Functions and Regulation of Endogenous Retrovirus Elements during Zygotic Genome Activation: Implications for Improving Somatic Cell Nuclear Transfer Efficiency
Abstract
1. Introduction
2. ERV Functions during ZGA
3. ERV Activation during ZGA
4. Multi-Tiered Regulation of ERVs in Early Embryos
5. ERVs: Friend or Foe for Host
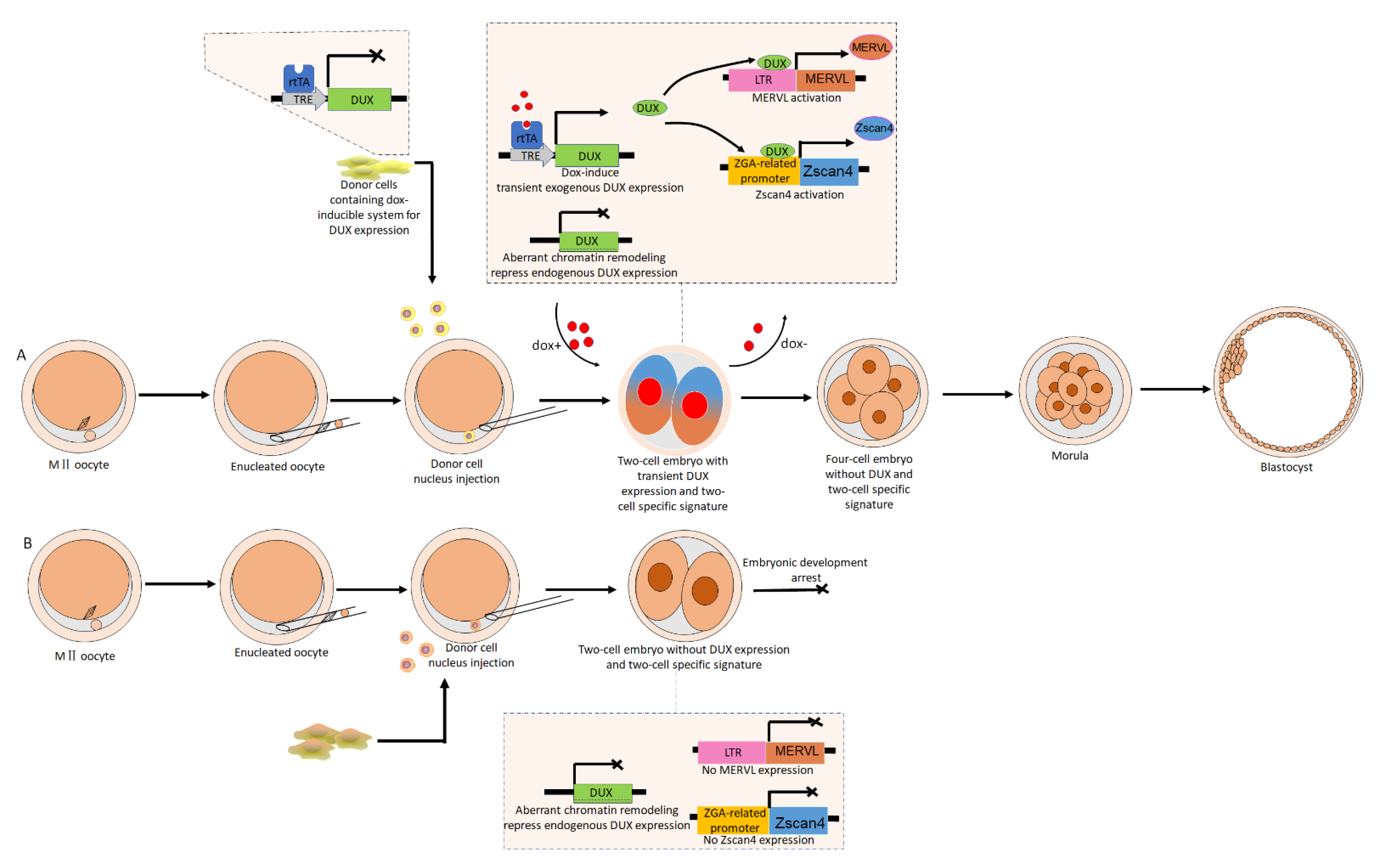
6. The Implications of ERVs Activation for Improving the Developmental Competence of Cloned Embryos
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Abe, K.; Yamamoto, R.; Franke, V.; Cao, M.; Suzuki, Y.; Suzuki, M.G.; Vlahovicek, K.; Svoboda, P.; Schultz, R.M.; Aoki, F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 2015, 34, 1523–1537. [Google Scholar] [CrossRef]
- Abe, K.-I.; Funaya, S.; Tsukioka, D.; Kawamura, M.; Suzuki, Y.; Suzuki, M.G.; Schultz, R.M.; Aoki, F. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. USA 2018, 115, E6780–E6788. [Google Scholar] [CrossRef]
- Johnson, W.E. Endogenous Retroviruses in the Genomics Era. Annu. Rev. Virol. 2015, 2, 135–159. [Google Scholar] [CrossRef] [PubMed]
- Dewannieux, M.; Heidmann, T. Endogenous retroviruses: Acquisition, amplification and taming of genome invaders. Curr. Opin. Virol. 2013, 3, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, X.; Rho, M.; Feschotte, C. Genome-wide characterization of endogenous retroviruses in the bat myotis iucifugus reveals recent and diverse infections. J. Virol. 2013, 87, 8493–8501. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Katzourakis, A. Marine origin of retroviruses in the early Palaeozoic Era. Nat. Commun. 2017, 8, 13954. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Gilbert, C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef]
- Metegnier, G.; Becking, T.; Chebbi, M.A.; Giraud, I.; Moumen, B.; Schaack, S.; Cordaux, R.; Gilbert, C. Comparative paleovi-rological analysis of crustaceans identifies multiple widespread viral groups. Mobile DNA 2015, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Cairns, J. The origins of human cancers. Nature 1981, 289, 353–357. [Google Scholar] [CrossRef]
- Nevers, P.; Saedler, H. Transposable genetic elements as agents of gene instability and chromosomal rearrangements. Nat. Cell Biol. 1977, 268, 109–115. [Google Scholar] [CrossRef]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J.; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of Somatic Retrotransposition in Human Cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Lopez, M.; Vilar-Astasio, R.; Tristan-Ramos, P.; Lopez-Ruiz, C.; Garcia-Pérez, J.L. Study of Transposable Elements and Their Genomic Impact. Adv. Struct. Saf. Stud. 2016, 1400, 1–19. [Google Scholar] [CrossRef]
- Doolittle, W.F.; Sapienza, C. Selfish genes, the phenotype paradigm and genome evolution. Nat. Cell Biol. 1980, 284, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Gu, T.-P.; Guo, F.; Yang, H.; Wu, H.-P.; Xu, G.-F.; Liu, W.; Xie, Z.-G.; Shi, L.; He, X.; Jin, S.-G.; et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nat. Cell Biol. 2011, 477, 606–610. [Google Scholar] [CrossRef]
