Kinetic Constraints in the Specific Interaction between Phosphorylated Ubiquitin and Proteasomal Shuttle Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. NMR Titration Experiments
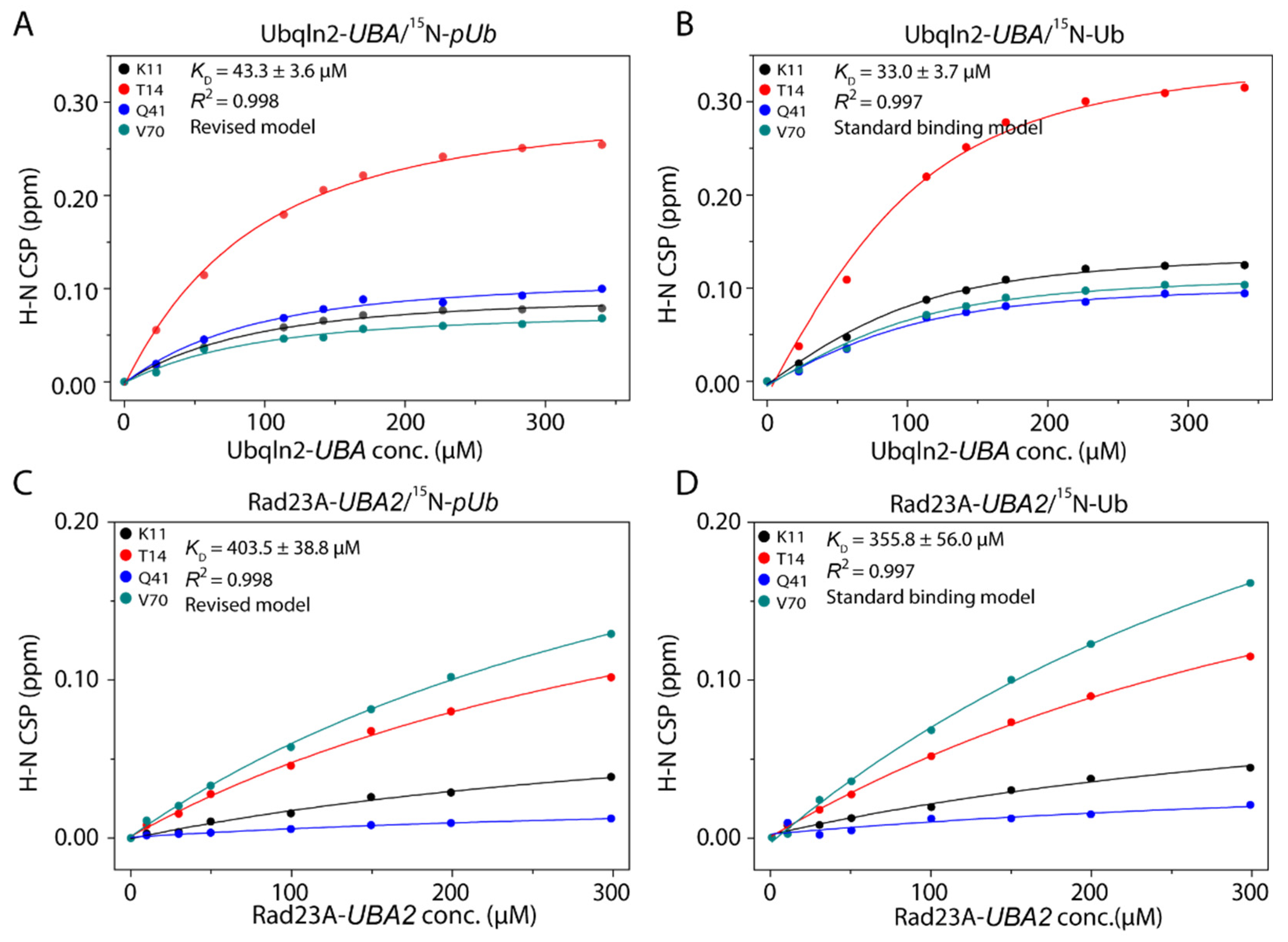
2.3. Determination of KD Value for pUb-UBA Complex
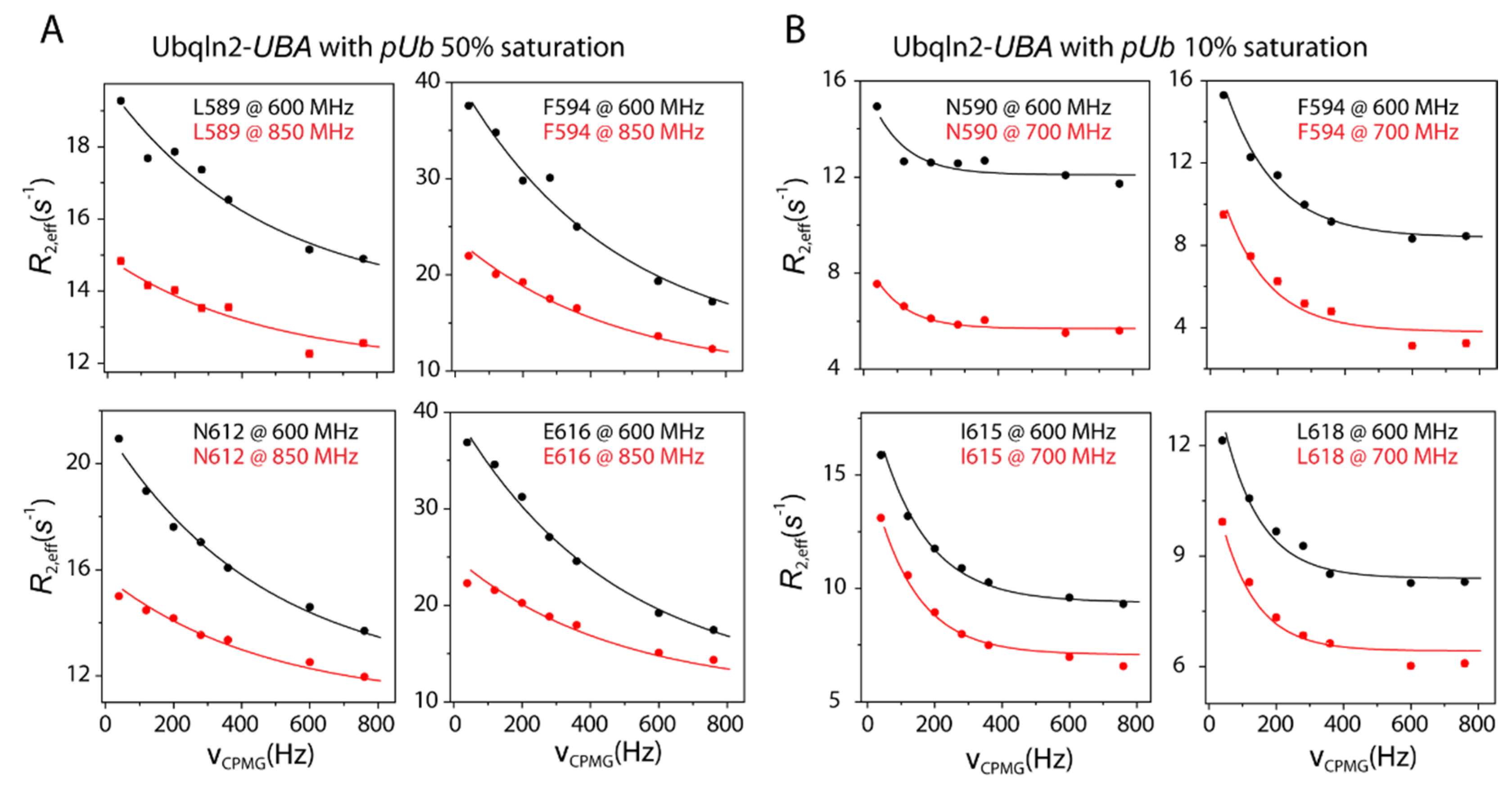
2.4. CPMG Relaxation Dispersion Experiment
2.5. Acquisition of NMR ZZ-Exchange Data
2.6. Calculation of the UBA-pUb Complex Structure
3. Results
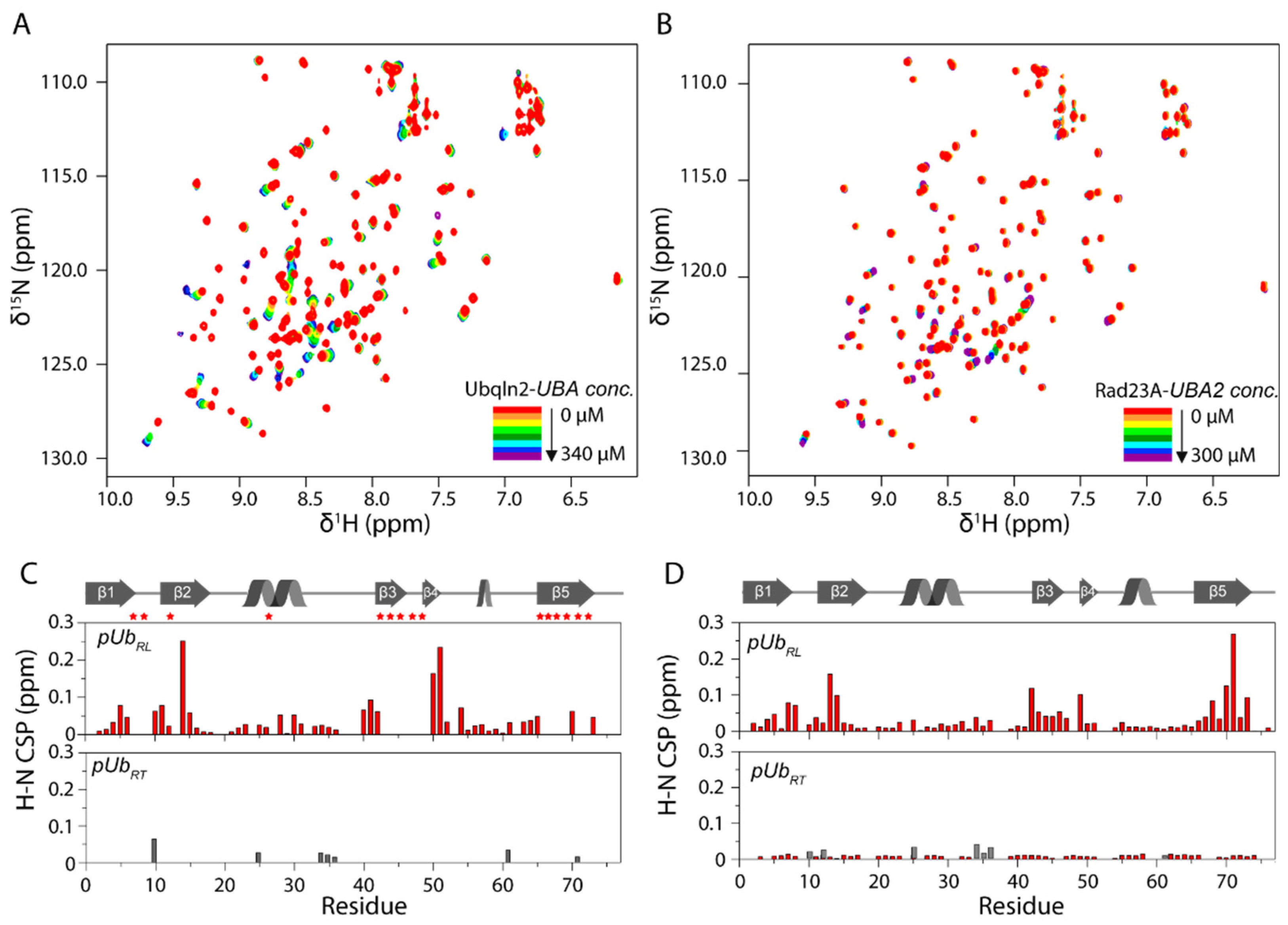
3.1. UBA Selectively Interacts with pUbRL
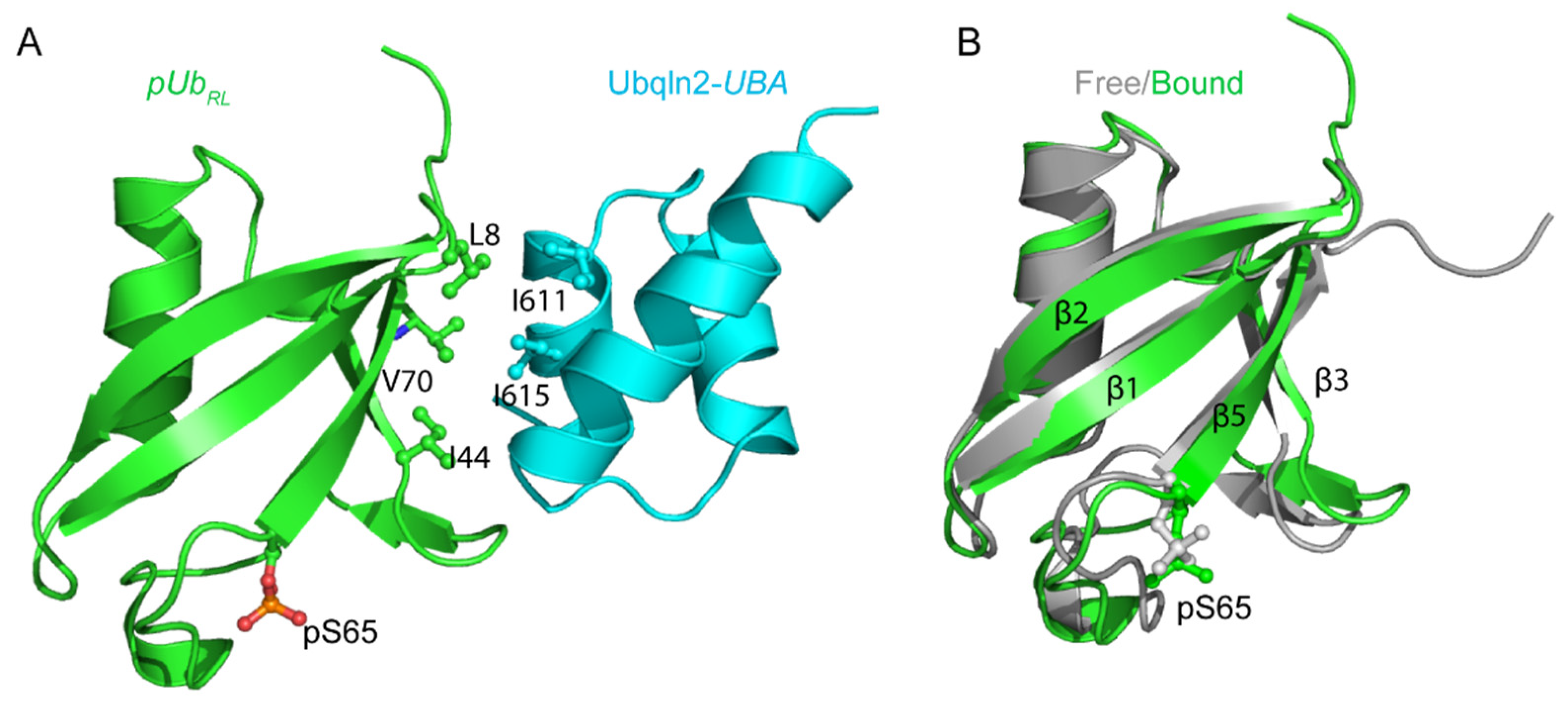
3.2. The Complex Structure Explains the Binding Preference for Ubqln2-UBA
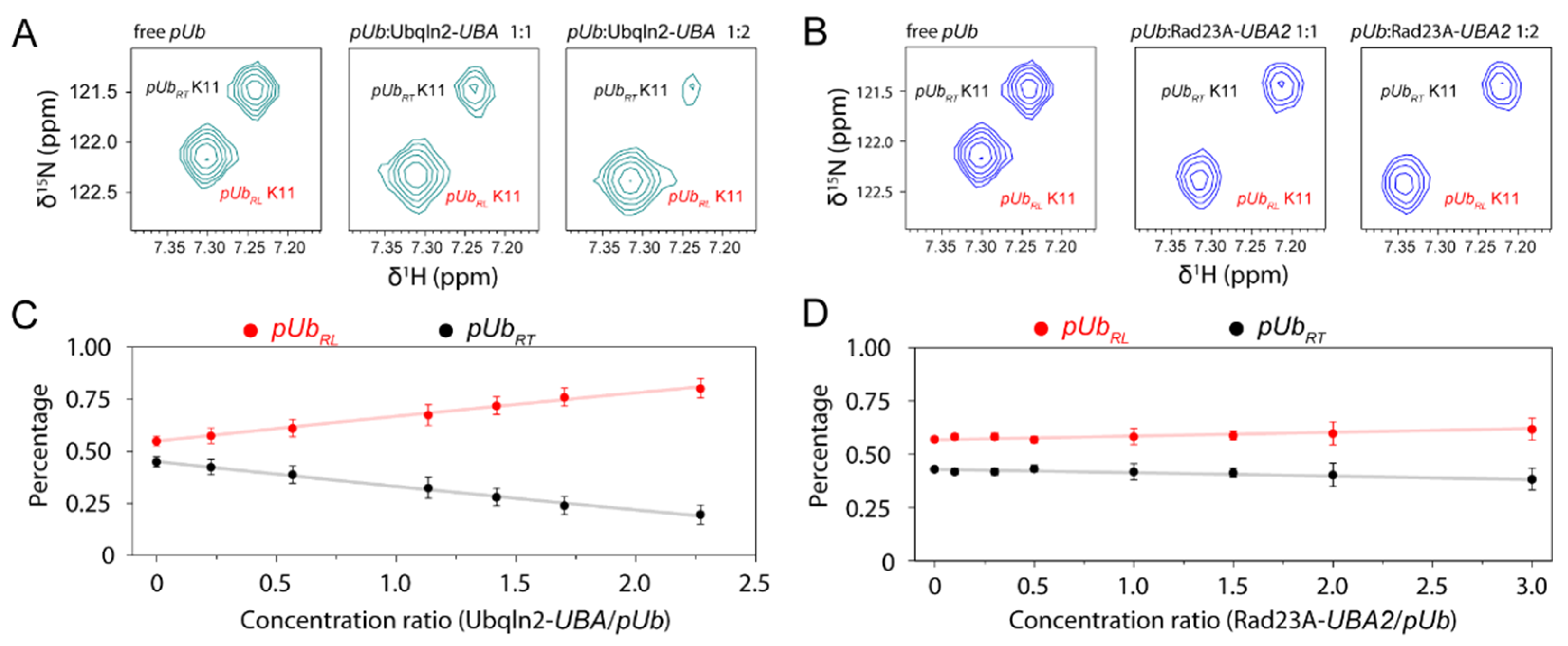
3.3. Kinetic Constraints for pUb Interaction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.X.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.H.; Shi, Y.; Zhang, N.; de Poot, S.A.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef]
- Liu, Z.; Dong, X.; Yi, H.W.; Yang, J.; Gong, Z.; Wang, Y.; Liu, K.; Zhang, W.P.; Tang, C. Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13. Cell Discov. 2019, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ebelle, D.L.; Wright, B.J.; Sridharan, V.; Hooper, E.; Walters, K.J. Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome. J. Mol. Biol. 2019, 431, 939–955. [Google Scholar] [CrossRef]
