MicroRNA Omics Analysis of Camellia sinesis Pollen Tubes in Response to Low-Temperature and Nitric Oxide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pollen Source and Culture
2.2. miRNA Library Construction and Sequencing
2.3. Fundamental Analysis for miRNA
2.4. Identification of Potential miRNA Target Genes and Annotation Analysis
2.5. RT-qPCR Analysis of miRNAs and Its Targets
2.6. Statistical Analysis
3. Results
3.1. Small RNA Sequencing and Annotation of miRNAs from C. sinensis Pollen Tubes
3.2. The Impact of Low-Temperature and NO on miRNA Expression
3.3. Validation of Differentially Expressed miRNAs by qPCR
3.4. Prediction and Functional Analysis of Target Genes of miRNA
3.5. Expression Analysis of miRNA and the Targets by qPCR
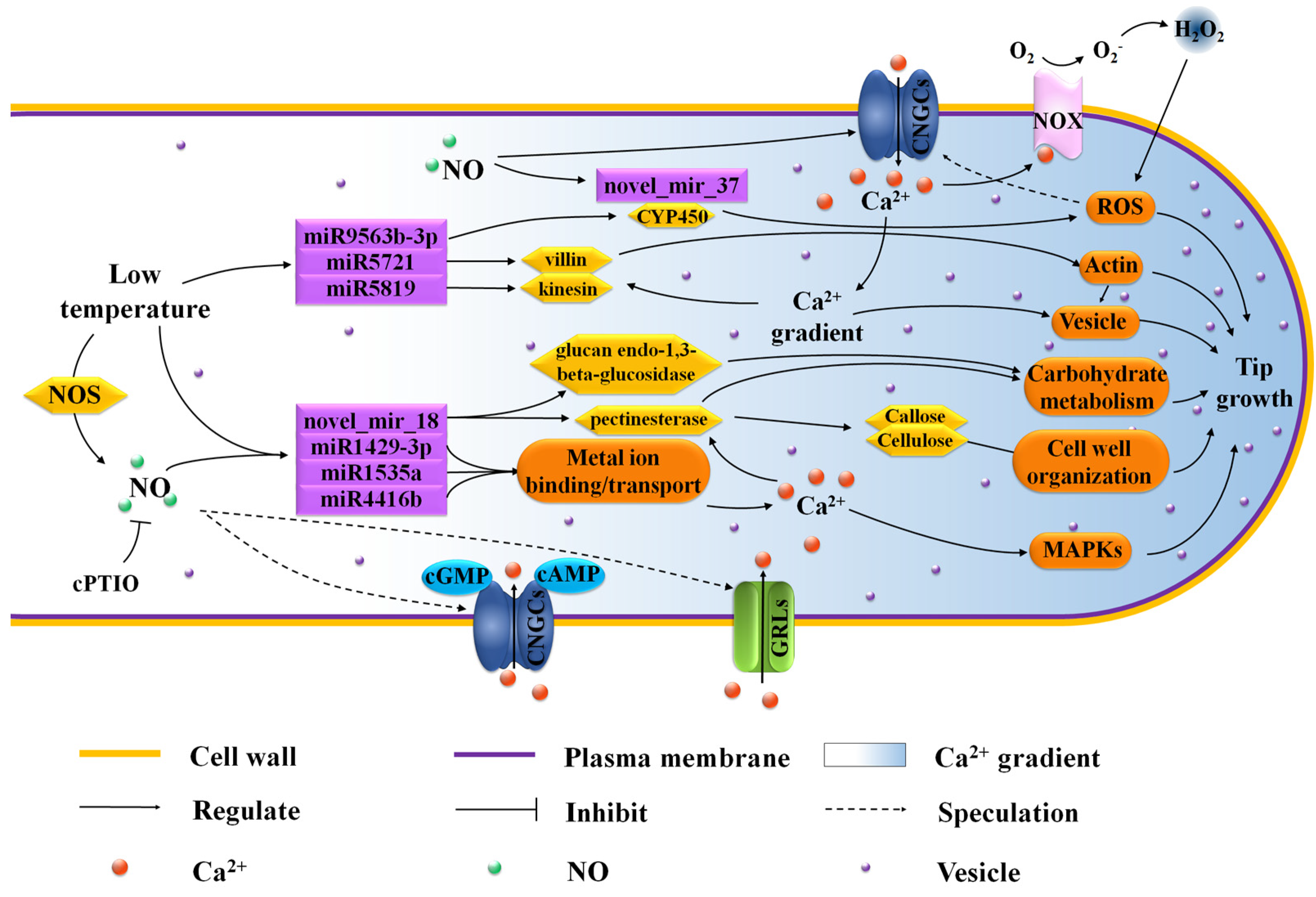
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Xiong, F.; Nong, S.; Liao, J.; Xing, A.; Shen, Q.; Ma, Y.; Fang, W.; Zhu, X. Effects of nitric oxide on the GABA, polyamines, and proline in tea (Camellia sinensis) roots under cold stress. Sci. Rep. 2020, 10, 12240. [Google Scholar] [CrossRef]
- Çetinbaş-Genç, A.; Cai, G.; Vardar, F.; Ünal, M. Differential effects of low and high temperature stress on pollen germination and tube length of hazelnut (Corylus avellana L.) genotypes. Sci. Hortic. 2019, 255, 61–69. [Google Scholar] [CrossRef]
- Parrotta, L.; Faleri, C.; Guerriero, G.; Cai, G. Cold stress affects cell wall deposition and growth pattern in tobacco pollen tubes. Plant. Sci. 2019, 283, 329–342. [Google Scholar] [CrossRef]
- Gao, Y.B.; Wang, C.L.; Wu, J.Y.; Zhou, H.S.; Jiang, X.T.; Wu, J.; Zhang, S.L. Low temperature inhibits pollen tube growth by disruption of both tip-localized reactive oxygen species and endocytosis in Pyrus bretschneideri Rehd. Plant. Physiol. Biochem. 2014, 74, 255–262. [Google Scholar] [CrossRef]
- Du, Y.Y.; Zhong, H.; Wu, W.L.; Lu, X.Q.; Liang, J.L. Effect of temperature on accumulation of chlorophylls and leaf ultrastructure of low temperature induced albino tea plant. Afr. J. Biotechnol. 2010, 7, 1881–1885. [Google Scholar] [CrossRef]
- Li, Q.h.; Xu, H.; Zhou, L.; Wang, M.; Zhu, X.; Fang, W. Effect of low temperature stress on chlorophyll fluorescence characteristics in leaf of two cultivars of Camellia sinensis. J. Plant. Resour. Environ. 2015, 24, 26–31. (In Chinese) [Google Scholar]
