Clinical and Immunological Biomarkers for Systemic Lupus Erythematosus
Abstract
1. Introduction
2. Characteristics of Biomarkers
3. Biomarkers for SLE Diagnosis and Classification
4. Non-Organ-Specific Biomarkers for SLE
4.1. Serum ANA
4.2. Serum Complement 3 (C3) and Complement 4 (C4)
4.3. Anti-Nucleosome Antibodies (ANuA)
4.4. Erythrocyte Sedimentation Rate (ESR) and C-Reactive Protein (CRP)
5. Organ-Specific SLE Biomarkers
5.1. Biomarkers in Lupus Nephritis (LN)
5.1.1. Serum Anti-dsDNA Antibodies
5.1.2. Serum Anti-SmAntibody
5.1.3. Anti-C1q Antibodies
5.1.4. Other Urinary Biomarkers
5.2. Biomarkers for Skin Lesions in SLE
5.3. Biomarkers in Neuropsychiatric SLE (NPSLE)
5.4. Biomarkers for Cardiovascular Involvement in SLE
6. Omics Approaches in SLE
6.1. Transcriptome in SLE
6.2. Epigenome in SLE
6.3. Genome in SLE
6.4. Metabolome in SLE
7. Challenges in Biomarker Discovery
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kiriakidou, M.; Ching, C. Systemic Lupus Erythematosus. Ann. Intern. Med. 2020, 172, ITC81–ITC96. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C. Systemic lupus erythematosus. N. Engl. J. Med. 2011, 365, 2110–2121. [Google Scholar] [CrossRef]
- Gergianaki, I.; Fanouriakis, A.; Repa, A.; Tzanakakis, M.; Adamichou, C.; Pompieri, A.; Spirou, G.; Bertsias, A.; Kabouraki, E.; Tzanakis, I.; et al. Epidemiology and burden of systemic lupus erythematosus in a Southern European population: Data from the community-based lupus registry of Crete, Greece. Ann. Rheum. Dis. 2017, 76, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Rees, F.; Doherty, M.; Grainge, M.; Lanyon, P.; Zhang, W. The worldwide incidence and prevalence of systemic lupus erythematosus: A systematic review of epidemiological studies. Rheumatology 2017, 56, 1945–1961. [Google Scholar] [CrossRef]
- Bertsias, G.; Karampli, E.; Sidiropoulos, P.; Gergianaki, I.; Drosos, A.; Sakkas, L.; Garyfallos, A.; Tzioufas, A.; Vassilopoulos, D.; Tsalapaki, C.; et al. Clinical and financial burden of active lupus in Greece: A nationwide study. Lupus 2016, 25, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Murimi-Worstell, I.B.; Lin, D.H.; Kan, H.; Tierce, J.; Wang, X.; Nab, H.; Desta, B.; Alexander, G.C.; Hammond, E.R. Healthcare Utilization and Costs of Systemic Lupus Erythematosus by Disease Severity in the United States. J. Rheumatol. 2021, 48, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Doria, A.; Amoura, Z.; Cervera, R.; Khamastha, M.A.; Schneider, M.; Richter, J.; Guillemin, F.; Kobelt, G.; Maurel, F.; Garofano, A.; et al. Annual direct medical cost of active systemic lupus erythematosus in five European countries. Ann. Rheum. Dis. 2014, 73, 154–160. [Google Scholar] [CrossRef]
- Szefler, S.J.; Wenzel, S.; Brown, R.; Erzurum, S.C.; Fahy, J.V.; Hamilton, R.G.; Hunt, J.F.; Kita, H.; Liu, A.H.; Panettieri, R.A., Jr.; et al. Asthma outcomes: Biomarkers. J. Allergy Clin. Immunol. 2012, 129, S9–S23. [Google Scholar] [CrossRef] [PubMed]
- González, L.A.; Ugarte-Gil, M.F.; Alarcón, G.S. Systemic lupus erythematosus: The search for the ideal biomarker. Lupus 2020, 961203320979051. [Google Scholar] [CrossRef]
- Bertolo, M.; Baumgart, S.; Durek, P.; Peddinghaus, A.; Mei, H.; Rose, T.; Enghard, P.; Grützkau, A. Deep Phenotyping of Urinary Leukocytes by Mass Cytometry Reveals a Leukocyte Signature for Early and Non-Invasive Prediction of Response to Treatment in Active Lupus Nephritis. Front. Immunol. 2020, 11, 256. [Google Scholar] [CrossRef]
- Narendra, D.; Blixt, J.; Hanania, N. Immunological biomarkers in severe asthma. Semin. Immunol. 2019, 46, 101332. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Ahearn, J.M. The search for lupus biomarkers. Best Pract. Res. Clin. Rheumatol. 2009, 23, 507–523. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Illei, G.G.; Tackey, E.; Lapteva, L.; Lipsky, P.E. Biomarkers in systemic lupus erythematosus. I. General overview of biomarkers and their applicability. Arthritis Rheum. 2004, 50, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zeng, J.; Yin, J.; Peng, Q.; Zhao, M.; Lu, Q. Organ-specific biomarkers in lupus. Autoimmun. Rev. 2017, 16, 391–397. [Google Scholar] [CrossRef]
- Liu, C.-C.; Kao, A.H.; Manzi, S.; Ahearn, J.M. Biomarkers in systemic lupus erythematosus: Challenges and prospects for the future. Ther. Adv. Musculoskelet. Dis. 2013, 5, 210–233. [Google Scholar] [CrossRef]
- Tan, E.M.; Cohen, A.S.; Fries, J.F.; Masi, A.T.; McShane, D.J.; Rothfield, N.F.; Schaller, J.G.; Talal, N.; Winchester, R.J. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982, 25, 1271–1277. [Google Scholar] [CrossRef]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef] [PubMed]
- Petri, M.; Orbai, A.-M.; Alarcón, G.S.; Gordon, C.; Merrill, J.T.; Fortin, P.R.; Bruce, I.N.; Isenberg, D.; Wallace, D.J.; Nived, O.; et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012, 64, 2677–2686. [Google Scholar]
