Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Transgenic Plants and Growth Conditions
2.2. DEX Treatment
2.3. cAMP Extraction and Measurement
2.4. RNA Extraction, cDNA Library Construction, and Transcriptome Sequencing
2.5. Transcripts Assembly, Annotation, and Quantification
2.6. Bioinformatics Analysis
2.7. GSEA
2.8. WGCNA and Network Visualization
2.9. Phytohormone Quantification
2.10. Phenotypic Characterization
2.11. Quantitative RT-PCR
3. Results
3.1. Establishment of an Inducible System for Adenylate Cyclase Activity and cAMP Elevation in Plants
3.2. Global Profile of Gene Expression Changes Associated with Cellular cAMP Elevation in plants
3.3. Functional Comparison between Up- and Down-Regulated CRGs in Plants
3.4. Identification of Biological Pathways Associated with Cellular cAMP Elevation in Plants
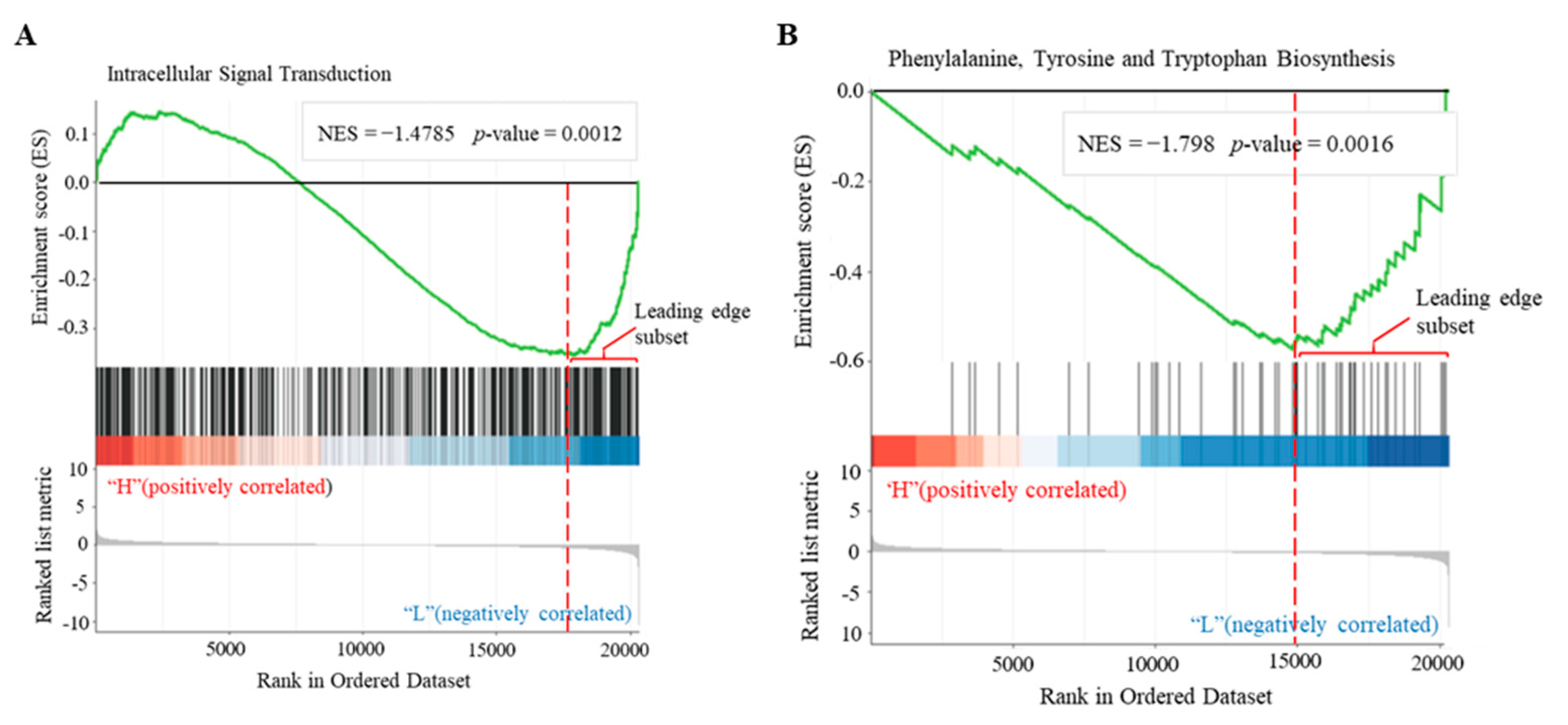
3.5. Identification of cAMP-Responsive Gene Sets in Plants
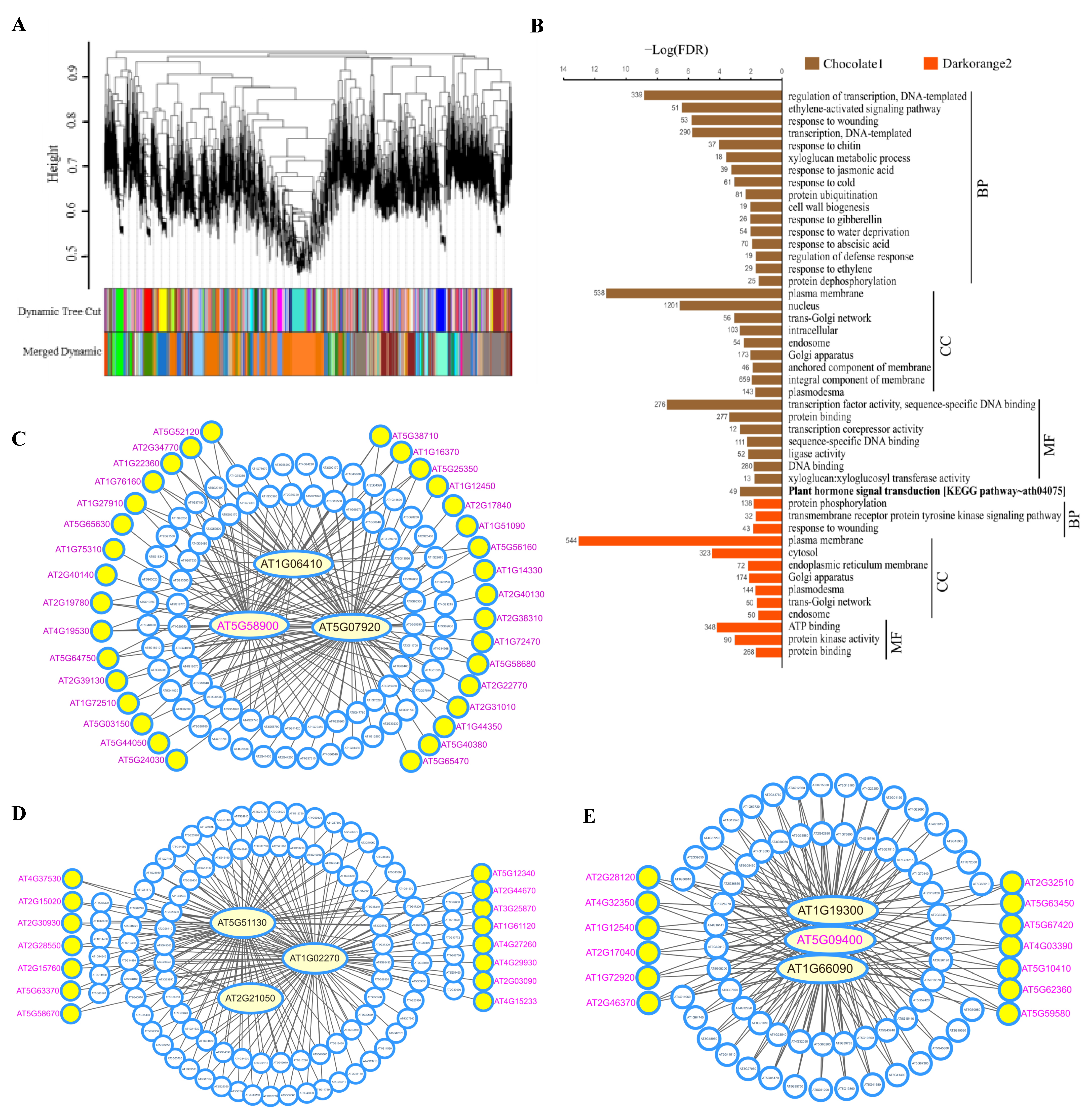
3.6. Co-Expression Networks and Hub Genes Associated with Cellular cAMP Elevation in Plants
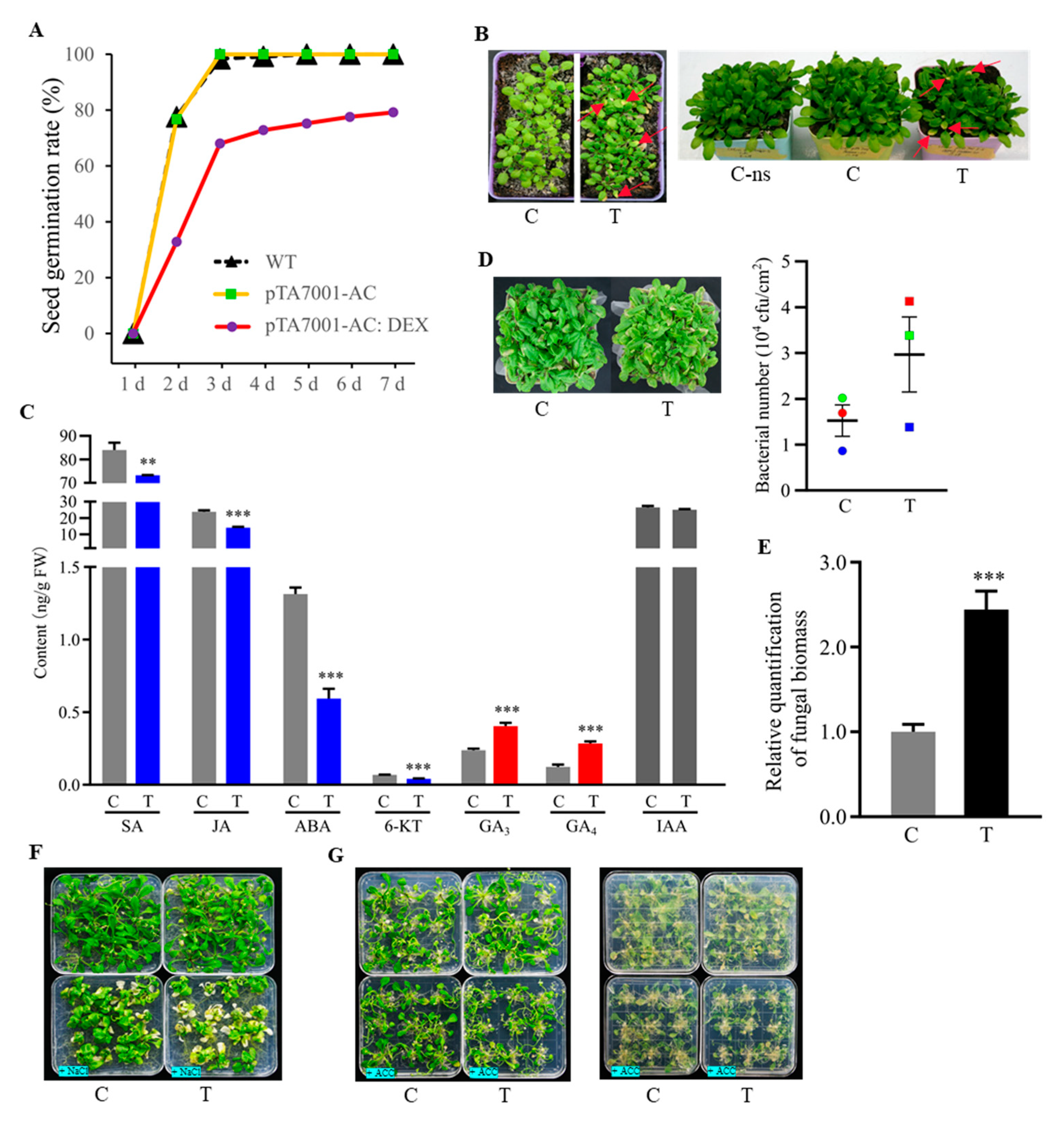
3.7. Phenotypic Effects of Cellular cAMP Elevation in Plants
4. Discussion
4.1. Interaction of cAMP and Hormones in Plants
4.2. Regulatory Roles of cAMP Signaling during Gene Expression in Plants
4.3. The Pathways and Interplay Network of cAMP Signaling in Plants
4.4. The Association of Ion Channels and Transporters with cAMP Signaling in Plants
4.5. cAMP-Responsive Gene Sets and Co-Expression Modules in Plants
4.6. Phenotypic Effects of cAMP Disturbance in Plants
4.7. A Role of cAMP Signaling in Plant Plasticity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sutherland, E.W. Studies on the mechanism of hormone action. Science 1972, 177, 401–408. [Google Scholar] [CrossRef]
- Biswas, A.; Bhattacharya, A.; Das, P.K. Role of cAMP Signaling in the survival and infectivity of the protozoan parasite, Leishmania donovani. Mol. Biol. Int. 2011, 2011, 782971. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Molenda, M.; Staab, J.F.; Lyman, C.A.; Gordon, L.J.; Sundstrom, P. Genome-wide transcriptional profiling of the cyclic AMP-dependent signaling pathway during morphogenic transitions of Candida albicans. Eukaryot. Cell 2007, 6, 2376–2390. [Google Scholar] [CrossRef] [PubMed]
- Bretschneider, T.; Vasiev, B.; Weijer, C.J. A model for dictyostelium slug movement. J. Theor. Biol. 1999, 199, 125–136. [Google Scholar] [CrossRef]
- Botsford, J.L.; Harman, J.G. Cyclic AMP in prokaryotes. Microbiol. Rev. 1992, 56, 100–122. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.P.; Smith, C.J. Cyclic nucleotides. Phytochemistry 2004, 65, 2423–2437. [Google Scholar] [CrossRef]
- Amrhein, N. The current status of cyclic AMP in higher plants. Ann. Rev. Plant Physiol. 1977, 28, 123–132. [Google Scholar] [CrossRef]
- Bolwell, G.P. Cyclic AMP, the reluctant messenger in plants. Trends Biochem. Sci. 1995, 20, 492–495. [Google Scholar] [CrossRef]
- Assmann, S.M. Cyclic AMP as a second messenger in higher plants—Status and future prospects. Plant Physiol. 1995, 108, 885–889. [Google Scholar] [CrossRef]
- Lemtiri-Chlieh, F.; Thomas, L.; Marondedze, C.; Irving, H.; Gehring, C. Cyclic nucleotides and nucleotide cyclases in plant stress responses. In Abiotic Stress Response in Plants—Physiological, Biochemical and Genetic Perspectives; Shanker, A., Venkateswarlu, B., Eds.; InTech: Shanghai, China, 2011; pp. 137–182. [Google Scholar]
- Gehring, C. Adenyl cyclases and cAMP in plant signaling—Past and present. Cell Commun. Signal. 2010, 8, 15. [Google Scholar] [CrossRef]
- Marondedze, C.; Wong, A.; Thomas, L.; Irving, H.; Gehring, C. Cyclic nucleotide monophosphates in plants and plant signaling. Handb. Exp. Pharmacol. 2017, 238, 87–103. [Google Scholar] [PubMed]
- Gehring, C.; Turek, I.S. Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front. Plant Sci. 2017, 8, 1704. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Fortunato, S.; Viggiano, L.; de Pinto, M.C. Cyclic AMP: A polyhedral signalling molecule in plants. Int. J. Mol. Sci. 2020, 21, 4862. [Google Scholar] [CrossRef]
