: Report of the First 65,000 Infants. Genet. Med. 2019, 21, 631–640. [Google Scholar] [CrossRef]
- Chien, Y.-H. Newborn Screening for Morquio Disease and Other Lysosomal Storage Diseases: Results from the 8-Plex Assay for 70,000 Newborns. Orphanet J. Rare Dis. 2020, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- International Fabry Disease Genotype-Phenotype Database. Available online: www.dbfgp.org (accessed on 24 May 2021).
- Germain, D.P.; Oliveira, J.P.; Bichet, D.G.; Yoo, H.-W.; Hopkin, R.J.; Lemay, R.; Politei, J.; Wanner, C.; Wilcox, W.R.; Warnock, D.G. Use of a Rare Disease Registry for Establishing Phenotypic Classification of Previously Unassigned GLA Variants: A Consensus Classification System by a Multispecialty Fabry Disease Genotype–Phenotype Workgroup. J. Med. Genet. 2020, 57, 542–551. [Google Scholar] [CrossRef]
- Fabry-Gen-Phen: The Fabry Working Group Genotype Phenotype Database. Available online: http://fabrygenphen.com/ (accessed on 24 May 2021).
- Lavalle, L.; Thomas, A.S.; Beaton, B.; Ebrahim, H.; Reed, M.; Ramaswami, U.; Elliott, P.; Mehta, A.B.; Hughes, D.A. Phenotype and Biochemical Heterogeneity in Late Onset Fabry Disease Defined by N215S Mutation. PLoS ONE 2018, 13, e0193550. [Google Scholar] [CrossRef] [PubMed]
- Lenders, M.; Weidemann, F.; Kurschat, C.; Canaan-Kühl, S.; Duning, T.; Stypmann, J.; Schmitz, B.; Reiermann, S.; Krämer, J.; Blaschke, D.; et al. Alpha-Galactosidase A p.A143T, a Non-Fabry Disease-Causing Variant. Orphanet J. Rare Dis. 2016, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Koca, S.; Tümer, L.; Okur, İ.; Erten, Y.; Bakkaloğlu, S.; Biberoğlu, G.; Kasapkara, Ç.; Küçükçongar, A.; Dalgıç, B.; Oktar, S.Ö.; et al. High Incidence of Co-Existing Factors Significantly Modifying the Phenotype in Patients with Fabry Disease. Gene 2019, 687, 280–288. [Google Scholar] [CrossRef]
- Elliott, P.; Baker, R.; Pasquale, F.; Quarta, G.; Ebrahim, H.; Mehta, A.B.; Hughes, D.A.; On Behalf of the ACES Study Group. Prevalence of Anderson-Fabry Disease in Patients with Hypertrophic Cardiomyopathy: The European Anderson-Fabry Disease Survey. Heart 2011, 97, 1957–1960. [Google Scholar] [CrossRef] [PubMed]
- Brabander, I.D. Phenotypical Characterization of α-Galactosidase A Gene Mutations Identified in a Large Fabry Disease Screening Program in Stroke in the Young. Clin. Neurol. Neurosurg. 2013, 115, 1088–1093. [Google Scholar] [CrossRef]
- Terryn, W.; Vanholder, R.; Hemelsoet, D.; Leroy, B.P.; Van Biesen, W.; De Schoenmakere, G.; Wuyts, B.; Claes, K.; De Backer, J.; De Paepe, G.; et al. Questioning the Pathogenic Role of the GLA p.Ala143Thr “Mutation” in Fabry Disease: Implications for Screening Studies and ERT. In JIMD Reports—Case and Research Reports, 2012/5; Zschocke, J., Gibson, K.M., Brown, G., Morava, E., Peters, V., Eds.; JIMD Reports; Springer: Berlin/Heidelberg, Germany, 2012; Volume 8, pp. 101–108. [Google Scholar] [CrossRef]
- Krüger, R.; Tholey, A.; Jakoby, T.; Vogelsberger, R.; Mönnikes, R.; Rossmann, H.; Beck, M.; Lackner, K.J. Quantification of the Fabry Marker LysoGb3 in Human Plasma by Tandem Mass Spectrometry. J. Chromatogr. B 2012, 883–884, 128–135. [Google Scholar] [CrossRef]
- The Genome Aggregation Database. Available online: https://gnomad.broadinstitute.org (accessed on 24 May 2021).