- Aringer, M.; Dörner, T.; Leuchten, N.; Johnson, S. Toward new criteria for systemic lupus erythematosus-a standpoint. Lupus 2016, 25, 805–811. [Google Scholar] [CrossRef]
- Aringer, M.; Costenbader, K.; Daikh, D.; Brinks, R.; Mosca, M.; Ramsey-Goldman, R.; Smolen, J.S.; Wofsy, D.; Boumpas, D.T.; Kamen, D.L.; et al. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019, 71, 1400–1412. [Google Scholar] [CrossRef]
- Adamichou, C.; Nikolopoulos, D.; Genitsaridi, I.; Bortoluzzi, A.; Fanouriakis, A.; Papastefanakis, E.; Kalogiannaki, E.; Gergianaki, I.; Sidiropoulos, P.; Boumpas, D.T.; et al. In an early SLE cohort the ACR-1997, SLICC-2012 and EULAR/ACR-2019 criteria classify non-overlapping groups of patients: Use of all three criteria ensures optimal capture for clinical studies while their modification earlier classification and treatment. Ann. Rheum. Dis. 2020, 79, 232–241. [Google Scholar] [CrossRef]
- Sacre, K.; Delaval, L.; Dossier, A.; Alexandra, J.-F.; Berleur, M.; Chauveheid, M.-P.; Ducrocq, G.; Goulenok, T.; van Gysel, D.; Rouzaud, D.; et al. New 2019 SLE EULAR/ACR classification criteria are valid for identifying patients with SLE among patients admitted for pericardial effusion. Ann. Rheum. Dis. 2019. [Google Scholar] [CrossRef]
- Nikolopoulos, D.; Kostopoulou, M.; Pieta, A.; Karageorgas, T.; Tseronis, D.; Chavatza, K.; Flouda, S.; Rapsomaniki, P.; Banos, A.; Kremasmenou, E.; et al. Evolving phenotype of systemic lupus erythematosus in Caucasians: Low incidence of lupus nephritis, high burden of neuropsychiatric disease and increased rates of late-onset lupus in the ’Attikon’ cohort. Lupus 2020, 29, 514–522. [Google Scholar] [CrossRef]
- Larosa, M.; Iaccarino, L.; Gatto, M.; Punzi, L.; Doria, A. Advances in the diagnosis and classification of systemic lupus erythematosus. Expert Rev. Clin. Immunol. 2016, 12, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Damoiseaux, J.; Andrade, L.E.C.; Carballo, O.G.; Conrad, K.; Francescantonio, P.L.C.; Fritzler, M.J.; Garcia de la Torre, I.; Herold, M.; Klotz, W.; Cruvinel, W.M.; et al. Clinical relevance of HEp-2 indirect immunofluorescent patterns: The International Consensus on ANA patterns (ICAP) perspective. Ann. Rheum. Dis. 2019, 78, 879–889. [Google Scholar] [CrossRef]
- Olsen, N.J.; Karp, D.R. Autoantibodies and SLE: The threshold for disease. Nat. Rev. Rheumatol. 2014, 10, 181–186. [Google Scholar] [CrossRef]
- Meroni, P.L.; Schur, P.H. ANA screening: An old test with new recommendations. Ann. Rheum. Dis. 2010, 69, 1420–1422. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S. Evolving story of autoantibodies in systemic lupus erythematosus. J. Autoimmun. 2020, 110, 102356. [Google Scholar] [CrossRef]
- Pisetsky, D.S. Antinuclear antibody testing—Misunderstood or misbegotten? Nat. Rev. Rheumatol. 2017, 13, 495–502. [Google Scholar] [CrossRef]
- Emlen, W.; O’Neill, L. Clinical significance of antinuclear antibodies: Comparison of detection with immunofluorescence and enzyme-linked immunosorbent assays. Arthritis Rheum. 1997, 40, 1612–1618. [Google Scholar] [CrossRef] [PubMed]
- Oke, V.; Wahren-Herlenius, M. Cutaneous lupus erythematosus: Clinical aspects and molecular pathogenesis. J. Intern. Med. 2013, 273, 544–554. [Google Scholar] [CrossRef]
- Sjöwall, C.; Sturm, M.; Dahle, C.; Bengtsson, A.A.; Jönsen, A.; Sturfelt, G.; Skogh, T. Abnormal antinuclear antibody titers are less common than generally assumed in established cases of systemic lupus erythematosus. J. Rheumatol. 2008, 35, 1994–2000. [Google Scholar]
- Choi, M.Y.; Clarke, A.E.; St Pierre, Y.; Hanly, J.G.; Urowitz, M.B.; Romero-Diaz, J.; Gordon, C.; Bae, S.-C.; Bernatsky, S.; Wallace, D.J.; et al. Antinuclear Antibody-Negative Systemic Lupus Erythematosus in an International Inception Cohort. Arthritis Care Res. 2019, 71, 893–902. [Google Scholar] [CrossRef]
- Pisetsky, D.S.; Rovin, B.H.; Lipsky, P.E. New Perspectives in Rheumatology: Biomarkers as Entry Criteria for Clinical Trials of New Therapies for Systemic Lupus Erythematosus: The Example of Antinuclear Antibodies and Anti-DNA. Arthritis Rheumatol. 2017, 69, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Furie, R.; Petri, M.; Zamani, O.; Cervera, R.; Wallace, D.J.; Tegzová, D.; Sanchez-Guerrero, J.; Schwarting, A.; Merrill, J.T.; Chatham, W.W.; et al. A phase III, randomized, placebo-controlled study of belimumab, a monoclonal antibody that inhibits B lymphocyte stimulator, in patients with systemic lupus erythematosus. Arthritis Rheum. 2011, 63, 3918–3930. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S.; Spencer, D.M.; Lipsky, P.E.; Rovin, B.H. Assay variation in the detection of antinuclear antibodies in the sera of patients with established SLE. Ann. Rheum. Dis. 2018, 77, 911–913. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.J.; Choi, M.Y.; Fritzler, M.J. Emerging technologies in autoantibody testing for rheumatic diseases. Arthrit. Res. Ther. 2017, 19, 172. [Google Scholar] [CrossRef]