- Calebiro, D.; Maiellaro, I. cAMP signaling microdomains and their observation by optical methods. Front. Cell. Neurosci. 2014, 8, 350. [Google Scholar] [CrossRef]
- Lefkimmiatis, K.; Zaccolo, M. cAMP signaling in subcellular compartments. Pharmacol. Ther. 2014, 143, 295–304. [Google Scholar] [CrossRef]
- Kamenetsky, M.; Middelhaufe, S.; Bank, E.M.; Levin, L.R.; Buck, J.; Steegborn, C. Molecular details of cAMP generation in mammalian cells: A tale of two systems. J. Mol. Biol. 2006, 362, 623–639. [Google Scholar] [CrossRef]
- Conti, M.; Beavo, J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: Essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 2007, 76, 481–511. [Google Scholar] [CrossRef] [PubMed]
- Beavo, J.A.; Brunton, L.L. Cyclic nucleotide research—Still expanding after half a century. Nat. Rev. Mol. Cell Biol. 2002, 3, 710–718. [Google Scholar] [CrossRef]
- Gancedo, J.M. Biological roles of cAMP: Variations on a theme in the different kingdoms of life. Biol. Rev. Camb. Philos. Soc. 2013, 88, 645–668. [Google Scholar] [CrossRef] [PubMed]
- Sabetta, W.; Vandelle, E.; Locato, V.; Costa, A.; Cimini, S.; Bittencourt Moura, A.; Luoni, L.; Graf, A.; Viggiano, L.; De Gara, L.; et al. Genetic buffering of cyclic AMP in Arabidopsis thaliana compromises the plant immune response triggered by an avirulent strain of Pseudomonas syringae pv. tomato. Plant J. 2019, 98, 590–606. [Google Scholar] [CrossRef]
- Steegborn, C. Structure, mechanism, and regulation of soluble adenylyl cyclases—Similarities and differences to transmembrane adenylyl cyclases. Biochim. Biophys. Acta 2014, 1842, 2535–2547. [Google Scholar] [CrossRef] [PubMed]
- Lomovatskaya, L.A.; Romanenko, A.S.; Filinova, N.V. Plant adenylate cyclases. J. Recept. Signal Transduct. Res. 2008, 28, 531–542. [Google Scholar] [CrossRef]
- Filinova, N.V.; Lomovatskaya, L.A.; Romanenko, A.S.; Salyaev, R.K. Calcium as a modulator of the adenylyl cyclase activity of potato cells in bacterial pathogenesis. Dokl. Biochem. Biophys. 2018, 483, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Pacini, B.; Petrigliano, A.; Diffley, P.; Paffetn, A.; Brown, E.G.; Martelli, P.; Trabalzini, L.; Bovalini, L.; Lhusini, P.; Newton, R.P. Adenylyl cyclase activity in roots of Pisum sativum. Phytochem. Anal. 1993, 34, 899–903. [Google Scholar] [CrossRef]
- Lusini, P.; Trabalzini, L.; Franchi, G.G.; Bovalini, L.; Martelli, P. Adenylate cyclase in roots of Ricinus communis: Stimulation by GTP and Mn2+. Phytochemistry 1991, 30, 109–111. [Google Scholar] [CrossRef]
- Carricart, V.C.; Bianchini, G.M.; Muschietti, J.P.; Téllez-Iñón, M.T.; Perticari, A.; Torres, N.; Flawiá, M.M. Adenylate cyclase activity in a higher plant, alfalfa (Medicago sativa). Biochem. J. 1988, 249, 807–811. [Google Scholar] [CrossRef]
- Witters, E.; Valcke, R.; Van Onckelen, H. Cytoenzymological analysis of adenylyl cyclase activity and 3′:5′-cAMP immunolocalization in chloroplasts of Nicotiana tabacum. New Phytol. 2005, 168, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, A.S.; Lomovatskaya, L.A.; Filinova, N.V.; Kopytchuk, V.N.; Salyaev, R.K. Intracellular localization of “soluble” adenylate cyclase in potato plants. Dokl. Biol. Sci. 2008, 422, 328–329. [Google Scholar] [CrossRef]
- Lomovatskaya, L.A.; Romanenko, A.S.; Filinova, N.V.; Salyaev, R.K. Detection of soluble adenylyl cyclase isoforms in plants. Dokl. Biochem. Biophys. 2008, 420, 124–126. [Google Scholar] [CrossRef]
- Moutinho, A.; Hussey, P.J.; Trewavas, A.J.; Malho, R. cAMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. USA 2001, 98, 10481–10486. [Google Scholar] [CrossRef] [PubMed]