- Oqvist, B.; Brenner, B.M.; Oliveira, J.P.; Ortiz, A.; Schaefer, R.; Svarstad, E.; Wanner, C.; Zhang, K.; Warnock, D.G. Nephropathy in Fabry Disease: The Importance of Early Diagnosis and Testing in High-Risk Populations. Nephrol. Dial. Transplant. 2009, 24, 1736–1743. [Google Scholar] [CrossRef]
- Ries, M.; Ramaswami, U.; Parini, R.; Lindblad, B.; Whybra, C.; Willers, I.; Gal, A.; Beck, M. The Early Clinical Phenotype of Fabry Disease: A Study on 35 European Children and Adolescents. Eur. J. Pediatr. 2003, 162, 767–772. [Google Scholar] [CrossRef]
- Germain, D.P.; Fouilhoux, A.; Decramer, S.; Tardieu, M.; Pillet, P.; Fila, M.; Rivera, S.; Deschênes, G.; Lacombe, D. Consensus recommendations for diagnosis, management and treatment of Fabry disease in paediatric patients. Clin. Genet. 2019, 96, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.M.; Groener, J.E.; Kuiper, S.; Donker-Koopman, W.E.; Strijland, A.; Ottenhoff, R.; van Roomen, C.; Mirzaian, M.; Wijburg, F.A.; Linthorst, G.E.; et al. Elevated Globotriaosylsphingosine Is a Hallmark of Fabry Disease. Proc. Natl. Acad. Sci. USA 2008, 105, 2812–2817. [Google Scholar] [CrossRef]
- Vedder, A.C.; Strijland, A.; Weerman, M.A.v.B.; Florquin, S.; Aerts, J.M.F.G.; Hollak, C.E.M. Manifestations of Fabry Disease in Placental Tissue. J. Inherit. Metab. Dis. 2006, 29, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Desnick, R.J. Prenatal Diagnosis of Fabry Disease. Prenat. Diagn. 2007, 27, 693–694. [Google Scholar] [CrossRef] [PubMed]
- Spada, M.; Kasper, D.; Pagliardini, V.; Biamino, E.; Giachero, S.; Porta, F. Metabolic Progression to Clinical Phenotype in Classic Fabry Disease. Ital. J. Pediatr. 2017, 43, 1. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, S.; Buchbinder, M. Patients-in-waiting: Living between sickness and health in the genomics era. J. Health Soc. Behav. J. Health Soc. Behav. 2010, 51, 408–423. [Google Scholar] [CrossRef]
- Lisi, E.; Ali, N. Opinions of adults affected with later-onset lysosomal storage diseases regarding newborn screening: A qualitative study. J. Genet. Couns. 2021. [Google Scholar] [CrossRef] [PubMed]
- Germain, D.P.; Moiseev, S.; Suárez-Obando, F.; Al Ismaili, F.; Al Khawaja, H.; Altarescu, G.; Barreto, F.C.; Haddoum, F.; Hadipour, F.; Maksimova, I.; et al. The benefits and challenges of family genetic testing in rare genetic diseases-lessons from Fabry disease. Mol. Genet. Genomic Med. 2021, e1666. [Google Scholar] [CrossRef]