- Pisetsky, D.S.; Lipsky, P.E. New insights into the role of antinuclear antibodies in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2020, 16, 565–579. [Google Scholar] [CrossRef]
- Pisetsky, D.S.; Bossuyt, X.; Meroni, P.L. ANA as an entry criterion for the classification of SLE. Autoimmun. Rev. 2019, 18, 102400. [Google Scholar] [CrossRef]
- Leffler, J.; Bengtsson, A.A.; Blom, A.M. The complement system in systemic lupus erythematosus: An update. Ann. Rheum. Dis. 2014, 73, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- Trouw, L.A.; Pickering, M.C.; Blom, A.M. The complement system as a potential therapeutic target in rheumatic disease. Nat. Rev. Rheumatol. 2017, 13, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lin, S.; Yang, S.; Chen, L.; Zheng, X. Diagnostic value of serum complement C3 and C4 levels in Chinese patients with systemic lupus erythematosus. Clin. Rheumatol. 2015, 34, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Petri, M.A.; van Vollenhoven, R.F.; Buyon, J.; Levy, R.A.; Navarra, S.V.; Cervera, R.; Zhong, Z.J.; Freimuth, W.W. Baseline predictors of systemic lupus erythematosus flares: Data from the combined placebo groups in the phase III belimumab trials. Arthritis Rheum. 2013, 65, 2143–2153. [Google Scholar] [CrossRef]
- Ho, A.; Barr, S.G.; Magder, L.S.; Petri, M. A decrease in complement is associated with increased renal and hematologic activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2001, 44, 2350–2357. [Google Scholar] [CrossRef]
- Gómez-Puerta, J.A.; Burlingame, R.W.; Cervera, R. Anti-chromatin (anti-nucleosome) antibodies: Diagnostic and clinical value. Autoimmun. Rev. 2008, 7, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Bizzaro, N.; Villalta, D.; Giavarina, D.; Tozzoli, R. Are anti-nucleosome antibodies a better diagnostic marker than anti-dsDNA antibodies for systemic lupus erythematosus? A systematic review and a study of metanalysis. Autoimmun. Rev. 2012, 12, 97–106. [Google Scholar] [CrossRef]
- Dima, A.; Opris, D.; Jurcut, C.; Baicus, C. Is there still a place for erythrocyte sedimentation rate and C-reactive protein in systemic lupus erythematosus? Lupus 2016, 25, 1173–1179. [Google Scholar] [CrossRef]
- Bertoli, A.M.; Vilá, L.M.; Reveille, J.D.; Alarcón, G.S. Systemic lupus erythematosus in a multiethnic US cohort (LUMINA): LXI. Value of C-reactive protein as a marker of disease activity and damage. J. Rheumatol. 2008, 35, 2355–2358. [Google Scholar] [CrossRef] [PubMed]
- Stojan, G.; Fang, H.; Magder, L.; Petri, M. Erythrocyte sedimentation rate is a predictor of renal and overall SLE disease activity. Lupus 2013, 22, 827–834. [Google Scholar] [CrossRef]
- Merrill, J.T.; Petri, M.A.; Buyon, J.; Ramsey-Goldman, R.; Kalunian, K.; Putterman, C.; Conklin, J.; Furie, R.A.; Dervieux, T. Erythrocyte-bound C4d in combination with complement and autoantibody status for the monitoring of SLE. Lupus Sci. Med. 2018, 5, e000263. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, G.S.; Calvo-Alén, J.; McGwin, G.; Uribe, A.G.; Toloza, S.M.A.; Roseman, J.M.; Fernández, M.; Fessler, B.J.; Vilá, L.M.; Ahn, C.; et al. Systemic lupus erythematosus in a multiethnic cohort: LUMINA XXXV. Predictive factors of high disease activity over time. Ann. Rheum. Dis. 2006, 65, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, A.F.; Solomon, D.H. Guidelines for immunologic laboratory testing in the rheumatic diseases: Anti-DNA antibody tests. Arthritis Rheum. 2002, 47, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Amezcua-Guerra, L.M.; Higuera-Ortiz, V.; Arteaga-García, U.; Gallegos-Nava, S.; Hübbe-Tena, C. Performance of the 2012 Systemic Lupus International Collaborating Clinics and the 1997 American College of Rheumatology classification criteria for systemic lupus erythematosus in a real-life scenario. Arthritis Care Res. 2015, 67, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Frodlund, M.; Wetterö, J.; Dahle, C.; Dahlström, Ö.; Skogh, T.; Rönnelid, J.; Sjöwall, C. Longitudinal anti-nuclear antibody (ANA) seroconversion in systemic lupus erythematosus: A prospective study of Swedish cases with recent-onset disease. Clin. Exp. Immunol. 2020, 199, 245–254. [Google Scholar] [CrossRef]
- Kwon, O.C.; Lee, J.S.; Ghang, B.; Kim, Y.G.; Lee, C.K.; Yoo, B.; Hong, S. Predicting eventual development of lupus nephritis at the time of diagnosis of systemic lupus erythematosus. Semin. Arthritis Rheum. 2018, 48, 462–466. [Google Scholar] [CrossRef]