- Dao, T.P.; Kolaitis, R.M.; Kim, H.J.; O’Donovan, K.; Martyniak, B.; Colicino, E.; Hehnly, H.; Taylor, J.P.; Castaneda, C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 2018, 69, 965–978 e966. [Google Scholar] [CrossRef]
- Sharkey, L.M.; Safren, N.; Pithadia, A.S.; Gerson, J.E.; Dulchavsky, M.; Fischer, S.; Patel, R.; Lantis, G.; Ashraf, N.; Kim, J.H.; et al. Mutant UBQLN2 promotes toxicity by modulating intrinsic self-assembly. Proc. Natl. Acad. Sci. USA 2018, 115, E10495–E10504. [Google Scholar] [CrossRef]
- Renaud, L.; Picher-Martel, V.; Codron, P.; Julien, J.P. Key role of UBQLN2 in pathogenesis of amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. Commun. 2019, 7, 103. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yue, H.W.; He, W.T.; Hong, J.Y.; Jiang, L.L.; Hu, H.Y. PolyQ-expanded huntingtin and ataxin-3 sequester ubiquitin adaptors hHR23B and UBQLN2 into aggregates via conjugated ubiquitin. FASEB J. 2018, 32, 2923–2933. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300. [Google Scholar] [CrossRef]
- Mueller, T.D.; Kamionka, M.; Feigon, J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 2004, 279, 11926–11936. [Google Scholar] [CrossRef]