- Burlina, A.B.; Polo, G.; Rubert, L.; Gueraldi, D.; Cazzorla, C.; Duro, G.; Salviati, L.; Burlina, A.P. Implementation of Second-Tier Tests in Newborn Screening for Lysosomal Disorders in North Eastern Italy. IJNS 2019, 5, 24. [Google Scholar] [CrossRef]
- Johnson, B.; Mascher, H.; Mascher, D.; Legnini, E.; Hung, C.Y.; Dajnoki, A.; Chien, Y.-H.; Maródi, L.; Hwu, W.-L.; Bodamer, O.A. Analysis of Lyso-Globotriaosylsphingosine in Dried Blood Spots. Ann. Lab. Med. 2013, 33, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.-R.; Niu, D.-M. Fabry Disease: Review and Experience during Newborn Screening. Trends Cardiovasc. Med. 2018, 28, 274–281. [Google Scholar] [CrossRef]
- Caudron, E.; Prognon, P.; Germain, D.P. Enzymatic diagnosis of Fabry disease using a fluorometric assay on dried blood spots: An alternative methodology. Eur. J. Med. Genet. 2015, 58, 681–684. [Google Scholar] [CrossRef]
- Tai, C.-L.; Liu, M.-Y.; Yu, H.-C.; Chiang, C.-C.; Chiang, H.; Suen, J.-H.; Kao, S.-M.; Huang, Y.-H.; Wu, T.J.-T.; Yang, C.-F.; et al. The Use of High Resolution Melting Analysis to Detect Fabry Mutations in Heterozygous Females via Dry Bloodspots. Clin. Chim. Acta 2012, 413, 422–427. [Google Scholar] [CrossRef]
- Lee, S.-H.; Li, C.-F.; Lin, H.-Y.; Lin, C.-H.; Liu, H.-C.; Tsai, S.-F.; Niu, D.-M. High-Throughput Detection of Common Sequence Variations of Fabry Disease in Taiwan Using DNA Mass Spectrometry. Mol. Genet. Metab. 2014, 111, 507–512. [Google Scholar] [CrossRef]
- Lu, Y.-H.; Huang, P.-H.; Wang, L.-Y.; Hsu, T.-R.; Li, H.-Y.; Lee, P.-C.; Hsieh, Y.-P.; Hung, S.-C.; Wang, Y.-C.; Chang, S.-K.; et al. Improvement in the Sensitivity of Newborn Screening for Fabry Disease among Females through the Use of a High-Throughput and Cost-Effective Method, DNA Mass Spectrometry. J. Hum. Genet. 2018, 63, 1–8. [Google Scholar] [CrossRef] [PubMed]
- MacDermot, K.D. Anderson-Fabry Disease: Clinical Manifestations and Impact of Disease in a Cohort of 60 Obligate Carrier Females. J. Med. Genet. 2001, 38, 769–775. [Google Scholar] [CrossRef]
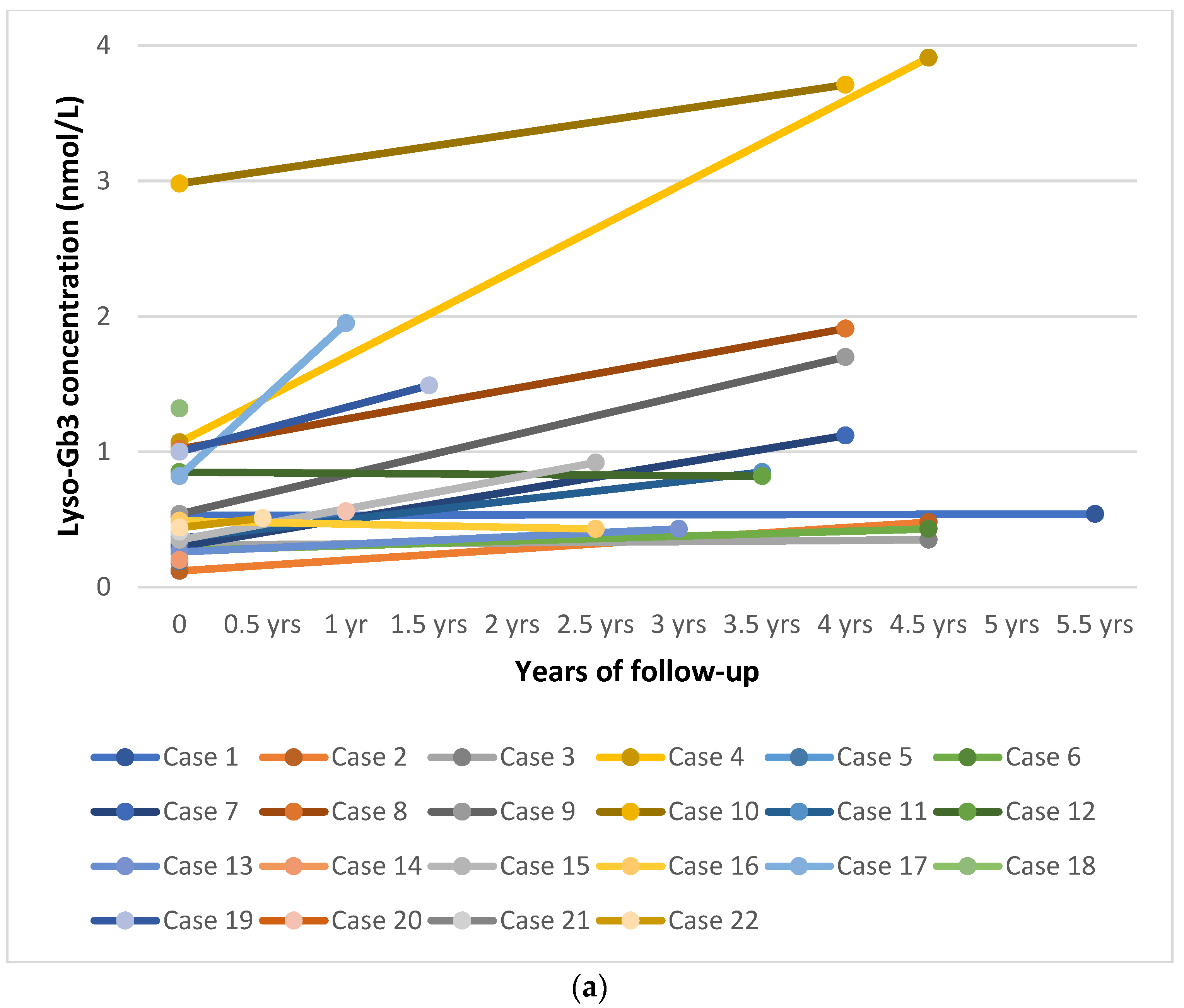
| Publication Year | Study Period | Region | Method | Number of NBS Samples | Positive NBS/ Patients Referred to Clinic | Confirmed Patients | Confirmed Male Patients | Reported Incidence * |
|---|---|---|---|---|---|---|---|---|
| 2006 | 2003–2005 | Italy [33] | Fluorometric enzyme assay | 37,104 (only males) | 12 (m) | 12 | 12 | 1:3,100 (m) |
| 2009 | 2006–2008 | Taiwan [34] | Fluorometric enzyme assay | 171,977 (m 90,288) | 94 (m 91) | 75 | 73 | 1:3821 (m 1:1237) |