- Ahn, S.S.; Yoo, B.W.; Song, J.J.; Park, Y.B.; Lee, S.K.; Lee, S.W. Anti-Sm is associated with the early poor outcome of lupus nephritis. Int. J. Rheum. Dis. 2016, 19, 897–902. [Google Scholar] [CrossRef]
- Ishizaki, J.; Saito, K.; Nawata, M.; Mizuno, Y.; Tokunaga, M.; Sawamukai, N.; Tamura, M.; Hirata, S.; Yamaoka, K.; Hasegawa, H.; et al. Low complements and high titre of anti-Sm antibody as predictors of histopathologically proven silent lupus nephritis without abnormal urinalysis in patients with systemic lupus erythematosus. Rheumatology 2015, 54, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Moroni, G.; Radice, A.; Giammarresi, G.; Quaglini, S.; Gallelli, B.; Leoni, A.; Li Vecchi, M.; Vecchi, M.L.; Messa, P.; Sinico, R.A. Are laboratory tests useful for monitoring the activity of lupus nephritis? A 6-year prospective study in a cohort of 228 patients with lupus nephritis. Ann. Rheum. Dis. 2009, 68, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Matrat, A.; Veysseyre-Balter, C.; Trolliet, P.; Villar, E.; Dijoud, F.; Bienvenu, J.; Fabien, N. Simultaneous detection of anti-C1q and anti-double stranded DNA autoantibodies in lupus nephritis: Predictive value for renal flares. Lupus 2011, 20, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Trendelenburg, M.; Marfurt, J.; Gerber, I.; Tyndall, A.; Schifferli, J.A. Lack of occurrence of severe lupus nephritis among anti-C1q autoantibody-negative patients. Arthritis Rheum. 1999, 42, 187–188. [Google Scholar] [CrossRef]
- Orbai, A.M.; Truedsson, L.; Sturfelt, G.; Nived, O.; Fang, H.; Alarcón, G.S.; Gordon, C.; Merrill, J.; Fortin, P.R.; Bruce, I.N.; et al. Anti-C1q antibodies in systemic lupus erythematosus. Lupus 2015, 24, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sun, L.; Jin, L. Spot urine protein/creatinine ratio is unreliable estimate of 24 h proteinuria in lupus nephritis when the histological scores of activity index are higher. Lupus 2015, 24, 943–947. [Google Scholar] [CrossRef]
- Guedes Marques, M.; Cotovio, P.; Ferrer, F.; Silva, C.; Botelho, C.; Lopes, K.; Maia, P.; Carreira, A.; Campos, M. Random spot urine protein/creatinine ratio: A reliable method for monitoring lupus nephritis? Clin. Kidney J. 2013, 6, 590–594. [Google Scholar] [CrossRef][Green Version]
- Schwartz, N.; Rubinstein, T.; Burkly, L.C.; Collins, C.E.; Blanco, I.; Su, L.; Hojaili, B.; Mackay, M.; Aranow, C.; Stohl, W.; et al. Urinary TWEAK as a biomarker of lupus nephritis: A multicenter cohort study. Arthrit. Res. Ther. 2009, 11, R143. [Google Scholar] [CrossRef] [PubMed]
- Howe, H.S.; Kong, K.O.; Thong, B.Y.H.; Law, W.G.; Chia, F.L.A.; Lian, T.Y.; Lau, T.C.; Chng, H.H.; Leung, B.P.L. Urine sVCAM-1 and sICAM-1 levels are elevated in lupus nephritis. Int. J. Rheum. Dis. 2012, 15, 13–16. [Google Scholar] [CrossRef]
- Guo Liu, R.-N.; Cheng, Q.-Y.; Zhou, H.-Y.; Li, B.-Z.; Ye, D.-Q. Elevated Blood and Urinary ICAM-1 is a Biomarker for Systemic Lupus Erythematosus: A Systematic Review and Meta-Analysis. Immunol. Investig. 2020, 49, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Vanarsa, K.; Soomro, S.; Zhang, T.; Strachan, B.; Pedroza, C.; Nidhi, M.; Cicalese, P.; Gidley, C.; Dasari, S.; Mohan, S.; et al. Quantitative planar array screen of 1000 proteins uncovers novel urinary protein biomarkers of lupus nephritis. Ann. Rheum. Dis. 2020, 79, 1349–1361. [Google Scholar] [CrossRef]
- Yu, H.; Jiang, L.; Liu, R.; Yang, A.; Yang, X.; Wang, L.; Zhang, W.; Che, T. Association between the ratio of aryl hydrocarbon receptor (AhR) in Th17 cells to AhR in Treg cells and SLE skin lesions. Int. Immunopharmacol. 2019, 69, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Pelkum, J.; Fritzler, M.; Mahler, M. Latest update on the Ro/SS-A autoantibody system. Autoimmun. Rev. 2009, 8, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Billi, A.C.; Gharaee-Kermani, M.; Fullmer, J.; Tsoi, L.C.; Hill, B.D.; Gruszka, D.; Ludwig, J.; Xing, X.; Estadt, S.; Wolf, S.J.; et al. The female-biased factor VGLL3 drives cutaneous and systemic autoimmunity. JCI Insight 2019, 4, e127291. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch-David, H.; Muller, S. Neuropsychiatric systemic lupus erythematosus: Pathogenesis and biomarkers. Nat. Rev. Neurol. 2014, 10, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Ho, R.C.; Thiaghu, C.; Ong, H.; Lu, Y.; Ho, C.S.; Tam, W.W.; Zhang, M.W. A meta-analysis of serum and cerebrospinal fluid autoantibodies in neuropsychiatric systemic lupus erythematosus. Autoimmun. Rev. 2016, 15, 124–138. [Google Scholar] [CrossRef]
- Choi, M.Y.; FitzPatrick, R.D.; Buhler, K.; Mahler, M.; Fritzler, M.J. A review and meta-analysis of anti-ribosomal P autoantibodies in systemic lupus erythematosus. Autoimmun. Rev. 2020, 19, 102463. [Google Scholar] [CrossRef] [PubMed]