- Zhang, D.; Raasi, S.; Fushman, D. Affinity makes the difference: Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 2008, 377, 162–180. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.X.; Gong, Z.; Yang, J.; An, Y.X.; Liu, Z.; Zhao, Q.; Lescop, E.; Dong, X.; Tang, C. Ubiquitin is double-phosphorylated by PINK1 for enhanced pH-sensitivity of conformational switch. Protein Cell 2019, 10, 908–913. [Google Scholar] [CrossRef]
- Tang, C.; Zhang, W.P. How Phosphorylation by PINK1 Remodels the Ubiquitin System: A Perspective from Structure and Dynamics. Biochemistry 2020, 59, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef]
- Gladkova, C.; Maslen, S.L.; Skehel, J.M.; Komander, D. Mechanism of parkin activation by PINK1. Nature 2018, 559, 410–414. [Google Scholar] [CrossRef]
- Fiesel, F.C.; Ando, M.; Hudec, R.; Hill, A.R.; Castanedes-Casey, M.; Caulfield, T.R.; Moussaud-Lamodiere, E.L.; Stankowski, J.N.; Bauer, P.O.; Lorenzo-Betancor, O.; et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015, 16, 1114–1130. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Fiesel, F.C.; Truban, D.; Castanedes Casey, M.; Lin, W.L.; Soto, A.I.; Tacik, P.; Rousseau, L.G.; Diehl, N.N.; Heckman, M.G.; et al. Age- and disease-dependent increase of the mitophagy marker phospho-ubiquitin in normal aging and Lewy body disease. Autophagy 2018, 14, 1404–1418. [Google Scholar] [CrossRef]