- Peaston, A.E.; Evsikov, A.V.; Graber, J.H.; de Vries, W.N.; Holbrook, A.E.; Solter, D.; Knowles, B.B. Retrotransposons Regulate Host Genes in Mouse Oocytes and Preimplantation Embryos. Dev. Cell 2004, 7, 597–606. [Google Scholar] [CrossRef]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nat. Cell Biol. 2012, 487, 57–63. [Google Scholar] [CrossRef]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. In Mobile DNA III; Craig, N., Chandler, M., Gellert, M., Lambowitz, A., Rice, P., Sandmeyer, S., Eds.; ASM Press: Washington, DC, USA, 2015; pp. 1079–1100. [Google Scholar] [CrossRef]
- Leung, D.C.; Lorincz, M.C. Silencing of endogenous retroviruses: When and why do histone marks predominate? Trends Biochem. Sci. 2012, 37, 127–133. [Google Scholar] [CrossRef]
- Jumpei, I.; Ryota, S.; Hirofumi, N.; Shiro, Y.; Tetsuaki, K.; Takahide, H.; Ituro, I.; Cédric, F. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 2017, 13, e1006883. [Google Scholar]
- Malfavon-Borja, R.; Feschotte, C. Fighting Fire with Fire: Endogenous Retrovirus Envelopes as Restriction Factors. J. Virol. 2015, 89, 4047–4050. [Google Scholar] [CrossRef]
- Zhou, L.-Q.; Dean, J. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol. 2015, 25, 82–91. [Google Scholar] [CrossRef]
- Ishiuchi, T.; Enriquez-Gasca, R.; Mizutani, E.; Bošković, A.; Ziegler-Birling, C.; Rodriguez-Terrones, D.; Wakayama, T.; Vaquerizas, J.M.; Torres-Padilla, M.-E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015, 22, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.A.; Svensson, V.; Krueger, C.; Stubbs, T.M.; Giehr, P.; Krueger, F.; Miragaia, R.J.; Kyriakopoulos, C.; Berrens, R.V.; Milagre, I.; et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 2016, 17, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lin, C.P.; Risso, D.; Chen, S.; Kim, T.A.; Tan, M.H.; Li, J.B.; Wu, Y.; Chen, C.; Xuan, Z. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 2017, 355, 596. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kim, J.K.; Do, D.V.; Lee, C. Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. Elife 2017, 6, e22345. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, P.G.; Doráis, J.; Grow, E.J.; Whiddon, J.L.; Lim, J.W.; Wike, C.L.; Weaver, B.D.; Pflueger, C.; Emery, B.R.; Wilcox, A.L. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotrans-posons. Nature Genet. 2017, 49, 925–934. [Google Scholar] [CrossRef]
- Whiddon, J.L.; Langford, A.T.; Wong, C.-J.; Zhong, J.W.; Tapscott, S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017, 49, 935–940. [Google Scholar] [CrossRef]
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945. [Google Scholar] [CrossRef]
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H.S. Viable offspring derived from fetal and adult mammalian cells. Nat. Cell Biol. 1997, 385, 810–813. [Google Scholar] [CrossRef]
- Rodriguez-Osorio, N.; Urrego, R.; Cibelli, J.B.; Eilertsen, K.; Memili, E. Reprogramming mammalian somatic cell. Theriogenology 2012, 78, 1869–1886. [Google Scholar] [CrossRef]
- Teperek, M.; Miyamoto, K. Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes. Reprod. Med. Biol. 2013, 12, 133–149. [Google Scholar] [CrossRef]