- Hirohata, S.; Sakuma, Y.; Yanagida, T.; Yoshio, T. Association of cerebrospinal fluid anti-Sm antibodies with acute confusional state in systemic lupus erythematosus. Arthrit. Res. Ther. 2014, 16, 450. [Google Scholar] [CrossRef]
- Fujii, T. Direct and indirect pathogenic roles of autoantibodies in systemic autoimmune diseases. Allergol. Int. 2014, 63, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Arinuma, Y.; Yanagida, T.; Hirohata, S. Association of cerebrospinal fluid anti-NR2 glutamate receptor antibodies with diffuse neuropsychiatric systemic lupus erythematosus. Arthritis Rheum. 2008, 58, 1130–1135. [Google Scholar] [CrossRef]
- Fragoso-Loyo, H.; Cabiedes, J.; Orozco-Narváez, A.; Dávila-Maldonado, L.; Atisha-Fregoso, Y.; Diamond, B.; Llorente, L.; Sánchez-Guerrero, J. Serum and cerebrospinal fluid autoantibodies in patients with neuropsychiatric lupus erythematosus. Implications for diagnosis and pathogenesis. PLoS ONE 2008, 3, e3347. [Google Scholar] [CrossRef] [PubMed]
- Bertsias, G.K.; Boumpas, D.T. Pathogenesis, diagnosis and management of neuropsychiatric SLE manifestations. Nat. Rev. Rheumatol. 2010, 6, 358–367. [Google Scholar] [CrossRef]
- Fragoso-Loyo, H.; Richaud-Patin, Y.; Orozco-Narváez, A.; Dávila-Maldonado, L.; Atisha-Fregoso, Y.; Llorente, L.; Sánchez-Guerrero, J. Interleukin-6 and chemokines in the neuropsychiatric manifestations of systemic lupus erythematosus. Arthritis Rheum. 2007, 56, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- López, P.; Rodríguez-Carrio, J.; Martínez-Zapico, A.; Pérez-Álvarez, Á.I.; Suárez-Díaz, S.; Mozo, L.; Benavente, L.; Caminal-Montero, L.; Suárez, A. Low-density granulocytes and monocytes as biomarkers of cardiovascular risk in systemic lupus erythematosus. Rheumatology 2020, 59, 1752–1764. [Google Scholar] [CrossRef]
- Chezel, J.; Costedoat-Chalumeau, N.; Laouénan, C.; Rouzaud, D.; Chenevier-Gobeaux, C.; Le Guern, V.; Mathian, A.; Belhadi, D.; de Almeida Chaves, S.; Duhaut, P.; et al. Highly sensitive serum cardiac troponin T and cardiovascular events in patients with systemic lupus erythematosus (TROPOPLUS study). Rheumatology 2021, 60, 1210–1215. [Google Scholar] [CrossRef]
- Winau, L.; Hinojar Baydes, R.; Braner, A.; Drott, U.; Burkhardt, H.; Sangle, S.; D’Cruz, D.P.; Carr-White, G.; Marber, M.; Schnoes, K.; et al. High-sensitive troponin is associated with subclinical imaging biosignature of inflammatory cardiovascular involvement in systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Yu, M.; Morin, E.E.; Kang, J.; Kaplan, M.J.; Schwendeman, A. High-Density Lipoprotein in Lupus: Disease Biomarkers and Potential Therapeutic Strategy. Arthritis Rheumatol. 2020, 72, 20–30. [Google Scholar] [CrossRef]
- López, P.; Rodríguez-Carrio, J.; Martínez-Zapico, A.; Pérez-Álvarez, Á.I.; López-Mejías, R.; Benavente, L.; Mozo, L.; Caminal-Montero, L.; González-Gay, M.A.; Suárez, A. Serum Levels of Anti-PON1 and Anti-HDL Antibodies as Potential Biomarkers of Premature Atherosclerosis in Systemic Lupus Erythematosus. Thromb. Haemost. 2017, 117, 2194–2206. [Google Scholar] [CrossRef] [PubMed]
- Domingues, V.; Magder, L.S.; Petri, M. Assessment of the independent associations of IgG, IgM and IgA isotypes of anticardiolipin with thrombosis in SLE. Lupus Sci. Med. 2016, 3, e000107. [Google Scholar] [CrossRef]
- Skeoch, S.; Haque, S.; Pemberton, P.; Bruce, I.N. Cell adhesion molecules as potential biomarkers of nephritis, damage and accelerated atherosclerosis in patients with SLE. Lupus 2014, 23, 819–824. [Google Scholar] [CrossRef]
- Stanley, S.; Mok, C.C.; Vanarsa, K.; Habazi, D.; Li, J.; Pedroza, C.; Saxena, R.; Mohan, C. Identification of Low-Abundance Urinary Biomarkers in Lupus Nephritis Using Electrochemiluminescence Immunoassays. Arthritis Rheumatol. 2019, 71, 744–755. [Google Scholar] [CrossRef]
- Rovin, B.H.; Parikh, S.V.; Alvarado, A. The kidney biopsy in lupus nephritis: Is it still relevant? Rheum. Dis. Clin. N. Am. 2014, 40, 537–552. [Google Scholar] [CrossRef]
- Caster, D.J.; Merchant, M.L.; Klein, J.B.; Powell, D.W. Precision medicine in lupus nephritis: Can biomarkers get us there? Transl. Res. 2018, 201, 26–39. [Google Scholar] [CrossRef]
- Aragón, C.C.; Tafúr, R.-A.; Suárez-Avellaneda, A.; Martínez, M.T.; Salas, A.L.; Tobón, G.J. Urinary biomarkers in lupus nephritis. J. Transl. Autoimmun. 2020, 3, 100042. [Google Scholar] [CrossRef] [PubMed]
- Giannico, G.; Fogo, A.B. Lupus nephritis: Is the kidney biopsy currently necessary in the management of lupus nephritis? Clin. J. Am. Soc. Nephrol. CJASN 2013, 8, 138–145. [Google Scholar] [CrossRef]
- Pisetsky, D.S. Anti-DNA antibodies--quintessential biomarkers of SLE. Nat. Rev. Rheumatol. 2016, 12, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Bombardier, C.; Gladman, D.D.; Urowitz, M.B.; Caron, D.; Chang, C.H. Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992, 35, 630–640. [Google Scholar] [CrossRef] [PubMed]