| 2017 | 2008 | Spain [27] | Fluorometric enzyme assay | 14,600 (m 7575) | 106 (m 68) | 37 | 20 | 1:394 (m 1:378) ** |
| 2009 | 2008–2009 | Taiwan [35] | Fluorometric enzyme assay | 110,027 (m 57,451) | 67 (m 58) | 45 | 42 | 1:2445 (m 1:1368) |
| 2013 | 2007–2010 | Japan [30] | Fluorometric enzyme assay | 21,170 (m 10,827) | 7 (m 5) | 6 | 5 | 1:3,024 (m 1:2166) |
| 2012 | 2010 | Austria [26] | MS/MS | 34,736 (deidentified) | 28 | 9 | 6 | 1:3860 |
| 2012 | 2010–2011 | Illinois [36] | Digital microfluidics | 8012 | 11 | 7 | 6 | 1:1145 |
| 2012 | 2011 | Hungary [25] | MS/MS | 40,024 | 34 | 14 | 6 | 1:2858 |
| 2012 | 2010–2012 | Italy [37] | Fluorometric enzyme assay | 3403 (m 1702) | 0 | 0 | 0 | 0 |
| 2014 | 2010–2013 | Taiwan [38] | MS/MS | 191,767 | 79 | 64 | 61 | 1:2996 |
| 2020 | 2011–2013 | California [39] | MS/MS, immunocapture assay, digital microfluidics (comparative) | 89,508 (de-identified) (m 44,664) | Variable based on method | 50 | 46 | 1:1790(m 1:970) |
| 2013 | 2013 | Washington State [22] | MS/MS | 108,905 (deidentified) (m 54,800) | 16 (m 13) | 7 | 7 | 1:15558 (m 1:7800) |
| 2015 | 2013 | Missouri [23] | Digital microfluidics | 43,701 | 28 | 15 | 15 | 1:2913 |
| 2017 | 2007–2014 | Japan [40] | Fluorometric enzyme assay | 2443 | 2 | 2 | 2 | 1:1222 |
| 2018 | 2008–2014 | Taiwan [41] | Fluorometric enzyme assay, then MS/MS | 792,247 (m 412,299) | 764 (m 425) | 324 | 272 | 1:2445 (m 1:1515) |
| 2016 | 2008–2015 | Taiwan [42] | Fluorometric enzyme assay, then MS/MS | 916,383 (m 476,909) | 936 (m 505) | 441 | 324 | 1:2078 (m 1:1472) |
| 2017 | 2012–2016 | Petroleos Mexicanos Health Services [43] | MS/MS | 20,018 (m 10,241) | 5 | 5 | 5 | 1:4003 (m 1:2048) |
| 2017 | 2014–2016 | Illinois [24] | MS/MS | 219,793 | 107 | 32 | 32 | 1:6968 |
| 2016 | 2016 | Washington [44] | MS/MS | 43,000 (deidentified) | 8 | 5 | NA | 1:8600 |
| 2018 | 2015–2017 | Italy [28] | MS/MS | 44,411 | 5 | 5 | 5 | 1:8882 |
| 2018 | 2017 | Brazil [45] | Digital microfluidics | 10,527 | 0 | 0 | 0 | 0 |
| 2020 | 2006–2018 | Japan [46] | Fluorometric enzyme assay | 599,711 | 138 | 108 | 64 | 1:5552 |
| 2019 | 2013–2019 | New York [47] | MS/MS | 65,605 | 31 | 7 | 7 | 1:9372 |
| 2020 | 2018–2019 | Taiwan [48] | MS/MS | 73,743 | 4 | 4 | NA | 1:18,436 |
| Males | Females | Total | |
|---|---|---|---|
| Screened newborns | 89,485 | 83,857 | 173,342 |
| Newborns with decreased enzyme activity in the 1st DBS, after retesting in duplicate | 44 | 9 | 53 |
| Recall % | 0.05% | 0.01% | 0.03% |