- Li, Y.; Wang, X.; Ban, Q.; Zhu, X.; Jiang, C.; Wei, C.; Bennetzen, J.L. Comparative transcriptomic analysis reveals gene expression associated with cold adaptation in the tea plant Camellia sinensis. BMC Genom. 2019, 20, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, C.; Cao, H.-L.; Wang, L.; Zhou, Y.-H.; Huang, Y.-T.; Hao, X.-Y.; Wang, Y.-C.; Wang, B.; Yang, Y.-J.; Wang, X.-C. Effects of cold acclimation on sugar metabolism and sugar-related gene expression in tea plant during the winter season. Plant. Mol. Biol. 2015, 88, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.Y.; Shin, S.; Wang, K.R.; Lu, J.L.; Liang, Y.R. Effect of temperature on the expression of genes related to the accumulation of chlorophylls and carotenoids in albino tea. J. Pomol. Hortic. Sci. 2009, 84, 365–369. [Google Scholar] [CrossRef]
- Yin, Y.; Ma, Q.-P.; Zhu, Z.-X.; Cui, Q.-Y.; Chen, C.-S.; Chen, X.; Fang, W.-P.; Li, X.-H. Functional analysis of CsCBF3 transcription factor in tea plant (Camellia sinensis) under cold stress. Plant Growth Regul. 2016, 80, 335–343. [Google Scholar] [CrossRef]
- Çetinbaş-Genç, A.; Cai, G.; Del Duca, S. Treatment with spermidine alleviates the effects of concomitantly applied cold stress by modulating Ca(2+), pH and ROS homeostasis, actin filament organization and cell wall deposition in pollen tubes of Camellia sinensis. Plant. Physiol. Biochem. 2020, 156, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Quesada, M.J.; Carmona, R.; Lima-Cabello, E.; Traverso, J.A.; Castro, A.J.; Claros, M.G.; Alche, J.D. Generation of nitric oxide by olive (Olea europaea L.) pollen during in vitro germination and assessment of the S-nitroso- and nitro-proteomes by computational predictive methods. Nitric Oxide 2017, 68, 23–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Lamattina, L.; Spoel, S.; Loake, G. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Prado, A.M.; Colaco, R.; Moreno, N.; Silva, A.C.; Feijó, J.A. Targeting of pollen tubes to ovules is dependent on nitric oxide (NO) signaling. Mol. Plant 2008, 1, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijo, J.A. Nitric Oxide: A Multitasked signaling gas in plants. Mol. Plant. 2015, 8, 506–520. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Li, X.C.; Zhu-Ge, Q.; Jiang, X.; Wang, W.D.; Fang, W.P.; Chen, X.; Li, X.H. Nitric oxide participates in cold-inhibited Camellia sinensis pollen germination and tube growth partly via cGMP In Vitro. PLoS ONE 2012, 7, e52436. [Google Scholar] [CrossRef] [Green Version]
- Sirova, J.; Sedlarova, M.; Piterkova, J.; Luhova, L.; Petrivalsky, M. The role of nitric oxide in the germination of plant seeds and pollen. Plant. Sci. 2011, 181, 560–572. [Google Scholar] [CrossRef]