- ter Borg, E.J.; Horst, G.; Hummel, E.J.; Limburg, P.C.; Kallenberg, C.G. Measurement of increases in anti-double-stranded DNA antibody levels as a predictor of disease exacerbation in systemic lupus erythematosus. A long-term, prospective study. Arthritis Rheum. 1990, 33, 634–643. [Google Scholar] [CrossRef]
- de Leeuw, K.; Bungener, L.; Roozendaal, C.; Bootsma, H.; Stegeman, C.A. Auto-antibodies to double-stranded DNA as biomarker in systemic lupus erythematosus: Comparison of different assays during quiescent and active disease. Rheumatology 2017, 56, 698–703. [Google Scholar] [CrossRef][Green Version]
- Steiman, A.J.; Urowitz, M.B.; Ibañez, D.; Li, T.T.; Gladman, D.D.; Wither, J. Anti-dsDNA and Antichromatin Antibody Isotypes in Serologically Active Clinically Quiescent Systemic Lupus Erythematosus. J. Rheumatol. 2015, 42, 810–816. [Google Scholar] [CrossRef]
- Schejbel, L.; Skattum, L.; Hagelberg, S.; Åhlin, A.; Schiller, B.; Berg, S.; Genel, F.; Truedsson, L.; Garred, P. Molecular basis of hereditary C1q deficiency--revisited: Identification of several novel disease-causing mutations. Genes Immun. 2011, 12, 626–634. [Google Scholar] [CrossRef][Green Version]
- Stojan, G.; Petri, M. Anti-C1q in systemic lupus erythematosus. Lupus 2016, 25, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Sinico, R.A.; Rimoldi, L.; Radice, A.; Bianchi, L.; Gallelli, B.; Moroni, G. Anti-C1q autoantibodies in lupus nephritis. Ann. N. Y. Acad. Sci. 2009, 1173, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Chamberlain, C.; Alarcón-Riquelme, M.E. Omics studies: Their use in diagnosis and reclassification of SLE and other systemic autoimmune diseases. Rheumatolog 2017, 56, i78–i87. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, C.; Lahesmaa, R.; Bonifacio, E.; Achenbach, P.; Tree, T. Immunological biomarkers for the development and progression of type 1 diabetes. Diabetologia 2018, 61, 2252–2258. [Google Scholar] [CrossRef]
- Arriens, C.; Mohan, C. Systemic lupus erythematosus diagnostics in the ’omics’ era. Int. J. Clin. Rheumtol. 2013, 8, 671–687. [Google Scholar] [CrossRef]
- Wang, T.-Y.; Wang, Y.-F.; Zhang, Y.; Shen, J.J.; Guo, M.; Yang, J.; Lau, Y.L.; Yang, W. Identification of Regulatory Modules That Stratify Lupus Disease Mechanism through Integrating Multi-Omics Data. Mol. Ther. Nucleic Acids 2020, 19, 318–329. [Google Scholar] [CrossRef]
- Der, E.; Suryawanshi, H.; Morozov, P.; Kustagi, M.; Goilav, B.; Ranabothu, S.; Izmirly, P.; Clancy, R.; Belmont, H.M.; Koenigsberg, M.; et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 2019, 20, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Nehar-Belaid, D.; Hong, S.; Marches, R.; Chen, G.; Bolisetty, M.; Baisch, J.; Walters, L.; Punaro, M.; Rossi, R.J.; Chung, C.-H.; et al. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat. Immunol. 2020, 21, 1094–1106. [Google Scholar] [CrossRef] [PubMed]
- Arazi, A.; Rao, D.A.; Berthier, C.C.; Davidson, A.; Liu, Y.; Hoover, P.J.; Chicoine, A.; Eisenhaure, T.M.; Jonsson, A.H.; Li, S.; et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 2019, 20, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Jourde-Chiche, N.; Whalen, E.; Gondouin, B.; Speake, C.; Gersuk, V.; Dussol, B.; Burtey, S.; Pascual, V.; Chaussabel, D.; Chiche, L. Modular transcriptional repertoire analyses identify a blood neutrophil signature as a candidate biomarker for lupus nephritis. Rheumatolog 2017, 56, 477–487. [Google Scholar] [CrossRef]
- Banchereau, R.; Hong, S.; Cantarel, B.; Baldwin, N.; Baisch, J.; Edens, M.; Cepika, A.M.; Acs, P.; Turner, J.; Anguiano, E.; et al. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell 2016, 165, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Scharer, C.D.; Blalock, E.L.; Mi, T.; Barwick, B.G.; Jenks, S.A.; Deguchi, T.; Cashman, K.S.; Neary, B.E.; Patterson, D.G.; Hicks, S.L.; et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat. Immunol. 2019, 20, 1071–1082. [Google Scholar] [CrossRef]
- Pernis, A.B.; Ivashkiv, L.B. ’-Omics’ shed light on B cells in lupus. Nat. Immunol. 2019, 20, 946–948. [Google Scholar] [CrossRef]
- Zhao, M.; Zhou, Y.; Zhu, B.; Wan, M.; Jiang, T.; Tan, Q.; Liu, Y.; Jiang, J.; Luo, S.; Tan, Y.; et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016, 75, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Zhang, Y.; Lin, Z.; Zhang, H.; Wang, T.Y.; Cao, Y.; Morris, D.L.; Sheng, Y.; Yin, X.; Zhong, S.L.; et al. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nat. Commun. 2021, 12, 772. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.; Alexsson, A.; Frodlund, M.; Morris, D.; Sandling, J.; Bolin, K.; Svenungsson, E.; Jönsen, A.; Bengtsson, C.; Gunnarsson, I.; et al. High genetic risk score is associated with early disease onset, damage accrual and decreased survival in systemic lupus erythematosus. Ann. Rheum. Dis. 2020, 79, 363–369. [Google Scholar] [CrossRef]