- Swiezawska, B.; Jaworski, K.; Pawelek, A.; Grzegorzewska, W.; Szewczuk, P.; Szmidt-Jaworska, A. Molecular cloning and characterization of a novel adenylyl cyclase gene, HpAC1, involved in stress signaling in Hippeastrum x hybridum. Plant Physiol. Biochem. 2014, 80, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Moeder, W.; Garcia-Petit, C.; Ung, H.; Fucile, G.; Samuel, M.A.; Christendat, D.; Yoshioka, K. Crystal structure and biochemical analyses reveal that the Arabidopsis triphosphate tunnel metalloenzyme AtTTM3 is a tripolyphosphatase involved in root development. Plant J. 2013, 76, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Takahashi, H.; Sawasaki, T.; Ohnishi, K.; Hikichi, Y.; Kiba, A. Novel type of adenylyl cyclase participates in tabtoxinine-beta-lactam-induced cell death and occurrence of wildfire disease in Nicotiana benthamiana. Plant Signal. Behav. 2014, 9, e27420. [Google Scholar] [CrossRef]
- Ruzvidzo, O.; Gehring, C.; Wong, A. New perspectives on plant adenylyl cyclases. Front. Mol. Biosci. 2019, 6, 136. [Google Scholar] [CrossRef] [PubMed]
- Bianchet, C.; Wong, A.; Quaglia, M.; Alqurashi, M.; Gehring, C.; Ntoukakis, V.; Pasqualini, S. An Arabidopsis thaliana leucine-rich repeat protein harbors an adenylyl cyclase catalytic center and affects responses to pathogens. J. Plant Physiol. 2019, 232, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Gehring, C. Computational identification of candidate nucleotide cyclases in higher plants. Methods Mol. Biol. 2013, 1016, 195–205. [Google Scholar]
- Al-Younis, I.; Wong, A.; Lemtiri-Chlieh, F.; Schmockel, S.; Tester, M.; Gehring, C.; Donaldson, L. The Arabidopsis thaliana K+-uptake permease 5 (AtKUP5) contains a functional cytosolic adenylate cyclase essential for K+ transport. Front. Plant Sci. 2018, 9, 1645. [Google Scholar] [CrossRef]
- Al-Younis, I.; Wong, A.; Gehring, C. The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett. 2015, 589, 3848–3852. [Google Scholar] [CrossRef]
- Chatukuta, P.; Dikobe, T.B.; Kawadza, D.T.; Sehlabane, K.S.; Takundwa, M.M.; Wong, A.; Gehring, C.; Ruzvidzo, O. An Arabidopsis clathrin assembly protein with a predicted role in plant defense can function as an adenylate cyclase. Biomolecules 2018, 8, 15. [Google Scholar] [CrossRef]
- Ruzvidzo, O.; Dikobe, B.T.; Kawadza, D.T.; Mabadahanye, G.H.; Chatukuta, P.; Kwezi, L. Recombinant expression and functional testing of candidate adenylate cyclase domains. Methods Mol. Biol. 2013, 1016, 13–25. [Google Scholar]
- Uematsu, K.; Fukui, Y. Role and regulation of cAMP in seed germination of Phacelia tanacetifolia. Plant Physiol. Biochem. 2008, 46, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.A.; Galsky, A.G. The action of cyclic-AMP on GA3 controlled responses IV. Characteristics of the promotion of seed germination in Lactuca satira variety ‘Spartan Lake’ by gibberellic acid and cyclic 3′,5′-adenosine monophosphate. Plant Cell Physiol. 1973, 14, 565–571. [Google Scholar]
- Salomon, D.; Mascarenhas, J.P. Auxin-induced synthesis of cyclic 3′, 5′-adenosine monophosphate in Avena coleoptiles. Life Sci. II 1971, 10, 879–885. [Google Scholar] [CrossRef]
- Truelsen, T.A.; Delaughe, E.; Verbeek-Wyndaele, R. Cyclic AMP induced growth promotion in sunflower callus tissue. Arch. Int. Physiol. Biochim. 1974, 82, 190–214. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.N.; Braun, A.C. 8-bromoadenosine 3′:5′-cyclic monophosphate as a promoter of cell division in excised tobacco pith parenchyma tissue. Proc. Natl. Acad. Sci. USA 1973, 70, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Rast, D.; Skrivainova, R.; Bachofen, R. Replacemnent of light by dibutyryl-cAMP and cAMP in betacyanin synthesis. Phytochemistry 1973, 12, 2669–2672. [Google Scholar] [CrossRef]
- Molchan, O.; Sokolovsky, S.; Volotovsky, I.D. The phytochrome control of the cAMP endogenous level in oat seedlings. Russ. J. Plant Physiol. 2000, 47, 463–467. [Google Scholar]
- Ehsan, H.; Roef, L.; Witters, E.; Reichheld, J.P.; Van Bockstaele, D.; Inze, D.; Van Onckelen, H. Indomethacin-induced G1/S phase arrest of the plant cell cycle. FEBS Lett. 1999, 458, 349–353. [Google Scholar] [CrossRef]
- Sabetta, W.; Vannini, C.; Sgobba, A.; Marsoni, M.; Paradiso, A.; Ortolani, F.; Bracale, M.; Viggiano, L.; Blanco, E.; de Pinto, M.C. Cyclic AMP deficiency negatively affects cell growth and enhances stress-related responses in tobacco Bright Yellow-2 cells. Plant Mol. Biol. 2016, 90, 467–483. [Google Scholar] [CrossRef]