- Matoba, S.; Liu, Y.; Lu, F.; Iwabuchi, K.A.; Shen, L.; Inoue, A.; Zhang, Y. Embryonic Development following Somatic Cell Nuclear Transfer Impeded by Persisting Histone Methylation. Cell 2014, 159, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Minami, N.; Imai, H. 89 Zygotically activated genes are suppressed in mouse nuclear-transferred embryos. Reprod. Fertil. Dev. 2007, 19, 162. [Google Scholar] [CrossRef]
- Liu, W.; Liu, X.; Wang, C.; Gao, Y.; Gao, R.; Kou, X.; Zhao, Y.; Li, J.; Wu, Y.; Xiu, W.; et al. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2016, 2, 16010. [Google Scholar] [CrossRef] [PubMed]
- Mda, B.; Zla, B.; Bc, B.; Ywa, B.; Hua, Y.; Yza, B.; Yu, C.; Jzb, C.; Feng, W. Aberrant DNA and histone methylation during zygotic genome activation in goat cloned embryos. Theriogenology 2020, 148, 27–36. [Google Scholar]
- Yang, L.; Song, L.; Lishuang, S.; Xuefei, L.; Lige, B.; Guangpeng, L. KDM6A and KDM6B play contrasting roles in nuclear transfer embryos revealed by MERVL reporter system. EMBO Rep. 2018, 19, e46240. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, F.; Zhang, L.; Wu, X.; Li, D.; Xin, J.; Xie, J.; Kong, F.; Wang, W.; Wu, Q.; et al. Transcriptional defects and reprogramming barriers in somatic cell nuclear reprogramming as revealed by single-embryo RNA sequencing. BMC Genom. 2018, 19, 734. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, L.; Liu, W.; Qiao, Z.; Shen, S.; Zhu, Q.; Gao, R.; Wang, M.; Wang, M.; Li, C.; et al. Dux-Mediated Corrections of Aberrant H3K9ac during 2-Cell Genome Activation Optimize Efficiency of Somatic Cell Nuclear Transfer. Cell Stem Cell 2021, 28, 150–163.e5. [Google Scholar] [CrossRef]
- Rowe, H.; Trono, D. Dynamic control of endogenous retroviruses during development. Virology 2011, 411, 273–287. [Google Scholar] [CrossRef]
- Qin, C.; Wang, Z.; Jin, S.; Bekkari, K.; Rong, L.; Pacchione, S.; Mcnulty, K.A.; Ng, A.; Barnum, J.E.; Storer, R.D. Intracisternal A particle genes: Distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of suscep-tibility to cancer. Mol. Carcinog. 2010, 49, 54–67. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Franke, V.; Ganesh, S.; Karlic, R.; Malik, R.; Pasulka, J.; Horvat, F.; Kuzman, M.; Fulka, H.; Cernohorska, M.; Urbanova, J.; et al. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 2017, 27, 1384–1394. [Google Scholar] [CrossRef]
- Cohen, C.J.; Lock, W.M.; Mager, D.L. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 2009, 448, 105–114. [Google Scholar] [CrossRef]
- Conley, A.B.; Jittima, P.; King, J.I. Retroviral promoters in the human genome. Bioinformatics 2008, 24, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, R.; Farivar, S.; Mager, D.L. C-GATE - catalogue of genes affected by transposable elements. Mobile DNA 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Grow, E.J.; Flynn, R.A.; Chavez, S.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nat. Cell Biol. 2015, 522, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Buckler-White, A.; Wollenberg, K.; Kozak, C.A. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl. Acad. Sci. USA 2009, 106, 3259–3263. [Google Scholar] [CrossRef]
- Yap, M.W.; Colbeck, E.; Ellis, S.A.; Stoye, J.P. Evolution of the Retroviral Restriction Gene Fv1: Inhibition of Non-MLV Retroviruses. PLoS Pathog. 2014, 10, e1003968. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Wang, L.; Li, J.; Zhao, Y.; Bou, G.; Li, Y.; Jiao, G.; Shen, X.; Wei, R.; et al. A novel long intergenic noncoding RNA indispensable for the cleavage of mouse two-cell embryos. EMBO Rep. 2016, 17, 1452–1470. [Google Scholar] [CrossRef]