- Wauer, T.; Swatek, K.N.; Wagstaff, J.L.; Gladkova, C.; Pruneda, J.N.; Michel, M.A.; Gersch, M.; Johnson, C.M.; Freund, S.M.; Komander, D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015, 34, 307–325. [Google Scholar] [CrossRef]
- Swaney, D.L.; Rodriguez-Mias, R.A.; Villen, J. Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover. EMBO Rep. 2015, 16, 1131–1144. [Google Scholar] [CrossRef]
- Huguenin-Dezot, N.; De Cesare, V.; Peltier, J.; Knebel, A.; Kristaryianto, Y.A.; Rogerson, D.T.; Kulathu, Y.; Trost, M.; Chin, J.W. Synthesis of Isomeric Phosphoubiquitin Chains Reveals that Phosphorylation Controls Deubiquitinase Activity and Specificity. Cell Rep. 2016, 16, 1180–1193. [Google Scholar] [CrossRef]
- Wauer, T.; Simicek, M.; Schubert, A.; Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 2015, 524, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Gong, Z.; Lu, Y.B.; Liu, K.; Qin, L.-Y.; Ran, M.L.; Zhang, C.L.; Liu, Z.; Zhang, W.P.; Tang, C. Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch. Proc. Natl. Acad. Sci. USA 2017, 114, 6770–6775. [Google Scholar] [CrossRef] [PubMed]