- Reichler, S.A.; Torres, J.; Rivera, A.L.; Cintolesi, V.A.; Clark, G.; Roux, S.J. Intersection of two signalling pathways: Extracellular nucleotides regulate pollen germination and pollen tube growth via nitric oxide. J. Exp. Bot. 2009, 60, 2129–2138. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, T.; Zhang, C.; Hao, H.; Liu, P.; Zheng, M.; Baluka, F.; Amaj, J.; Lin, J. Nitric oxide modulates the influx of extracellular Ca2+ and actin filament organization during cell wall construction in Pinus bungeana pollen tubes. New Phytol. 2009, 182, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Napieraj, N.; Reda, M.; Janicka, M. The role of NO in plant response to salt stress: Interactions with polyamines. Funct. Plant. Biol. 2020, 47, 865–879. [Google Scholar] [CrossRef]
- Rai, K.K.; Pandey, N.; Rai, S.P. Salicylic acid and nitric oxide signaling in plant heat stress. Physiol. Plant. 2019, 168, 241–255. [Google Scholar] [CrossRef]
- Majeed, S.; Nawaz, F.; Naeem, M.; Ashraf, M.Y.; Ejaz, S.; Ahmad, K.S.; Tauseef, S.; Farid, G.; Khalid, I.; Mehmood, K. Nitric oxide regulates water status and associated enzymatic pathways to inhibit nutrients imbalance in maize (Zea mays L.) under drought stress. Plant Physiol. Biochem. 2020, 155, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Costa-Broseta, Á.; Perea-Resa, C.; Castillo, M.-C.; Ruíz, M.F.; Salinas, J.; León, J. Nitric Oxide controls constitutive freezing tolerance in arabidopsis by attenuating the levels of osmoprotectants, stress-related hormones and anthocyanins. Sci. Rep. 2018, 8, 9268. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, A.; Abat, J.K.; Deswal, R. RuBisCO depletion improved proteome coverage of cold responsive S-nitrosylated targets in Brassica juncea. Front. Plant Sci. 2013, 4, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.D.; Sheng, X.Y.; Shu, Z.F.; Li, D.Q.; Pan, J.T.; Ye, X.L.; Chang, P.P.; Li, X.H.; Wang, Y.H. Combined cytological and transcriptomic analysis reveals a nitric oxide signaling pathway involved in cold-inhibited Camellia sinensis pollen tube growth. Front. Plant. Sci. 2016, 7, 456. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Wang, W.; Li, D.; Shu, Z.; Ye, X.; Chang, P.; Wang, Y. Gene expression profile indicates involvement of NO in Camellia sinensis pollen tube growth at low temperature. BMC Genom. 2016, 17, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B. MicroRNAs in control of plant development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Abla, M.; Sun, H.; Li, Z.; Wei, C.; Gao, F.; Zhou, Y.; Feng, J. Identification of miRNAs and their response to cold stress in Astragalus Membranaceus. Biomolecules 2019, 9, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Zhang, Y.; Tang, R.; Qu, H.; Duan, X.; Jiang, Y. Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress. BMC Genom. 2019, 20, 33. [Google Scholar] [CrossRef] [Green Version]