- Zhou, W.; Chung, Y.J.; Castellar, E.R.P.; Zheng, Y.; Chung, H.-J.; Bandle, R.; Liu, J.; Tessarollo, L.; Batchelor, E.; Aplan, P.D.; et al. Far Upstream Element Binding Protein Plays a Crucial Role in Embryonic Development, Hematopoiesis, and Stabilizing Myc Expression Levels. Am. J. Pathol. 2016, 186, 701–715. [Google Scholar] [CrossRef]
- Ke, Y.; Xu, Y.; Chen, X.; Feng, S.; Liu, Z.; Sun, Y.; Yao, X.; Li, F.; Zhu, W.; Gao, L.; et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 2017, 170, 367–381.e20. [Google Scholar] [CrossRef] [PubMed]
- Gassler, J.; Brando, H.; Imakaev, M.; Flyamer, I.M.; Tachibana-Konwalski, K.A. Mechanism of cohesin-dependent loop ex-trusion organizes zygotic genome architecture. EMBO J. 2017, 36, 3600–3618. [Google Scholar] [CrossRef]
- Du, Z.; Zheng, H.; Huang, B.; Ma, R.; Wu, J.; Zhang, X.; He, J.; Xiang, Y.; Wang, Q.; Li, Y.; et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nat. Cell Biol. 2017, 547, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Tachibana-Konwalski, K.; Flyamer, I.M.; Mirny, L.A.; Imakaev, M. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar]
- Adam, B.; Maria-Elena, T.P. Epigenetic reprogramming and development: A unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genom. 2010, 9, 444–454. [Google Scholar]
- Casser, E.; Israel, S.; Witten, A.; Schulte, K.; Schlatt, S.; Nordhoff, V.; Boiani, M. Totipotency segregates between the sister blastomeres of two-cell stage mouse embryos. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tarkowski, A.K. Experiments on the Development of Isolated Blastomeres of Mouse Eggs. Nat. Cell Biol. 1959, 184, 1286–1287. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Nakagawa, S.; Ueda, M.T.; Imanishi, T.; Frith, M.C.; Mitsuhashi, H. Nanopore-based single molecule sequencing of the D4Z4 array responsible for facioscapulohumeral muscular dystrophy. Sci. Rep. 2017, 7, 14789. [Google Scholar] [CrossRef]
- Clapp, J.; Mitchell, L.M.; Bolland, D.J.; Fantes, J.; Corcoran, A.E.; Scotting, P.J.; Armour, J.A.; Hewitt, J.E. Evolutionary Conservation of a Coding Function for D4Z4, the Tandem DNA Repeat Mutated in Facioscapulohumeral Muscular Dystrophy. Am. J. Hum. Genet. 2007, 81, 264–279. [Google Scholar] [CrossRef]
- Sugie, K.; Funaya, S.; Kawamura, M.; Nakamura, T.; Suzuki, M.G.; Aoki, F. Expression of Dux family genes in early preimplantation embryos. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Geng, L.N.; Yao, Z.; Snider, L.; Fong, A.P.; Cech, J.N.; Young, J.M.; van der Maarel, S.; Ruzzo, W.L.; Gentleman, R.C.; Tawil, R.; et al. DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Dev. Cell 2012, 22, 38–51. [Google Scholar] [CrossRef]
- Hewitt, J. Loss of epigenetic silencing of the DUX4 transcription factor gene in facioscapulohumeral muscular dystrophy. Human Mol. Genet. 2015, 24, 17–23. [Google Scholar] [CrossRef]
- Ooga, M.; Fulka, H.; Hashimoto, S.; Suzuki, M.G.; Aoki, F. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics 2016, 11, 85–94. [Google Scholar] [CrossRef]