- Sato, S.; Tabata, S.; Hotta, Y. Changes in intracellular cAMP level and activities of adenyl cyclase and phosphodiesterase during meiosis of lily microsporocytes. Cell Struct. Funct. 1992, 17, 335–339. [Google Scholar] [CrossRef][Green Version]
- Curvetto, N.; Darjania, L.; Delmastro, S. Effect of two cAMP analogues on stomatal opening in Vicia faba. Possible relationship with cytosolic calcium concentration. Plant Physiol. Biochem. 1994, 32, 365–372. [Google Scholar]
- Kurosaki, F.; Nishi, A. Stimulation of calcium influx and calcium cascade by cyclic AMP in cultured carrot cells. Arch. Biochem. Biophys. 1993, 302, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.J.; Smith, C.J.; Walton, T.J.; Newton, R.P. Evidence that cyclic AMP is involved in the hypersensitive response of Medicago sativa to a fungal elicitor. Phytochemistry 1994, 35, 889–895. [Google Scholar] [CrossRef]
- Duszyn, M.; Swiezawska, B.; Szmidt-Jaworska, A.; Jaworski, K. Cyclic nucleotide gated channels (CNGCs) in plant signalling—Current knowledge and perspectives. J. Plant Physiol. 2019, 241, 153035. [Google Scholar] [CrossRef]
- Reggiani, R. Alteration of levels of cyclic nucleotides in response to anaerobiosis in rice seedlings. Plant Cell Physiol. 1997, 38, 740–742. [Google Scholar] [CrossRef]
- Van Damme, T.; Blancquaert, D.; Couturon, P.; Van Der Straeten, D.; Sandra, P.; Lynen, F. Wounding stress causes rapid increase in concentration of the naturally occurring 2′,3′-isomers of cyclic guanosine- and cyclic adenosine monophosphate (cGMP and cAMP) in plant tissues. Phytochemistry 2014, 103, 59–66. [Google Scholar] [CrossRef]
- Maathuis, F.J.; Sanders, D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001, 127, 1617–1625. [Google Scholar] [CrossRef]
- Gao, F.; Han, X.; Wu, J.; Zheng, S.; Shang, Z.; Sun, D.; Zhou, R.; Li, B. A heat-activated calcium-permeable channel--Arabidopsis cyclic nucleotide-gated ion channel 6--is involved in heat shock responses. Plant J. 2012, 70, 1056–1069. [Google Scholar] [CrossRef]
- Ordoñez, N.M.; Marondedze, C.; Thomas, L.; Pasqualini, S.; Shabala, L.; Shabala, S.; Gehring, C. Cyclic mononucleotides modulate potassium and calcium flux responses to H2O2 in Arabidopsis roots. FEBS Lett. 2014, 588, 1008–1015. [Google Scholar] [CrossRef]
- Ma, W.; Qi, Z.; Smigel, A.; Walker, R.K.; Verma, R.; Berkowitz, G.A. Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. USA 2009, 106, 20995–21000. [Google Scholar] [CrossRef]
- Chung, C.H.; Lim, S.U.; Song, P.S.; Berlin, J.D.; Goodin, J.R. Swelling of etiolated oat protoplasts induced by phytochrome, cAMP, and gibberellic acid: A kinetic study. Plant Cell Physiol. 1988, 29, 855–860. [Google Scholar]
- Thomas, L.; Marondedze, C.; Ederli, L.; Pasqualini, S.; Gehring, C. Proteomic signatures implicate cAMP in light and temperature responses in Arabidopsis thaliana. J. Proteom. 2013, 83, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Alqurashi, M.; Gehring, C.; Marondedze, C. Changes in the Arabidopsis thaliana proteome implicate cAMP in biotic and abiotic stress responses and changes in energy metabolism. Int. J. Mol. Sci. 2016, 17, 852. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.K.; Sharma, M.; Pandey, G.K. Role of cyclic nucleotide gated channels in stress management in plants. Curr. Genom. 2016, 17, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Fortuna, A.; Lee, J.; Ung, H.; Chin, K.; Moeder, W.; Yoshioka, K. Crossroads of stress responses, development and flowering regulation—The multiple roles of Cyclic Nucleotide Gated Ion Channel 2. Plant Signal. Behav. 2015, 10, e989758. [Google Scholar] [CrossRef] [PubMed]
- Talke, I.N.; Blaudez, D.; Maathuis, F.J.; Sanders, D. CNGCs: Prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 2003, 8, 286–293. [Google Scholar] [CrossRef]