| Newborns with decreased enzyme activity in the 2nd DBS and referred to Clinic Unit for confirmatory testing | 22 | 1 | 23 |
| Newborns confirmed by low enzyme activity in lymphocytes and GLA gene mutation | 22 | 0 | 22 |
| Pathogenic classical variants | 0 | 0 | 0 |
| Pathogenic later-onset variants | 13 | 0 | 13 |
| Benign variants | 1 | 0 | 1 |
| False-positive results | 0 | 1 | 1 |
| Unclassified variants | 4 | 0 | 4 |
| p.Ala143Thr variant | 4 | 0 | 4 |
| Overall incidence | 1:4068 | 0 | 1:7879 |
| Pathogenic variants incidence | 1:6883 | 0 | 1:13,334 |
| Case | Year of Birth | Gender | Ethnic Origin | DBS AGAL Activity * | DBS LysoGb3 (nv < 1.13 nmol/L) | Lymphocytes AGAL Activity ** | Plasma LysoGb3 at First Visit (nv < 0.43 nmol/L) | cDNA Variation (Protein Variation) | Classification International Fabry Disease Genotype-Phenotype Database [49] *** | Age at Last Visit | Clinical Manifestations | Plasma LysoGb3 at the Last Visit (nv < 0.43 nmol/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2015 | M | Europe | 3.21 | NA | 100 | 0.53 | c.427G>A (p.Ala143Thr) | Benign | 5.5 years | No | 0.54 |
| 2 | 2015 | M | Europe | 2.76 | NA | 9 | 0.12 | c.427G>A (p.Ala143Thr) | Benign | 4.5 years | No | 0.48 |
| 3 | 2015 | M | Europe | 2.93 | NA | 354 | 0.31 | c.427G>A (p.Ala143Thr) | Benign | 4.5 years | No | 0.35 |
| 4 | 2016 | M | Europe | 0.64 | NA | 0 | 1.07 | c.644 A>G (p.Asn215Ser) + IVS2-77_81del5; IVS4-16A>G; IVS6-22C>T | Later-onset + NA **** | 4.5 years | No | 3.91 |
| 5 | 2016 | M | Europe | 2.25 | 1.02 | 355 | 0.19 | -10C>T; IVS2-77_81del5; IVS4-16A>G; IVS6-22C>T | NA **** | Lost to follow-up | ||
| 6 | 2016 | M | Europe | 3.45 | NA | 346 | 0.27 | c.737C>T (p.Thr246Ile) | NA | 4.5 years | No | 0.43 |
| 7 | 2016 | M | East Asia | 0.77 | 0.79 | 143 | 0.3 | IVS4 + 919G>A | Later-onset | 4 years | No | 1.12 |
| 8 | 2016 | M | North Africa | 0.72 | 1.79 | 66 | 1.02 | c.1088G>A (p.Arg363His) | Later-onset | 4 years | No | 1.91 |
| 9 | 2016 | M | East Asia | 1.16 | 0.62 | 222 | 0.54 | IVS4 + 919G>A | Later-onset | 4 years | No | 1.7 |
| 10 | 2016 | M | Europe | 0.73 | 2.17 | 27 | 2.98 | c.1066 C>G (p.Arg356Gly) | Likely later-onset | 4 years | No | 3.71 |
| 11 | 2017 | M | Europe | 2.05 | 0.54 | 316 | 0.36 | c.427G>A (p.Ala143Thr) + IVS4-61_60delGT | Benign + NA | 3.5 years | No | 0.85 |
| 12 | 2017 | M | Europe | 1.37 | 1.25 | NA | 0.85 | c.153G>A (p.Met51Ile) | Later-onset | 3.5 years | No | 0.82 |
| 13 | 2017 | M | West Africa | 1.51 | 0.96 | 0.73 | 0.26 | c.1067G>A (p.Arg356Gln) | Later-onset | 3 years | No | 0.43 |