- Akiyama, T.; Suzuki, O.; Matsuda, J.; Aoki, F.; Bickmore, W.A. Dynamic replacement of histone H3 variants reprograms epi-genetic marks in early mouse embryos. PLoS Genet. 2011, 7, e1002279. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.; Okamoto, I.; Casanova, M.; El Marjou, F.; Le Baccon, P.; Almouzni, G. A Strand-Specific Burst in Transcription of Pericentric Satellites Is Required for Chromocenter Formation and Early Mouse Development. Dev. Cell 2010, 19, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Ryoma, Y.; Fugaku, A. A unique mechanism regulating gene expression in 1-cell embryos. J. Reprod. Dev. 2016, 63, 9–11. [Google Scholar]
- Yamamoto, R.; Abe, K.-I.; Suzuki, Y.; Suzuki, M.G.; Aoki, F. Characterization of gene expression in mouse embryos at the 1-cell stage. J. Reprod. Dev. 2016, 62, 87–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aoki, F.; Worrad, D.M.; Schultz, R.M. Regulation of transcriptional activity during the first and second cell cycles in the pre-implantation mouse embryo. Dev. Biol. 1997, 181, 296–307. [Google Scholar] [CrossRef]
- Ho, C.S.; Gearhart, M.; Cui, Z.; Darko, B.; Minjee, K.; Natalie, S.; Michael, K. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 2016, 44, 5161–5173. [Google Scholar]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nat. Cell Biol. 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, Y.; Zhou, J.; Bi, Y.; Xu, J.; Xu, C.; Kou, X.; Zhao, Y.; Li, Y.; Tu, Z.; et al. Precise temporal regulation of Dux is important for embryo development. Cell Res. 2019, 29, 956–959. [Google Scholar] [CrossRef]
- Yang, L.; Liu, X.; Song, L.; Di, A.; Su, G.; Bai, C.; Wei, Z.; Li, G. Transient Dux expression facilitates nuclear transfer and induced pluripotent stem cell reprogramming. EMBO Rep. 2020, 21, 50054. [Google Scholar] [CrossRef]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71. [Google Scholar] [CrossRef]
- Mak, J.; Kleiman, L. Primer tRNAs for reverse transcription. J. Virol. 1997, 71, 8087–8095. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Gaughan, J.; Midic, U.; Han, Z.; Liang, C.-G.; Patel, B.G.; Latham, K.E. Systems Genetics Implicates Cytoskeletal Genes in Oocyte Control of Cloned Embryo Quality. Genetics 2013, 193, 877–896. [Google Scholar] [CrossRef]
- Midic, U.; Vincent, K.A.; Wang, K.; Lokken, A.; Severance, A.L.; Ralston, A.; Knott, J.G.; Latham, K.E. Novel key roles for structural maintenance of chromosome flexible domain containing 1 (Smchd1 ) during preimplantation mouse development. Mol. Reprod. Dev. 2018, 85, 635–648. [Google Scholar] [CrossRef]
- Ruebel, M.L.; Vincent, K.A.; Schall, P.Z.; Wang, K.; Latham, K.E. SMCHD1 terminates the first embryonic genome activation event in mouse two-cell embryos and contributes to a transcriptionally repressive state. Am. J. Physiol. Physiol. 2019, 317, C655–C664. [Google Scholar] [CrossRef]