- Jiang, J.; Fan, L.W.; Wu, W.H. Evidences for involvement of endogenous cAMP in Arabidopsis defense responses to Verticillium toxins. Cell Res. 2005, 15, 585–592. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Y.; Tang, S.; Pan, J.; Yu, Y.; Han, J.; Li, Y.; Du, X.; Nan, Z.; Sun, Q. AtCNGC2 is involved in jasmonic acid-induced calcium mobilization. J. Exp. Bot. 2016, 67, 809–819. [Google Scholar] [CrossRef]
- Donaldson, L.; Meier, S.; Gehring, C. The arabidopsis cyclic nucleotide interactome. Cell Commun. Signal. 2016, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Chua, N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997, 11, 605–612. [Google Scholar] [CrossRef]
- Xu, R.; Zhao, H.; Dinkins, R.D.; Cheng, X.; Carberry, G.; Li, Q.Q. The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol. Biol. 2006, 61, 799–815. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.; Huckelhoven, R.; Schafer, P.; Imani, J.; Sharma, M.; Weiss, M.; Waller, F.; Kogel, K.H. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc. Natl. Acad. Sci. USA 2006, 103, 18450–18457. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Chua, N.H. Chemical-inducible systems for regulated expression of plant genes. Curr. Opin. Biotechnol. 2000, 11, 146–151. [Google Scholar] [CrossRef]
- Park, C.; Canlas, P.E.; Ronald, P.C. Establishment of glucocorticoid-mediated transcriptional induction of the rice XA21 pattern recognition receptor. J. Plant Biol. 2012, 55, 43–49. [Google Scholar] [CrossRef]
- Qin, X.; Zeevaart, J.A. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef]
- Neumann, S.; Malik, S.S.; Marcus-Samuels, B.; Eliseeva, E.; Jang, D.; Klubo-Gwiezdzinska, J.; Krieger, C.C.; Gershengorn, M.C. Thyrotropin causes dose-dependent biphasic regulation of cAMP production mediated by Gs and Gi/o proteins. Mol. Pharmacol. 2020, 97, 2–8. [Google Scholar] [CrossRef]
- Halls, M.L.; Bathgate, R.A.; Summers, R.J. Relaxin family peptide receptors RXFP1 and RXFP2 modulate cAMP signaling by distinct mechanisms. Mol. Pharmacol. 2006, 70, 214–226. [Google Scholar] [CrossRef]
- Hussain, J.; Chen, J.; Locato, V.; Sabetta, W.; Behera, S.; Cimini, S.; Griggio, F.; Martinez-Jaime, S.; Graf, A.; Bouneb, M.; et al. Constitutive cyclic GMP accumulation in Arabidopsis thaliana compromises systemic acquired resistance induced by an avirulent pathogen by modulating local signals. Sci. Rep. 2016, 6, 36423. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Mitsuda, N.; Ohme-Takagi, M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009, 50, 1232–1248. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Sassone-Corsi, P. The cyclic AMP pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011148. [Google Scholar] [CrossRef] [PubMed]
- Pietrowska-Borek, M.; Nuc, K. Both cyclic-AMP and cyclic-GMP can act as regulators of the phenylpropanoid pathway in Arabidopsis thaliana seedlings. Plant Physiol. Biochem. 2013, 70, 142–149. [Google Scholar] [CrossRef] [PubMed]
- DiMario, R.J.; Clayton, H.; Mukherjee, A.; Ludwig, M.; Moroney, J.V. Plant carbonic anhydrases: Structures, locations, evolution, and physiological roles. Mol. Plant 2017, 10, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Fabre, N.; Reiter, I.M.; Becuwe-Linka, N.; Genty, B.; Rumeau, D. Characterization and expression analysis of genes encoding α and β carbonic anhydrases in Arabidopsis. Plant Cell Environ. 2007, 30, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- Ramon, M.; De Smet, I.; Vandesteene, L.; Naudts, M.; Leyman, B.; Van Dijck, P.; Rolland, F.; Beeckman, T.; Thevelein, J.M. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1015–1032. [Google Scholar] [CrossRef]
- Iordachescu, M.; Imai, R. Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 2008, 50, 1223–1229. [Google Scholar] [CrossRef]