| 14 | 2018 | M | Europe | 0.79 | 0.41 | NA | 0.2 | c.868A>C (p.Met290Leu) + -10C>T; IVS2-77_81del5; IVS4-16A>G; IVS6-22C>T | Later-onset + NA **** | Lost to follow-up | ||
| 15 | 2018 | M | Europe | 0.87 | 0.73 | 0.82 | 0.35 | c.347G>C (p.Gly116Ala) + c.376A>G (p.Ser126Gly) + -10C>T; IVS2-77_81del5; IVS4-16A>G; IVS6-22C>T | NA + likely benign + NA **** | 2.5 years | No | 0.92 |
| 16 | 2018 | M | Europe | 1.28 | 0.22 | 3.44 | 0.49 | c.856C>G (p.Leu286Val) | NA | 2 years | No | 0.43 |
| 17 | 2019 | M | Europe | 0.63 | 0.5 | 1.84 | 0.82 | c.644A>G (p.Asn215Ser) | Later-onset | 1 year | No | 1.95 |
| 18 | 2019 | M | Europe | 1.4 | 1.1 | 3.35 | 1.32 | c.644A>G (p.Asn215Ser) | Later-onset | 1.5 years | No | NA |
| 19 | 2019 | M | North Africa | 0.77 | 2.7 | 2.41 | 1 | c.1088G>A (p.Arg363His) | Later-onset | 1.5 years | No | 1.49 |
| 20 | 2019 | M | West Africa | 1.63 | 1.07 | 1.05 | NA | c.1067G>A (p.Arg356Gln) | Later-onset | 1 year | No | 0.56 |
| 21 | 2020 | M | Europe | 1.12 | 1.07 | 2.18 | 0.41 | c.856C>G (p.Leu286Val) | NA | 10 d | No | 0.41 |
| 22 | 2020 | M | Europe | 1.88 | 0.77 | 1.94 | 0.44 | c.868A>C (p.Met290Leu) | Later-onset | 6 m | No | 0.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gragnaniello, V.; Burlina, A.P.; Polo, G.; Giuliani, A.; Salviati, L.; Duro, G.; Cazzorla, C.; Rubert, L.; Maines, E.; Germain, D.P.; et al. Newborn Screening for Fabry Disease in Northeastern Italy: Results of Five Years of Experience. Biomolecules 2021, 11, 951. https://doi.org/10.3390/biom11070951
Gragnaniello V, Burlina AP, Polo G, Giuliani A, Salviati L, Duro G, Cazzorla C, Rubert L, Maines E, Germain DP, et al. Newborn Screening for Fabry Disease in Northeastern Italy: Results of Five Years of Experience. Biomolecules. 2021; 11(7):951. https://doi.org/10.3390/biom11070951
Chicago/Turabian StyleGragnaniello, Vincenza, Alessandro P Burlina, Giulia Polo, Antonella Giuliani, Leonardo Salviati, Giovanni Duro, Chiara Cazzorla, Laura Rubert, Evelina Maines, Dominique P Germain, and et al. 2021. "Newborn Screening for Fabry Disease in Northeastern Italy: Results of Five Years of Experience" Biomolecules 11, no. 7: 951. https://doi.org/10.3390/biom11070951
APA StyleGragnaniello, V., Burlina, A. P., Polo, G., Giuliani, A., Salviati, L., Duro, G., Cazzorla, C., Rubert, L., Maines, E., Germain, D. P., & Burlina, A. B. (2021). Newborn Screening for Fabry Disease in Northeastern Italy: Results of Five Years of Experience. Biomolecules, 11(7), 951. https://doi.org/10.3390/biom11070951