- Vitullo, P.; Sciamanna, I.; Baiocchi, M.; Sinibaldi-Vallebona, P.; Spadafora, C. LINE-1 retrotransposon copies are amplified during murine early embryo development. Mol. Reprod. Dev. 2012, 79, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Sciamanna, I.; Vitullo, P.; Curatolo, A.; Spadafora, C. A Reverse Transcriptase-Dependent Mechanism Is Essential for Murine Preimplantation Development. Genes 2011, 2, 360–373. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Bošković, A.; Rando, O.J.; Torres-Padilla, M.-E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Chamani, I.J.; Wang, F.; Luo, D.; Navarro, P.A.; Cortes, V.L.; Keefe, D.L. Inhibition of line-1 transposition blocks telomere elongation and downregulates totipotency genes during mouse embryo development. Fertil. Steril. 2019, 112, e126. [Google Scholar] [CrossRef]
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e19. [Google Scholar] [CrossRef] [PubMed]
- Wolf, G.; Yang, P.; Füchtbauer, A.C.; Füchtbauer, E.-M.; Silva, A.M.; Park, C.; Wu, W.; Nielsen, A.L.; Pedersen, F.S.; Macfarlan, T.S. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015, 29, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, R.; Krebs, J.R. Arms races between and within species. Proc. R. Soc. London. Ser. B Boil. Sci. 1979, 205, 489–511. [Google Scholar] [CrossRef]
- Evsikov, A.; De Vries, W.; Peaston, A.; Radford, E.; Fancher, K.; Chen, F.; Blake, J.; Bult, C.; Latham, K.; Solter, D.; et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 2004, 105, 240–250. [Google Scholar] [CrossRef]
- Evsikov, A.V.; Graber, J.H.; Brockman, J.M.; Hampl, A.; Holbrook, A.E.; Singh, P.; Eppig, J.J.; Solter, D.; Knowles, B.B. Cracking the egg: Molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006, 20, 2713–2727. [Google Scholar] [CrossRef]
- Bui, L.C.; Evsikov, A.V.; Khan, D.R.; Archilla, C.; Peynot, N.; Henaut, A.; Le Bourhis, D.; Vignon, X.; Renard, J.P.; Duranthon, V. Retrotransposon expression as a defining event of genome reprograming in fertilized and cloned bovine embryos. Reproduction 2009, 138, 289–299. [Google Scholar] [CrossRef]
- Faulkner, G.J.; Kimura, Y.; O Daub, C.; Wani, S.; Plessy, C.; Irvine, K.M.; Schroder, K.; Cloonan, N.; Steptoe, A.L.; Lassmann, T.; et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009, 41, 563–571. [Google Scholar] [CrossRef]
- Fischer, M.G.; Hackl, T. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nat. Cell Biol. 2016, 540, 288–291. [Google Scholar] [CrossRef]
- Britten, R.J.; Davidson, E.H. Gene Regulation for Higher Cells: A Theory. Science 1969, 165, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 1971, 46, 111–138. [Google Scholar]
- Blewitt, M.E.; Vickaryous, N.K.; Paldi, A.; Koseki, H.; Whitelaw, E. Dynamic reprogramming of DNA methylation at an epi-genetically sensitive allele in mice. PLoS Genetics 2006, 2, e49. [Google Scholar] [CrossRef]
- Cosby, R.L.; Chang, N.-C.; Feschotte, C. Host–transposon interactions: Conflict, cooperation, and cooption. Genes Dev. 2019, 33, 1098–1116. [Google Scholar] [CrossRef]
- Yang, X.; Smith, S.L.; Tian, X.C.; A Lewin, H.; Renard, J.-P.; Wakayama, T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 2007, 39, 295–302. [Google Scholar] [CrossRef]
- Li, S.; Chen, X.; Fang, Z.; Shi, J.; Sheng, H.Z. Rabbits generated from fibroblasts through nuclear transfer. Reproduction 2006, 131, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ross, J.W.; Hao, Y.; Spate, L.D.; Walters, E.M.; Samuel, M.S.; Rieke, A.; Murphy, C.N.; Prather, R.S. Significant im-provement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol. Reprod. 2009, 81, 525–530. [Google Scholar] [CrossRef]