- Derevyanchuk, M.; Kretynin, S.; Kolesnikov, Y.; Litvinovskaya, R.; Martinec, J.; Khripach, V.; Kravets, V. Seed germination, respiratory processes and phosphatidic acid accumulation in Arabidopsis diacylglycerol kinase knockouts—The effect of brassinosteroid, brassinazole and salinity. Steroids 2019, 147, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Bhosale, R. Developmental roles of AUX1/LAX auxin influx carriers in plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Bae, M.S.; Cho, E.J.; Choi, E.Y.; Park, O.K. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003, 36, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, B.A.; Weretilnyk, E.A. Sustaining S-adenosyl-L-methionine-dependent methyltransferase activity in plant cells. Physiol. Plant 2001, 113, 435–442. [Google Scholar] [CrossRef]
- Nandety, R.S.; Caplan, J.L.; Cavanaugh, K.; Perroud, B.; Wroblewski, T.; Michelmore, R.W.; Meyers, B.C. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013, 162, 1459–1472. [Google Scholar] [CrossRef]
- Lee, C.; Zhong, R.; Richardson, E.A.; Himmelsbach, D.S.; McPhail, B.T.; Ye, Z.H. The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 2007, 48, 1659–1672. [Google Scholar] [CrossRef]
- Han, M.; Wu, W.; Wu, W.H.; Wang, Y. Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol. Plant 2016, 9, 437–446. [Google Scholar] [CrossRef]
- Mangat, B.S.; Janjua, S. Cyclic nucleotides and in vitro plant cultures: I. Induction of organogenesis in tobacco (Nicotiana tabacum) callus cultures. J. Exp. Biol. 1987, 38, 2059–2067. [Google Scholar]
- Azhar, S.; Murti, C.R. Effect of indole-3-acetic acid on the synthesis of cyclic 3′-5′ adenosine phosphate by Bengal gram seeds. Biochem. Biophys. Res. Commun. 1971, 43, 58–64. [Google Scholar] [CrossRef]
- Jin, X.C.; Wu, W.H. Involvement of cyclic AMP in ABA- and Ca2+-mediated signal transduction of stomatal regulation in Vicia faba. Plant Cell Physiol. 1999, 40, 1127–1133. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Barbez, E.; Kleine-Vehn, J. Cellular auxin homeostasis: Gatekeeping is housekeeping. Mol. Plant 2012, 5, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Booker, M.A.; DeLong, A. Producing the ethylene signal: Regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 2015, 169, 42–50. [Google Scholar] [CrossRef]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012, 69, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Li, X.G.; Gao, X.Q.; Zhao, X.Y.; Dong, Y.X.; Zhou, C. The Arabidopsis phytohormone crosstalk network involves a consecutive metabolic route and circular control units of transcription factors that regulate enzyme-encoding genes. BMC Syst. Biol. 2016, 10, 87. [Google Scholar] [CrossRef]
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Elorduy Vergara, L.; Palme, J.; Marin-de la Rosa, N.; Sauer, M.; Wenig, M.; Villaecija-Aguilar, J.A.; et al. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Liu, C.; Yu, H.; Broun, P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 2007, 134, 2073–2081. [Google Scholar] [CrossRef]
- Hughes, P.; Landoulsi, A.; Kohiyama, M. A novel role for cAMP in the control of the activity of the E. coli chromosome replication initiator protein, DnaA. Cell 1988, 55, 343–350. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Azhar, S.; Murti, C.R. Stimulation of RNA synthesis in Cicer arietinum seedlings by cyclic 3′:5′ adenosine monophosphate. FEBS Lett. 1973, 33, 239–240. [Google Scholar] [CrossRef][Green Version]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed]
- Delghandi, M.P.; Johannessen, M.; Moens, U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell. Signal. 2005, 17, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Krishnaswamy, S.; Verma, S.; Rahman, M.H.; Kav, N.N. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 2011, 75, 107–127. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef]
- Long, Y.; Smet, W.; Cruz-Ramirez, A.; Castelijns, B.; de Jonge, W.; Mahonen, A.P.; Bouchet, B.P.; Perez, G.S.; Akhmanova, A.; Scheres, B.; et al. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 2015, 27, 1185–1199. [Google Scholar] [CrossRef]
- de Marcos, A.; Houbaert, A.; Trivino, M.; Delgado, D.; Martin-Trillo, M.; Russinova, E.; Fenoll, C.; Mena, M. A mutation in the bHLH domain of the SPCH transcription factor uncovers a BR-dependent mechanism for stomatal development. Plant Physiol. 2017, 174, 823–842. [Google Scholar] [CrossRef] [PubMed]