- Jatan, R.; Chauhan, P.S.; Lata, C. Pseudomonas putida modulates the expression of miRNAs and their target genes in response to drought and salt stresses in chickpea (Cicer arietinum L.). BMC Genom. 2019, 111, 509–519. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, P.; Kang, H.; Liu, L.; Cao, Q.; Sun, J.; Dong, T.; Zhu, M.; Li, Z.; Xu, T. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genom. 2020, 21, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Zheng, S.; Li, Y.; Liu, R.; Zhang, L.; Wu, Y. Comparative profiling of roots small RNA expression and corresponding gene ontology and pathway analyses for low- and high-cadmium-accumulating genotypes of wheat in response to cadmium stress. Funct. Integr. Genom. 2020, 20, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zhao, L.; Wang, Y.; Shen, J.; Zhang, Y.; Jia, S.; Li, Y.; Ding, Z. Integrated RNA-Seq and sRNA-Seq analysis identifies chilling and freezing responsive key molecular players and pathways in tea plant (Camellia sinensis). PLoS ONE 2015, 10, e0125031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, X.; Chen, X.; Song, C.; Zou, Z.; Wang, Y.; Wang, M.; Fang, W.; Li, X. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014, 14, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, M.; Landgraf, P.; Ludwig, J.; Rice, A.; Ojo, T.; Lin, C.; Holoch, D.; Lim, C.; Tuschl, T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 2008, 44, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2004, 33, D192–D200. [Google Scholar] [CrossRef] [Green Version]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.-J.; Zhang, P.-J.; Chen, W.-J.; Jie, D.; Dan, F.; Jia, Y.-H.; Xie, L.-X. Characterization and identification of novel serum microRNAs in Sepsis Patients with different outcomes. Shock 2013, 39, 480–487. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, E.-H.; Li, F.-D.; Tong, W.; Li, P.-H.; Wu, Q.; Zhao, H.-J.; Ge, R.-H.; Li, R.-P.; Li, Y.-Y.; Zhang, Z.-Z.; et al. Tea Plant information archive: A comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 2019, 17, 1938–1953. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wang, S.; Chen, M.; Yin, Y.; Storey, K.B. MiR-200-3p is potentially involved in cell cycle arrest by regulating cyclin A during aestivation in Apostichopus japonicus. Cells 2019, 8, 843. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Zhu, X.; Shen, J.; Xing, H.; Zou, Z.; Ma, Y.; Wang, Y.; Fang, W. Genome-wide identification, characterization and expression analysis of the amino acid permease gene family in tea plants (Camellia sinensis). Genomics 2020, 112, 2866–2874. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sehrawat, A.; Gupta, R.; Deswal, R. Nitric oxide-cold stress signalling cross-talk, evolution of a novel regulatory mechanism. Proteomics 2013, 13, 1816–1835. [Google Scholar] [CrossRef]
- Aydinoglu, F. Elucidating the regulatory roles of microRNAs in maize (Zea mays L.) leaf growth response to chilling stress. Planta 2020, 251, 38. [Google Scholar] [CrossRef]
- Jiang, W.; Shi, W.; Ma, X.; Zhao, J.; Wang, S.; Tan, L.; Sun, C.; Liu, F. Identification of microRNAs responding to cold stress in Dongxiang common wild rice. Genome 2019, 62, 635–642. [Google Scholar] [CrossRef]
- Maeda, S.; Sakazono, S.; Masuko-Suzuki, H.; Taguchi, M.; Yamamura, K.; Nagano, K.; Endo, T.; Saeki, K.; Osaka, M.; Nabemoto, M.; et al. Comparative analysis of microRNA profiles of rice anthers between cool-sensitive and cool-tolerant cultivars under cool-temperature stress. Genes Genet. Syst. 2016, 91, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Gupta, O.P.; Meena, N.L.; Sharma, I.; Sharma, P. Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat. Mol. Biol. Rep. 2014, 41, 4623–4629. [Google Scholar] [CrossRef]
- Seifert, F.; Bossow, S.; Kumlehn, J.; Gnad, H.; Scholten, S. Analysis of wheat microspore embryogenesis induction by transcriptome and small RNA sequencing using the highly responsive cultivar “Svilena”. BMC Plant Biol. 2016, 16, 97. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.E.; Zhao, Y.T.; Wang, X.J.; Croft, L.; Wang, Z.H.; Haerizadeh, F.; Mattick, J.S.; Singh, M.B.; Carroll, B.J.; Bhalla, P.L. MicroRNAs in the shoot apical meristem of soybean. J. Exp. Bot. 2011, 62, 2495–2506. [Google Scholar] [CrossRef] [Green Version]
- Kulcheski, F.R.; de Oliveira, L.F.; Molina, L.G.; Almerão, M.P.; Rodrigues, F.A.; Marcolino, J.; Barbosa, J.F.; Stolf-Moreira, R.; Nepomuceno, A.L.; Marcelino-Guimarães, F.C.; et al. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011, 12, 307. [Google Scholar] [CrossRef] [Green Version]
- Cai, G.; Parrotta, L.; Cresti, M. Organelle trafficking, the cytoskeleton, and pollen tube growth. J. Integr. Plant Biol. 2015, 57, 63–78. [Google Scholar] [CrossRef]