- Kazansky, Y.; Lai, M.Y.; Singh, R.K.; Fushman, D. Impact of different ionization states of phosphorylated Serine-65 on ubiquitin structure and interactions. Sci. Rep. 2018, 8, 2651. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Mittermaier, A.; Kay, L.E. New tools provide new insights in NMR studies of protein dynamics. Science 2006, 312, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Sugase, K.; Konuma, T.; Lansing, J.C.; Wright, P.E. Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J. Biomol. NMR 2013, 56, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Latham, M.P.; Zimmermann, G.R.; Pardi, A. NMR chemical exchange as a probe for ligand-binding kinetics in a theophylline-binding RNA aptamer. J. Am. Chem. Soc. 2009, 131, 5052–5053. [Google Scholar] [CrossRef][Green Version]
- Rücket, M.; Otting, G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000, 122, 7793–7797. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Bermejo, G.A.; Clore, G.M. Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 2018, 27, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.X.; Gu, X.H.; Dong, X.; Tang, C. Lanthanoid tagging via an unnatural amino acid for protein structure characterization. J. Biomol. NMR 2017, 67, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Massi, F.; Grey, M.J.; Palmer, A.G., 3rd. Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1rho relaxation experiments. Protein Sci. 2005, 14, 735–742. [Google Scholar] [CrossRef]