- Smith, R.D.; Reeves, A.; Azeez, A.; Levy, M.L. Human Oocytes Reprogram Somatic Cells to a Pluripotent State. World Neurosurg. 2012, 77, 9–11. [Google Scholar] [CrossRef]
- Kigami, D.; Minami, N.; Takayama, H.; Imai, H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 2003, 68, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Puschendorf, M.; Terranova, R.; Boutsma, E.; Mao, X.; Isono, K.-I.; Brykczynska, U.; Kolb, C.; Otte, A.P.; Koseki, H.; Orkin, S.H.; et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 2008, 40, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Bosnakovski, D.; Xu, Z.; Gang, E.J.; Galindo, C.L.; Liu, M.; Simsek, T.; Garner, H.R.; Agha-Mohammadi, S.; Tassin, A.; Coppée, F.; et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008, 27, 2766–2779. [Google Scholar] [CrossRef] [PubMed]
- Bosnakovski, D.; Daughters, R.S.; Xu, Z.; Slack, J.M.W.; Kyba, M. Biphasic Myopathic Phenotype of Mouse DUX, an ORF within Conserved FSHD-Related Repeats. PLoS ONE 2009, 4, e7003. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kou, Z.; Zhang, Y.; Gao, S. Dynamic Reprogramming of Histone Acetylation and Methylation in the First Cell Cycle of Cloned Mouse Embryos1. Biol. Reprod. 2007, 77, 1007–1016. [Google Scholar] [CrossRef]
- Yan, Y.L.; Zhang, C.; Hao, J.; Wang, X.L.; Wang, Y. DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic tran-scriptional program. PLoS Biol. 2019, 17, e3000324. [Google Scholar] [CrossRef] [PubMed]
- Loi, P.; Modlinski, J.; Ptak, G. Interspecies somatic cell nuclear transfer: A salvage tool seeking first aid. Theriogenology 2011, 76, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Mrowiec, P.; Bugno-Poniewierska, M.; Młodawska, W. The perspective of the incompatible of nucleus and mitochondria in interspecies somatic cell nuclear transfer for endangered species. Reprod. Domest. Anim. 2021, 56, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Leidenroth, A.; Clapp, J.; Mitchell, L.M.; Coneyworth, D.; Dearden, F.L.; Iannuzzi, L.; Hewitt, J.E. Evolution of DUX gene macrosatellites in placental mammals. Chromosoma 2012, 121, 489–497. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, B.; Ma, H.; Liu, D. Functions and Regulation of Endogenous Retrovirus Elements during Zygotic Genome Activation: Implications for Improving Somatic Cell Nuclear Transfer Efficiency. Biomolecules 2021, 11, 829. https://doi.org/10.3390/biom11060829
Fu B, Ma H, Liu D. Functions and Regulation of Endogenous Retrovirus Elements during Zygotic Genome Activation: Implications for Improving Somatic Cell Nuclear Transfer Efficiency. Biomolecules. 2021; 11(6):829. https://doi.org/10.3390/biom11060829
Chicago/Turabian StyleFu, Bo, Hong Ma, and Di Liu. 2021. "Functions and Regulation of Endogenous Retrovirus Elements during Zygotic Genome Activation: Implications for Improving Somatic Cell Nuclear Transfer Efficiency" Biomolecules 11, no. 6: 829. https://doi.org/10.3390/biom11060829
APA StyleFu, B., Ma, H., & Liu, D. (2021). Functions and Regulation of Endogenous Retrovirus Elements during Zygotic Genome Activation: Implications for Improving Somatic Cell Nuclear Transfer Efficiency. Biomolecules, 11(6), 829. https://doi.org/10.3390/biom11060829