- Ali, I.; Yang, W.-C. The functions of kinesin and kinesin-related proteins in eukaryotes. Cell Adhes. Migr. 2020, 14, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Cresti, M. Microtubule motors and pollen tube growth—Still an open question. Protoplasma 2010, 247, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Blanchoin, L.; Staiger, C.J. Signaling to actin stochastic dynamics. Annu. Rev. Plant. Biol. 2014, 66, 415–440. [Google Scholar] [CrossRef]
- Bao, C.; Wang, J.; Zhang, R.; Zhang, B.; Zhang, H.; Zhou, Y.; Huang, S. Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. Plant J. 2012, 71, 962–975. [Google Scholar] [CrossRef]
- Khurana, P.; Henty, J.L.; Huang, S.; Staiger, A.M.; Blanchoin, L.; Staiger, C.J. Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 2010, 22, 2727–2748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Qu, X.; Bao, C.; Khurana, P.; Wang, Q.; Xie, Y.; Zheng, Y.; Chen, N.; Blanchoin, L.; Staiger, C.J.; et al. Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 2010, 22, 2749–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Tomar, A.; Parrill, A.L.; Khurana, S. Functional Dissection and Molecular characterization of calcium-sensitive actin-capping and actin-depolymerizing sites in villin*. J. Biol. Chem. 2004, 279, 45036–45046. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Qu, X.; Zhuang, Y.; Wang, L.; Bosch, M.; Franklin-Tong, V.E.; Xue, Y.; Huang, S. Villin controls the formation and enlargement of punctate actin foci in pollen tubes. J. Cell Sci. 2020, 133, jcs237404. [Google Scholar] [CrossRef]
- Beck, E.H.; Fettig, S.; Knake, C.; Hartig, K.; Bhattarai, T. Specific and unspecific responses of plants to cold and drought stress. J. Biosci. 2007, 32, 501–510. [Google Scholar] [CrossRef]
- Zhao, C.; Lang, Z.; Zhu, J.K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 2015, 20, 466–468. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yuan, J.; Qin, L.; Shi, W.; Xia, G.; Liu, S. TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol. J. 2019, 18, 791–804. [Google Scholar] [CrossRef] [Green Version]
- Renault, H.; Bassard, J.-E.; Hamberger, B.; Werck-Reichhart, D. Cytochrome P450-mediated metabolic engineering: Current progress and future challenges. Curr. Opin. Plant Biol. 2014, 19, 27–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef]
- Bosch, M.; Cheung, A.Y.; Hepler, P.K. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005, 138, 1334–1346. [Google Scholar] [CrossRef] [Green Version]