- Lange, O.F.; Lakomek, N.A.; Fares, C.; Schroder, G.F.; Walter, K.F.; Becker, S.; Meiler, J.; Grubmuller, H.; Griesinger, C.; de Groot, B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 2008, 320, 1471–1475. [Google Scholar] [CrossRef]
- Ban, D.; Funk, M.; Gulich, R.; Egger, D.; Sabo, T.M.; Walter, K.F.; Fenwick, R.B.; Giller, K.; Pichierri, F.; de Groot, B.L.; et al. Kinetics of conformational sampling in ubiquitin. Angew. Chem. Int. Ed. Engl. 2011, 50, 11437–11440. [Google Scholar] [CrossRef]
- Wlodarski, T.; Zagrovic, B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. USA 2009, 106, 19346–19351. [Google Scholar] [CrossRef]
- Peters, J.H.; de Groot, B.L. Ubiquitin dynamics in complexes reveal molecular recognition mechanisms beyond induced fit and conformational selection. PLoS Comput. Biol. 2012, 8, e1002704. [Google Scholar] [CrossRef]
- Paul, F.; Weikl, T.R. How to Distinguish Conformational Selection and Induced Fit Based on Chemical Relaxation Rates. PLoS Comput. Biol. 2016, 12, e1005067. [Google Scholar] [CrossRef]
- Phillips, A.H.; Zhang, Y.; Cunningham, C.N.; Zhou, L.; Forrest, W.F.; Liu, P.S.; Steffek, M.; Lee, J.; Tam, C.; Helgason, E.; et al. Conformational dynamics control ubiquitin-deubiquitinase interactions and influence in vivo signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 11379–11384. [Google Scholar] [CrossRef]