- Ota, M.; Nagafuchi, Y.; Hatano, H.; Ishigaki, K.; Terao, C.; Takeshima, Y.; Yanaoka, H.; Kobayashi, S.; Okubo, M.; Shirai, H.; et al. Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell 2021, 184, 3006–3021. [Google Scholar] [CrossRef]
- Chung, S.A.; Brown, E.E.; Williams, A.H.; Ramos, P.S.; Berthier, C.C.; Bhangale, T.; Alarcon-Riquelme, M.E.; Behrens, T.W.; Criswell, L.A.; Graham, D.C.; et al. Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. J. Am. Soc. Nephrol. 2014, 25, 2859–2870. [Google Scholar] [CrossRef]
- Yang, W.; Zhao, M.; Hirankarn, N.; Lau, C.S.; Mok, C.C.; Chan, T.M.; Wong, R.W.S.; Lee, K.W.; Mok, M.Y.; Wong, S.N.; et al. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum. Mol. Genet. 2009, 18, 2063–2070. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Jiang, H.; Gu, S.; Feng, N.; Zhang, N.; Lv, L.; Liu, F. Fecal Metabolites Were Altered, Identified as Biomarkers and Correlated With Disease Activity in Patients With Systemic Lupus Erythematosus in a GC-MS-Based Metabolomics Study. Front. Immunol. 2020, 11, 2138. [Google Scholar] [CrossRef]
- Nicolaou, O.; Sokratous, K.; Makowska, Z.; Morell, M.; De Groof, A.; Montigny, P.; Hadjisavvas, A.; Michailidou, K.; Oulas, A.; Spyrou, G.M.; et al. Proteomic analysis in lupus mice identifies Coronin-1A as a potential biomarker for lupus nephritis. Arthrit. Res. Ther. 2020, 22, 147. [Google Scholar] [CrossRef]
- Kok, H.M.; van den Hoogen, L.L.; van Roon, J.A.G.; Adriaansen, E.J.M.; Fritsch-Stork, R.D.E.; Nguyen, T.Q.; Goldschmeding, R.; Radstake, T.R.D.J.; Bovenschen, N. Systemic and local granzyme B levels are associated with disease activity, kidney damage and interferon signature in systemic lupus erythematosus. Rheumatology 2017, 56, 2129–2134. [Google Scholar] [CrossRef]
- Brown, M.A.; Li, Z.; Cao, K.-A.L. Biomarker development for axial spondyloarthritis. Nat. Rev. Rheumatol. 2020, 16, 448–463. [Google Scholar] [CrossRef]
| Biomarkers | ACR-1997 Criteria | SLICC-2012 Criteria | EULAR/ACR-2019 Criteria |
|---|---|---|---|
| Proteinuria | Persistent proteinuria > 0.5 g/24 h or >3+, if quantitation not performed | Urine protein to creatinine ratio (or 24-h urine protein) representing 500 mg protein/24 h | Proteinuria > 0.5 g/24 h by 24-h urine or equivalent spot urine protein to creatinine ratio |
| Urinary casts | Cellular casts may be red cell, hemoglobin, granular, tubular, or mixed | Red blood cell casts | — |
| Hemolytic anemia | Hemolytic anemia with reticulocytosis | Direct Coombs’ test in the absence of hemolytic anemia | Evidence of hemolysis, such as reticulocytosis, low haptoglobin, elevated indirect bilirubin, elevated LDH, and positive Coombs’ (direct antiglobulin) test |
| White blood cell count | White blood cell count < 4000/mm3 on 2 or more occasions; OR Lymphocyte count < 1500/mm3 on 2 or more occasions | White blood cell count < 4000/mm3 at least once, in the absence of other known causes such as Felty’s syndrome, drugs, and portal hypertension; OR Lymphocyte count < 1000/mm3 at least once, in the absence of other known causes such as corticosteroids, drugs, and infection | White blood cell count < 4000/mm3 |
| Platelet count | Platelet count < 100,000/mm3 in the absence of offending drugs | Platelet count < 100,000/mm3 at least once, in the absence of other known causes such as drugs, portal hypertension, and thrombotic thrombocytopenic purpura immunologic criteria | Platelet count < 100,000/mm3 |
| Sm antibody | Presence of antibodies to Sm nuclear antigen | Presence of antibodies to Sm nuclear antigen | anti-Sm antibodies |
| Serologic text for syphilis | False positive serologic test for syphilis known to be positive for at least 6 months and confirmed by Treponernapallidun immobilization or fluorescent treponemal antibody absorption test | — | — |
| Antinuclear antibody levels | An abnormal titer of antinuclear antibody by immunofluorescence or an equivalent assay at any point in time and in the absence of drugs known to be associated with “drug-induced lupus” syndrome | ANA level above laboratory reference range | ANA at a titer of ≥1:80 on HEp-2 cells or an equivalent positive test at least once; testing by immunofluorescence on HEp-2 cells or a solid-phase ANA screening immunoassay with at least equivalent performance is highly recommended |
| DNA antibody | Antibody to native DNA in abnormal titer | Anti-dsDNA antibody level above laboratory reference range (or 2-fold the reference range if tested by ELISA) | Anti-dsDNA antibodies in an immunoassay with demonstrated ≥ 90% specificity for SLE against relevant disease controls |
| CH50 | CH50 | Low CH50 | — |