- Garrido, D.; Busscher, J.; van Tunen, A.J. Promoter activity of a putative pollen monosaccharide transporter in Petunia hybrida and characterisation of a transposon insertion mutant. Protoplasma 2006, 228, 3–11. [Google Scholar] [CrossRef]
- Gonzalez, M.V.; Coque, M.; Herrero, M. Pollen-pistil interaction in kiwifruit (Actinidia deliciosa; Actinidiaceae). Am. J. Bot. 1996, 148–154. [Google Scholar] [CrossRef] [Green Version]
- Moustacas, A.M.; Nari, J.; Borel, M.; Noat, G.; Ricard, J. Pectin methylesterase, metal ions and plant cell-wall extension. The role of metal ions in plant cell-wall extension. Biochem. J. 1991, 279, 351–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charnay, D.; Nari, J.; Noat, G. Regulation of plant cell-wall pectin methyl esterase by polyamines—Interactions with the effects of metal ions. Eur. J. Biochem. 1992, 205, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-F.; Munemasa, S.; Nishimura, N.; Ren, H.-M.; Robert, N. Identification of Cyclic GMP-Activated Nonselective Ca2+-Permeable Cation Channels and Associated CNGC5 and CNGC6 Genes in Arabidopsis Guard Cells. Plant. Physiol. 2013, 163, 578–590. [Google Scholar] [CrossRef] [Green Version]
- Konrad, K.R.; Wudick, M.M.; Feijó, J.A. Calcium regulation of tip growth: New genes for old mechanisms. Curr. Opin. Plant. Biol. 2011, 14, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.H.; Obermeyer, G.; Feijó, J.A. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Loka, D.A.; Fitzsimons, T.R.; Zhou, Z. Potassium deficiency limits reproductive success by altering carbohydrate and protein balances in cotton (Gossypium hirsutum L.). Environ. Exp. Bot. 2018, 145, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, S.; Hirata, S.; Yamamoto, K.; Nakajima, Y.; Kurahashi, K.; Tokumoto, H. ZnO nanoparticles effect on pollen grain germination and pollen tube elongation. Plant Cell Tissue Organ Cult. (PCTOC) 2021, 145, 405–415. [Google Scholar] [CrossRef]
- Guan, Y.; Lu, J.; Xu, J.; McClure, B.; Zhang, S. Two mitogen-activated protein kinases, MPK3 and MPK6, are required for funicular guidance of pollen tubes in arabidopsis. Plant. Physiol. 2014, 165, 528–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, T.; Wang, T.; Li, H.; Wassie, M.; Xu, H.; Chen, L. Genome-wide small RNA profiling reveals tiller development in tall fescue (Festuca arundinacea Schreb). BMC Genom. 2020, 21, 696. [Google Scholar] [CrossRef] [PubMed]
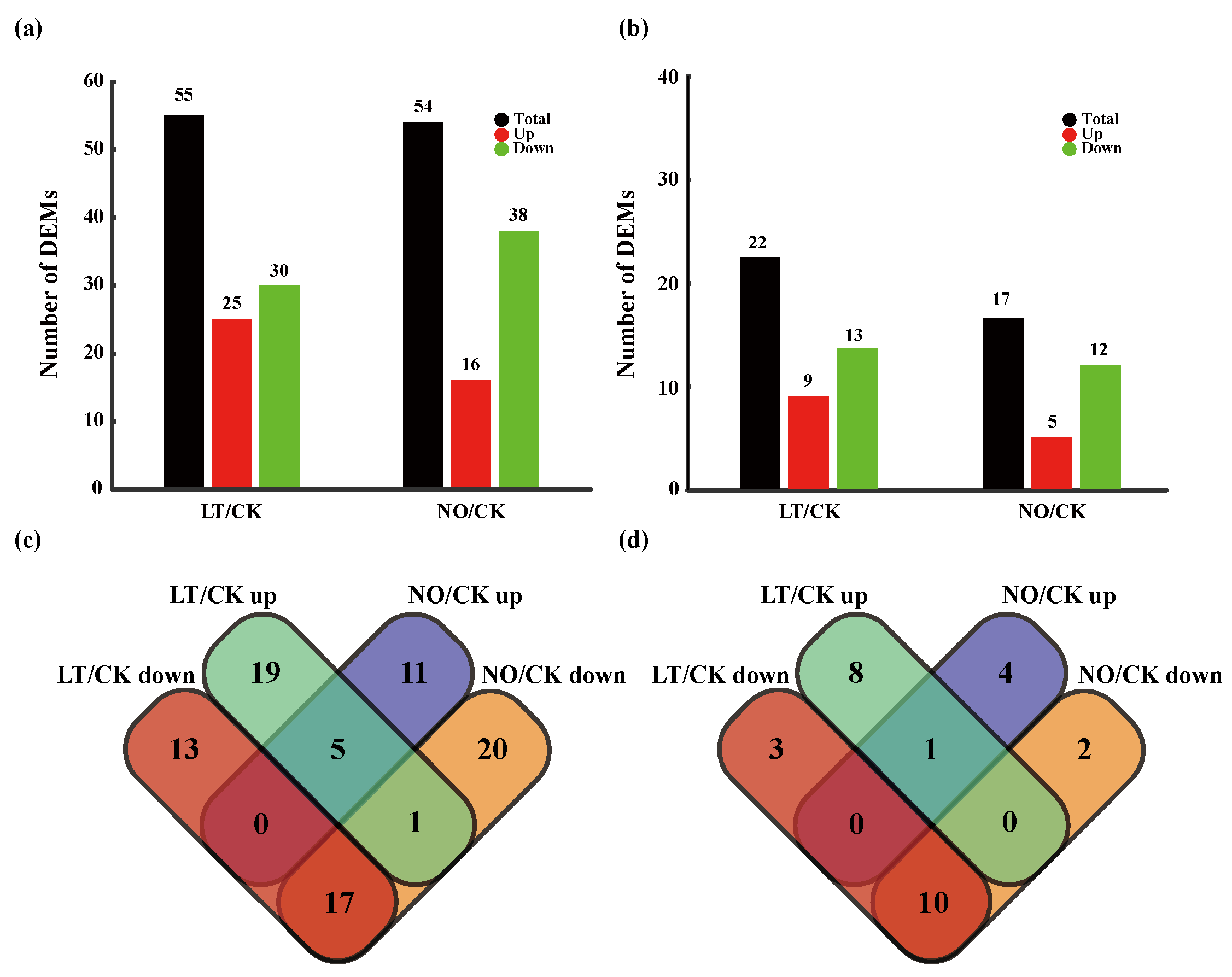

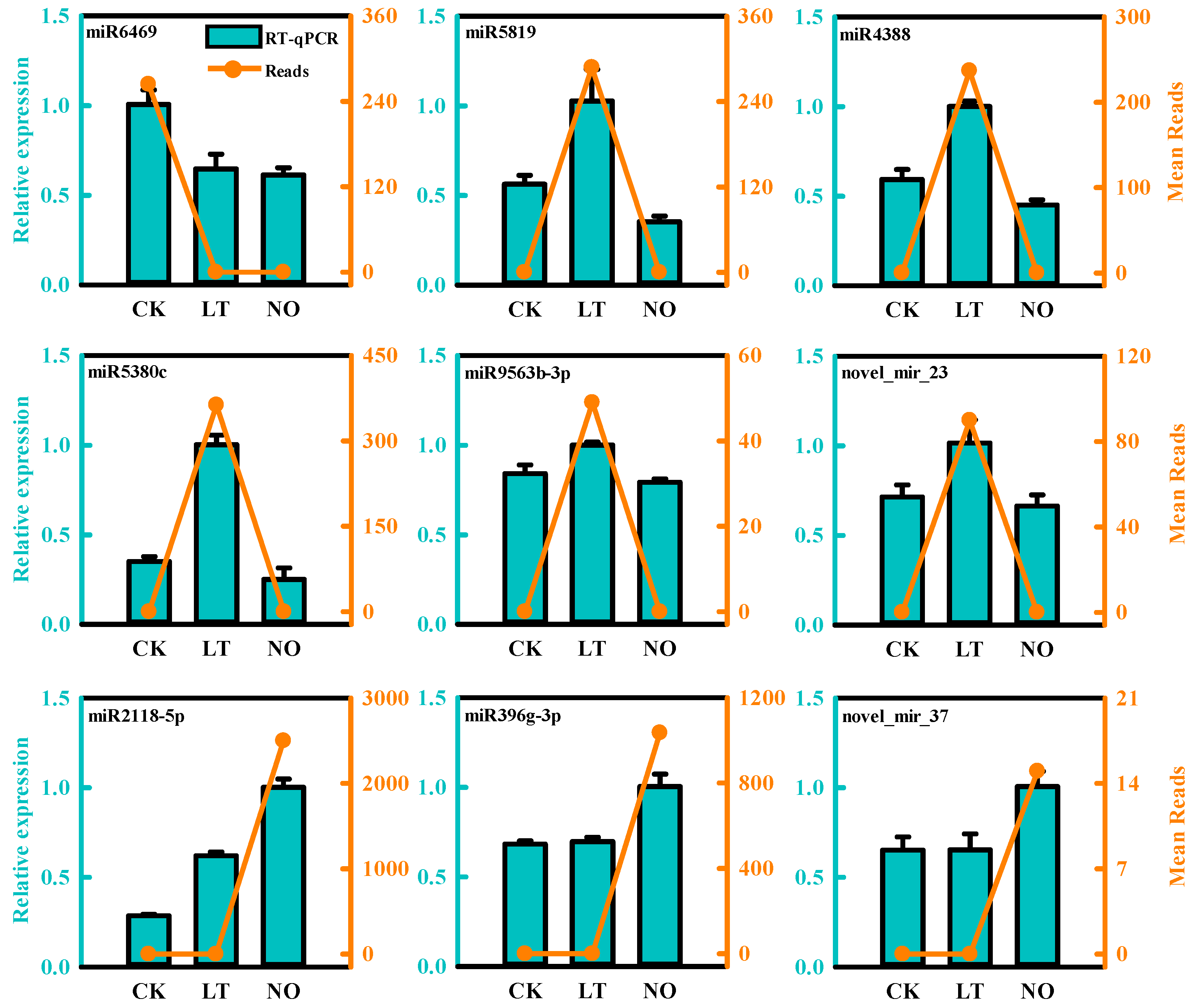
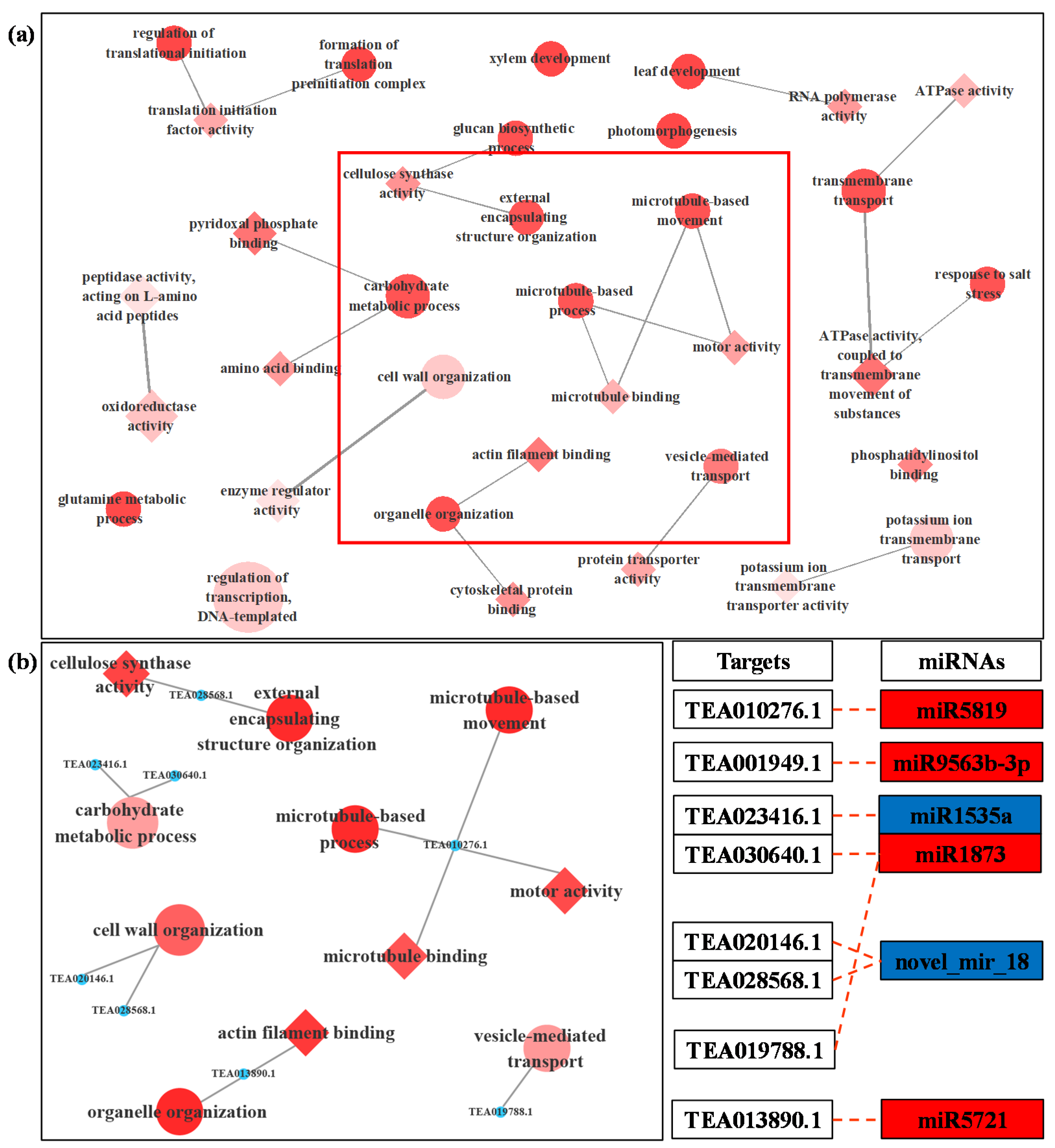


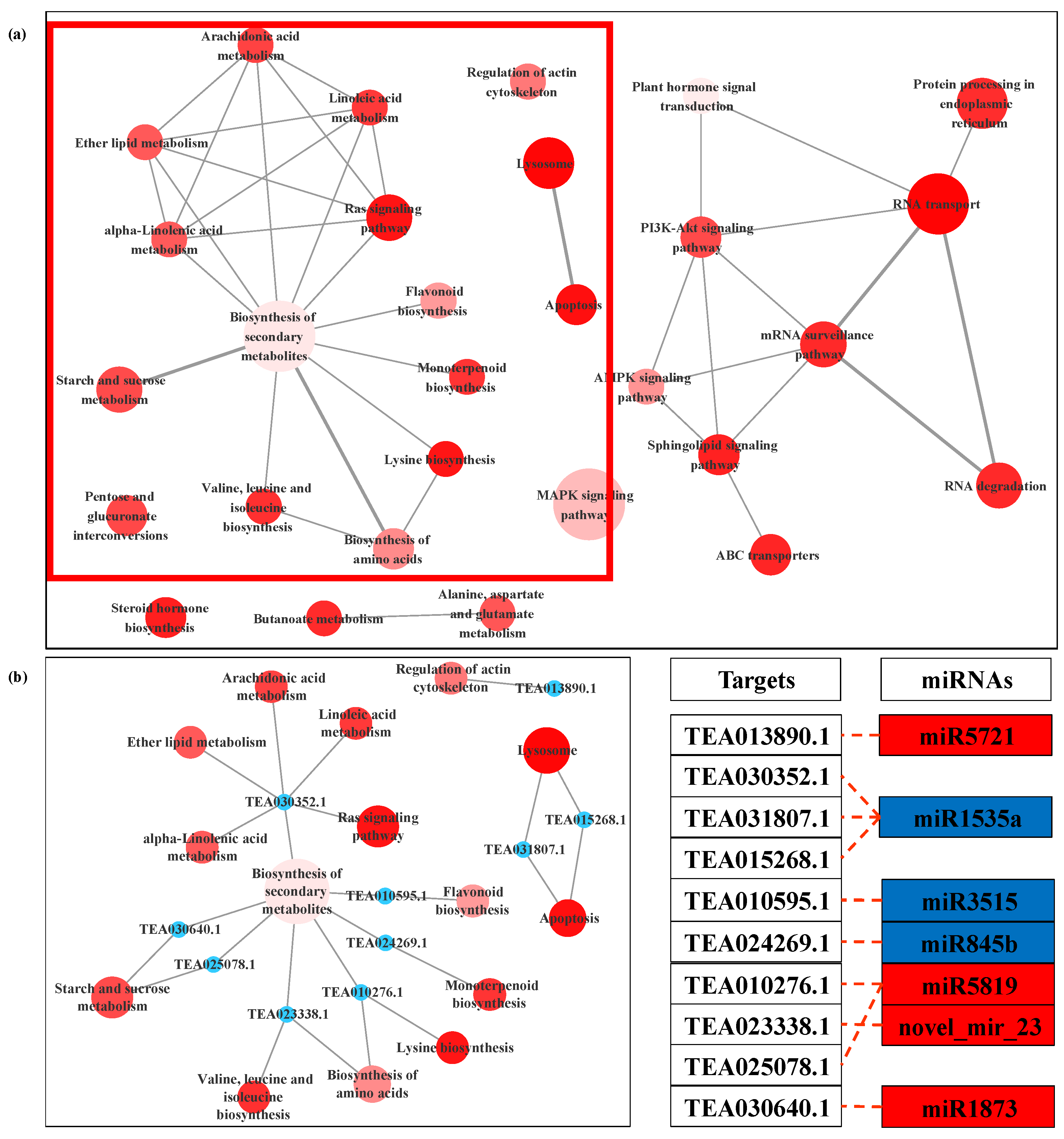

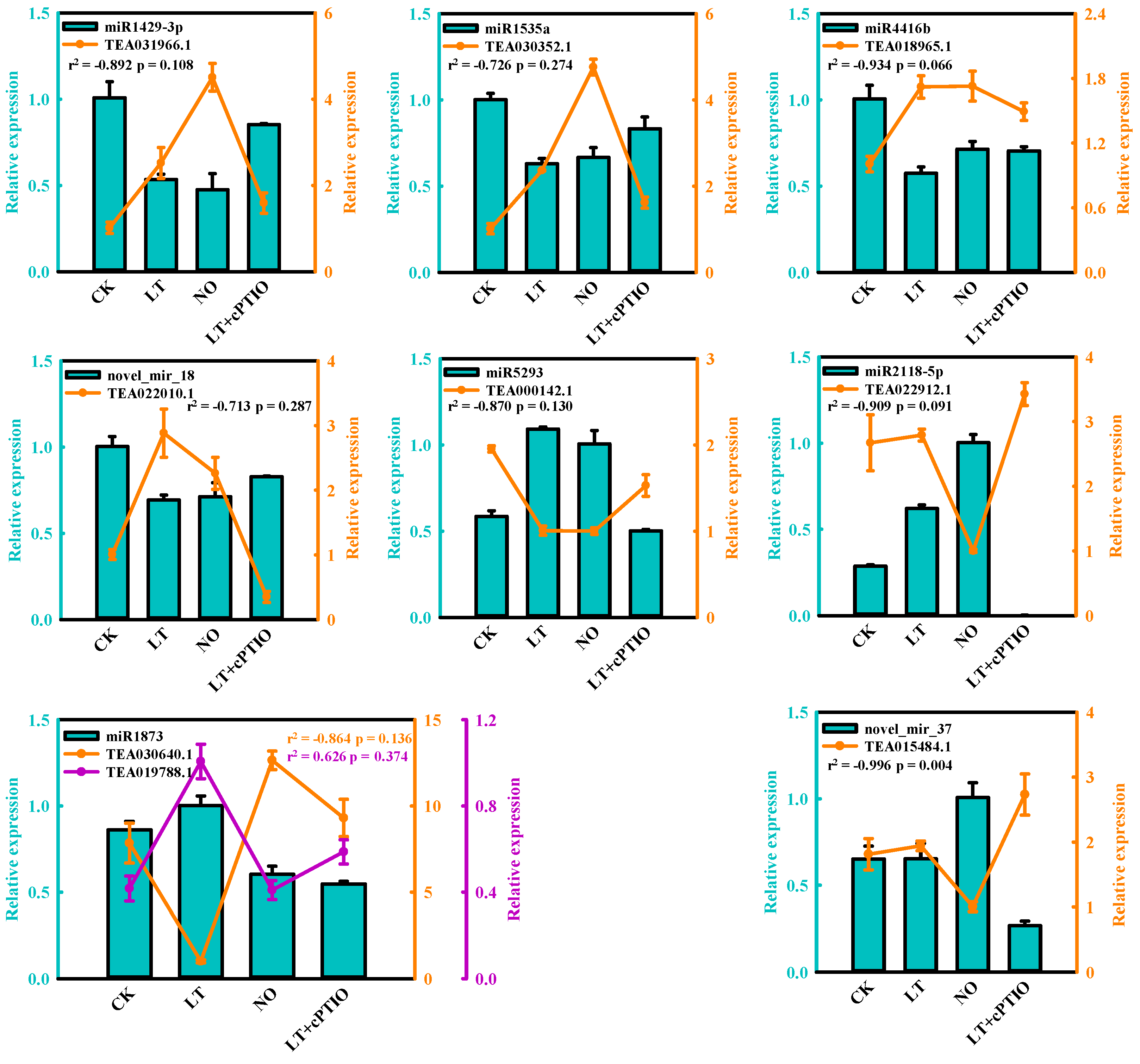