- Hammes, G.G.; Chang, Y.C.; Oas, T.G. Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA 2009, 106, 13737–13741. [Google Scholar] [CrossRef] [PubMed]
- Vogt, A.D.; Di Cera, E. Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 2012, 51, 5894–5902. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.-Y.; Gong, Z.; Liu, K.; Dong, X.; Tang, C. Kinetic Constraints in the Specific Interaction between Phosphorylated Ubiquitin and Proteasomal Shuttle Factors. Biomolecules 2021, 11, 1008. https://doi.org/10.3390/biom11071008
Qin L-Y, Gong Z, Liu K, Dong X, Tang C. Kinetic Constraints in the Specific Interaction between Phosphorylated Ubiquitin and Proteasomal Shuttle Factors. Biomolecules. 2021; 11(7):1008. https://doi.org/10.3390/biom11071008
Chicago/Turabian StyleQin, Ling-Yun, Zhou Gong, Kan Liu, Xu Dong, and Chun Tang. 2021. "Kinetic Constraints in the Specific Interaction between Phosphorylated Ubiquitin and Proteasomal Shuttle Factors" Biomolecules 11, no. 7: 1008. https://doi.org/10.3390/biom11071008
APA StyleQin, L.-Y., Gong, Z., Liu, K., Dong, X., & Tang, C. (2021). Kinetic Constraints in the Specific Interaction between Phosphorylated Ubiquitin and Proteasomal Shuttle Factors. Biomolecules, 11(7), 1008. https://doi.org/10.3390/biom11071008