| Complement 3 | Complement 3 | Low complement 3 | Low complement 3 |
| Complement 4 | Complement 4 | Low complement 4 | Low complement 4 |
| Complement 2 | Complement 2 | — | — |
| Antiphospholipid antibody | Antiphospholipid antibody positivity | Antiphospholipid antibody positivity as determined by any of the following: positive test result for lupus anticoagulant; false-positive test result for rapid plasma regain; medium- or high-titer anticardiolipin antibody level (IgA, IgG, or IgM); positive test result for anti-2-glycoprotein I (IgA, IgG, or IgM) | Anticardiolipin antibodies (IgA, IgG, or IgM) at medium or high titer (>40 APL, GPL, or MPL, or >the 99th percentile) or positive anti-β2GPI antibodies (IgA, IgG, or IgM) or positive lupus anticoagulant |
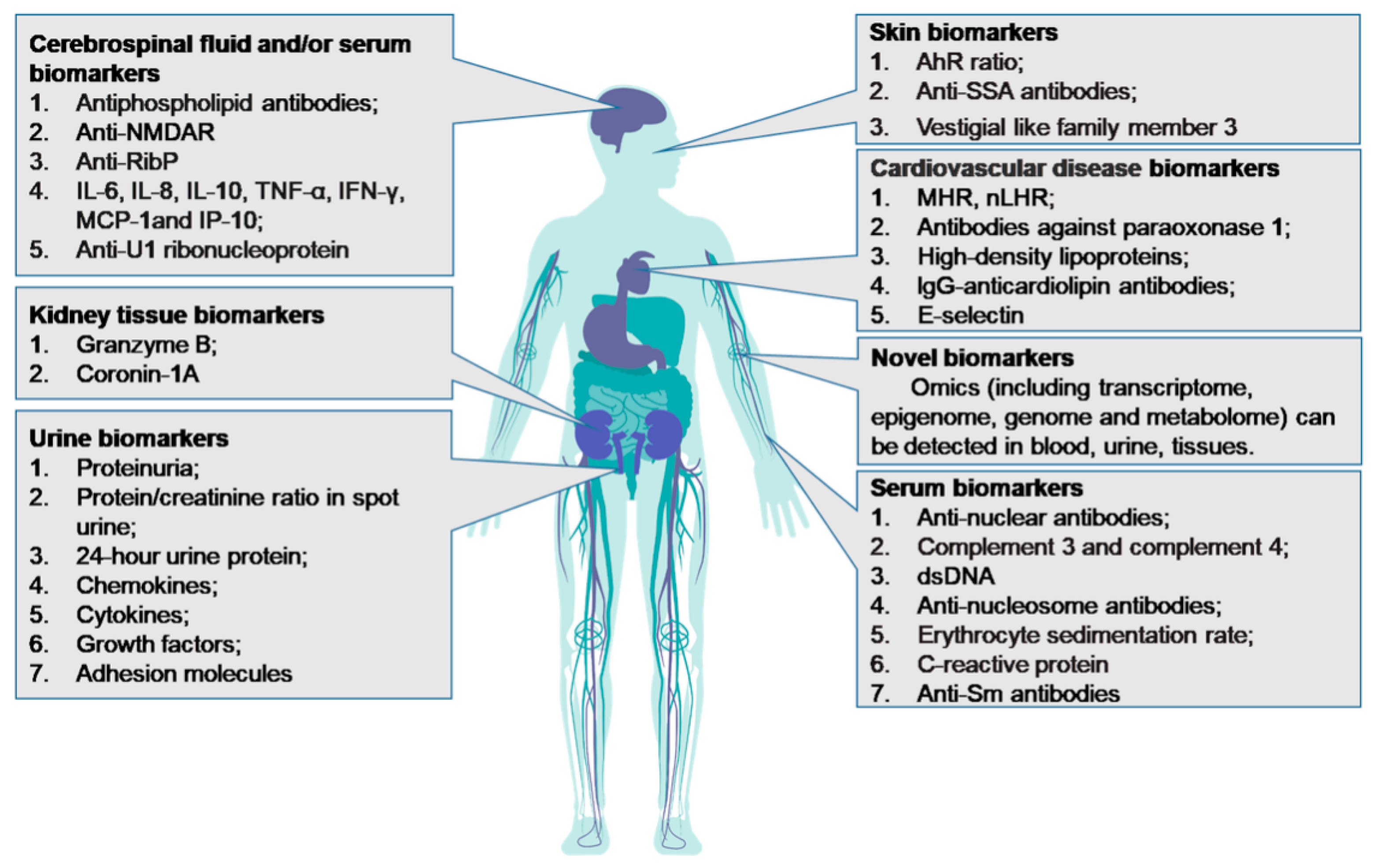
| Organ-Specific Damage in SLE | Biomarkers | Sample Type | Key Points | Refs |
|---|---|---|---|---|
| Lupus nephritis | Anti-dsDNA antibodies | Serum | Associated with SLE disease activity and can predict the development of LN; high specificity (96%), low diagnostic sensitivity (52–70%). | [18,50,51,52] |
| Anti-Sm antibodies | Serum | Correlates with SLE disease activity and LN; highly specific diagnostic biomarker for SLE with a specificity of 99% but with a low sensitivity of 5–30%; high titers of anti-Sm antibodies predict silent LN; predict early poor outcomes in LN. | [1,18,26,53,54,55,56,57] | |
| Anti-C1q antibodies | Serum | Increased anti-C1q antibody titers predict renal flares in LN with an 81–97% sensitivity and a 71% to 95% specificity; anti-C1q titerscorrelates with active LN, and the absence of anti-C1q is associated with a nearly 100% negative predictive value for the development of LN; Standardized laboratory assay has not been established. | [58,59,60,61] | |
| Proteinuria; Protein/creatinine ratio; 24-h urine protein | Urine | Conventional urinary biomarkers for LN. Spot urine protein/creatinine ratio is not always reliable estimate of 24-h proteinuria. | [62,63] | |
| Chemokines (MCP-1, IL-8, RANTES, IP-10, CXCL-16); Cytokines (TGF-β, IL-17,uTWEAK, adiponectin, IL-6, osteoprotegerin); Adhesionmolecules (VCAM-1, ICAM-1) | Urine | Evaluated as potential SLE biomarkers, but few of them have been independently validated. | [64,65,66] | |
| Angiopoetin-like 4; L-selectin; TGF-β1 | Urine | Biomarker candidates for tracking disease activity in LN. | [67] | |
| Skin lesions | AhR ratio | Serum | Associated with SLE activity and may be an independent risk factor for skin lesions in SLE. | [68] |
| Anti-SSA antibodies | Serum | Associated with subacute cutaneous lupus. | [69] | |
| VGLL-3 | Serum | Leads to cutaneous lupus. | [70] | |
| NPSLE | Lupus anticoagulant antibodies; Anticardiolipin antibodies; Anti-β2-glycoprotein I antibodies | Serum or CSF | Associated with NPSLE manifestations; diagnostic biomarkers of NPSLE and used to make treatment decisions. | [71,72] |
| Anti-RibP | Serum or CSF | Highly specific biomarker in the diagnosis of SLE and associated with NPSLE. | [73] | |
| Anti-U1 ribonucleoprotein antibodies | Serum or CSF | Elevated in CSF and sera of NPSLE patients; might cause NPSLE. | [74,75] | |
| Anti-NMDAR | CSF | Associated with central nervous system manifestations of NPSLE. | [76,77] | |
| IL-6, 8, 10, TNF-a; IFN-γ; MCP-1; IP-10 | CSF | Associated with NPSLE. | [78,79] | |
| CVD | LHR; MHR | Serum | Predicts CVD risk in SLE patients, even at the onset of disease. | [80] |
| Cardiac troponin T | Serum | Independently associated with incident cardiovascular events in SLE patients. | [81,82] | |
| Paraoxonase 1 and HDL | Serum | Key biomarker of accelerated atherosclerosis in lupus and may serve as a potential therapeutic biomarker for SLE patients with CVD; early biomarkers of endothelial damage and premature atherosclerosis in SLE; therapeutic targets for preventing CVD in SLE patients. | [83,84] | |
| IgG-anticardiolipin antibodies; E-selectin | Serum | Associated with CVD in SLE and correlated with disease activity. | [85,86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Nagafuchi, Y.; Fujio, K. Clinical and Immunological Biomarkers for Systemic Lupus Erythematosus. Biomolecules 2021, 11, 928. https://doi.org/10.3390/biom11070928
Yu H, Nagafuchi Y, Fujio K. Clinical and Immunological Biomarkers for Systemic Lupus Erythematosus. Biomolecules. 2021; 11(7):928. https://doi.org/10.3390/biom11070928
Chicago/Turabian StyleYu, Haitao, Yasuo Nagafuchi, and Keishi Fujio. 2021. "Clinical and Immunological Biomarkers for Systemic Lupus Erythematosus" Biomolecules 11, no. 7: 928. https://doi.org/10.3390/biom11070928
APA StyleYu, H., Nagafuchi, Y., & Fujio, K. (2021). Clinical and Immunological Biomarkers for Systemic Lupus Erythematosus. Biomolecules, 11(7), 928. https://doi.org/10.3390/biom11070928