| miRNA | Up/Down Regulation | Potential Target Gene | Function Annotation |
|---|---|---|---|
| miR1312 | Down | TEA029389.1 | Protein TRANSPARENT TESTA 12 |
| miR1429-3p | Down | TEA031966.1 | Terpene synthase |
| miR1508a | Down | TEA027146.1 | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase isoform X2 |
| miR1535a | Down | TEA005439.1 | BAG family molecular chaperone regulator 2 |
| TEA023416.1 | Glucan endo-1,3-beta-glucosidase 8 | ||
| TEA030352.1 | Phospholipase A2-alpha-like | ||
| TEA005524.1 | Regulator of chromosome condensation repeat-containing protein isoform 1 | ||
| miR2118a-3p | Down | TEA017334.1 | Cancer-related nucleoside-triphosphatase homolog isoform X2 |
| TEA028604.1 | Extensin-2-like | ||
| TEA016266.1 | Polyadenylate-binding protein 8-like | ||
| miR3515 | Down | TEA010595.1 | Protein SRG1 |
| miR4416b | Down | TEA018965.1 | Protease-associated RING/U-box zinc finger family protein |
| miR5293 | Up | TEA000142.1 | S-acyltransferase 19 isoform X1 |
| TEA023749.1 | Serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform | ||
| miR6469 | Down | TEA023847.1 | Dof zinc finger protein DOF5.6-like |
| TEA001762.1 | Hypothetical protein VITISV_027707 | ||
| TEA018038.1 | Putative membrane protein insertion efficiency factor isoform X2 | ||
| novel_mir_18 | Down | TEA020146.1 | Pectinesterase 2 |
| TEA028568.1 | Pectinesterase 2 | ||
| TEA022010.1 | Potassium transporter 2 | ||
| TEA004684.1 | Transport protein SEC13 homolog B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Wang, W.; Sun, Y.; Xing, A.; Wu, Z.; Tian, Z.; Li, X.; Wang, Y. MicroRNA Omics Analysis of Camellia sinesis Pollen Tubes in Response to Low-Temperature and Nitric Oxide. Biomolecules 2021, 11, 930. https://doi.org/10.3390/biom11070930
Xu X, Wang W, Sun Y, Xing A, Wu Z, Tian Z, Li X, Wang Y. MicroRNA Omics Analysis of Camellia sinesis Pollen Tubes in Response to Low-Temperature and Nitric Oxide. Biomolecules. 2021; 11(7):930. https://doi.org/10.3390/biom11070930
Chicago/Turabian StyleXu, Xiaohan, Weidong Wang, Yi Sun, Anqi Xing, Zichen Wu, Zhiqiang Tian, Xuyan Li, and Yuhua Wang. 2021. "MicroRNA Omics Analysis of Camellia sinesis Pollen Tubes in Response to Low-Temperature and Nitric Oxide" Biomolecules 11, no. 7: 930. https://doi.org/10.3390/biom11070930
APA StyleXu, X., Wang, W., Sun, Y., Xing, A., Wu, Z., Tian, Z., Li, X., & Wang, Y. (2021). MicroRNA Omics Analysis of Camellia sinesis Pollen Tubes in Response to Low-Temperature and Nitric Oxide. Biomolecules, 11(7), 930. https://doi.org/10.3390/biom11070930