- Kuusk, S.; Sohlberg, J.J.; Magnus Eklund, D.; Sundberg, E. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 2006, 47, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Ruhl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Lloyd, A.M.; Davis, R.W. The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev. 1993, 7, 367–379. [Google Scholar] [CrossRef]
- An, L.; Zhou, Z.; Sun, L.; Yan, A.; Xi, W.; Yu, N.; Cai, W.; Chen, X.; Yu, H.; Schiefelbein, J.; et al. A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 2012, 72, 474–490. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef]
- Soundappan, I.; Bennett, T.; Morffy, N.; Liang, Y.; Stanga, J.P.; Abbas, A.; Leyser, O.; Nelson, D.C. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 2015, 27, 3143–3159. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, H. mRNA decay in plants: Both quantity and quality matter. Curr. Opin. Plant Biol. 2017, 35, 138–144. [Google Scholar] [CrossRef]
- Sorenson, R.S.; Deshotel, M.J.; Johnson, K.; Adler, F.R.; Sieburth, L.E. Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc. Natl. Acad. Sci. USA 2018, 115, E1485–E1494. [Google Scholar] [CrossRef]
- Walley, J.W.; Kelley, D.R.; Savchenko, T.; Dehesh, K. Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal. Behav. 2010, 5, 802–805. [Google Scholar] [CrossRef][Green Version]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.P.; Bozhkov, P.V. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Abbasi, N.; Choi, S.B. Bruno-like proteins modulate flowering time via 3′ UTR-dependent decay of SOC1 mRNA. New Phytol. 2013, 198, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Miricescu, A.; Goslin, K.; Graciet, E. Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J. Exp. Bot. 2018, 69, 4511–4527. [Google Scholar] [CrossRef] [PubMed]
- Abd-Hamid, N.A.; Ahmad-Fauzi, M.I.; Zainal, Z.; Ismail, I. Diverse and dynamic roles of F-box proteins in plant biology. Planta 2020, 251, 68. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Tong, M.; Tian, L.; Zhu, C.; Liu, X.; Zhang, Y.; Li, X. Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J. 2020, 39, e104915. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M. News from the PUB: Plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 2018, 69, 371–384. [Google Scholar] [CrossRef]
- Kim, H.J.; Chiang, Y.H.; Kieber, J.J.; Schaller, G.E. SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. USA 2013, 110, 10028–10033. [Google Scholar] [CrossRef]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef]
- Nakamura, M.; Toyota, M.; Tasaka, M.; Morita, M.T. An Arabidopsis E3 ligase, SHOOT GRAVITROPISM9, modulates the interaction between statoliths and F-actin in gravity sensing. Plant Cell 2011, 23, 1830–1848. [Google Scholar] [CrossRef]
- Rinaldi, L.; Delle Donne, R.; Catalanotti, B.; Torres-Quesada, O.; Enzler, F.; Moraca, F.; Nistico, R.; Chiuso, F.; Piccinin, S.; Bachmann, V.; et al. Feedback inhibition of cAMP effector signaling by a chaperone-assisted ubiquitin system. Nat. Commun. 2019, 10, 2572. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Niu, J.; Sun, M.X.; Zhu, J.; Gao, J.F.; Yang, J.; Zhou, Q.; Yang, Z.N. Cyclin-dependent kinase G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell 2013, 25, 637–648. [Google Scholar] [CrossRef]
- Guo, Y.; Xiong, L.; Ishitani, M.; Zhu, J.K. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. USA 2002, 99, 7786–7791. [Google Scholar] [CrossRef] [PubMed]
- Vanstraelen, M.; Benkova, E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012, 28, 463–487. [Google Scholar] [CrossRef]
- Bigeard, J.; Hirt, H. Nuclear signaling of plant MAPKs. Front. Plant Sci. 2018, 9, 469. [Google Scholar] [CrossRef]
- D’Souza, C.A.; Heitman, J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 2001, 25, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Lemke, C.; Potter, K.C.; Schulte, S.; Peters, R.J. Conserved bases for the initial cyclase in gibberellin biosynthesis: From bacteria to plants. Biochem. J. 2019, 476, 2607–2621. [Google Scholar] [CrossRef]
- Tholl, D.; Sohrabi, R.; Huh, J.H.; Lee, S. The biochemistry of homoterpenes—Common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 2011, 72, 1635–1646. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.J.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A three-ring circus: Metabolism of the three proteogenic aromatic amino acids and their role in the health of plants and animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development—Active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites—Pathways, transcription factors and applied aspects—A brief review. New Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef]
- Jeandet, P.; Hebrard, C.; Deville, M.A.; Cordelier, S.; Dorey, S.; Aziz, A.; Crouzet, J. Deciphering the role of phytoalexins in plant-microorganism interactions and human health. Molecules 2014, 19, 18033–18056. [Google Scholar] [CrossRef]
- Vail, G.; Roepke, T.A. Membrane-initiated estrogen signaling via Gq-coupled GPCR in the central nervous system. Steroids 2019, 142, 77–83. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, I. Phosphoinositide signaling in plant development. Development 2016, 143, 2044–2055. [Google Scholar] [CrossRef]
- Desfougeres, Y.; Wilson, M.S.C.; Laha, D.; Miller, G.J.; Saiardi, A. ITPK1 mediates the lipid-independent synthesis of inositol phosphates controlled by metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 24551–24561. [Google Scholar] [CrossRef] [PubMed]
- Alford, S.R.; Rangarajan, P.; Williams, P.; Gillaspy, G.E. myo-Inositol oxygenase is required for responses to low energy conditions in Arabidopsis thaliana. Front. Plant Sci. 2012, 3, 69. [Google Scholar] [CrossRef]
- Laha, D.; Parvin, N.; Hofer, A.; Giehl, R.F.H.; Fernandez-Rebollo, N.; von Wiren, N.; Saiardi, A.; Jessen, H.J.; Schaaf, G. Arabidopsis ITPK1 and ITPK2 have an evolutionarily conserved phytic acid kinase activity. ACS Chem. Biol. 2019, 14, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Kanehara, K.; Yu, C.Y.; Cho, Y.; Cheong, W.F.; Torta, F.; Shui, G.; Wenk, M.R.; Nakamura, Y. Arabidopsis AtPLC2 is a primary phosphoinositide-specific phospholipase C in phosphoinositide metabolism and the endoplasmic reticulum stress response. PLoS Genet. 2015, 11, e1005511. [Google Scholar] [CrossRef] [PubMed]
- Montag, K.; Hornbergs, J.; Ivanov, R.; Bauer, P. Phylogenetic analysis of plant multi-domain SEC14-like phosphatidylinositol transfer proteins and structure-function properties of PATELLIN2. Plant Mol. Biol. 2020, 104, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Chen, X.; Li, G. Involvement of phospholipid signaling in plant growth and hormone effects. Curr. Opin. Plant Biol. 2007, 10, 483–489. [Google Scholar] [CrossRef]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135–170. [Google Scholar] [PubMed]
- Gou, M.; Zhang, Z.; Zhang, N.; Huang, Q.; Monaghan, J.; Yang, H.; Shi, Z.; Zipfel, C.; Hua, J. Opposing effects on two phases of defense responses from concerted actions of HEAT SHOCK COGNATE70 and BONZAI1 in Arabidopsis. Plant Physiol. 2015, 169, 2304–2323. [Google Scholar] [PubMed]
- Tang, J.; Han, Z.; Chai, J. Q&A: What are brassinosteroids and how do they act in plants? BMC Biol. 2016, 14, 113. [Google Scholar]
- Wheeler, J.I.; Wong, A.; Marondedze, C.; Groen, A.J.; Kwezi, L.; Freihat, L.; Vyas, J.; Raji, M.A.; Irving, H.R.; Gehring, C. The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant J. 2017, 91, 590–600. [Google Scholar] [CrossRef]
- Yu, X.; Xu, G.; Li, B.; de Souza Vespoli, L.; Liu, H.; Moeder, W.; Chen, S.; de Oliveira, M.V.V.; Ariadina de Souza, S.; Shao, W.; et al. The receptor kinases BAK1/SERK4 regulate Ca2+ channel-mediated cellular homeostasis for cell death containment. Curr. Biol. 2019, 29, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zaccolo, M.; Movsesian, M.A. cAMP and cGMP signaling cross-talk: Role of phosphodiesterases and implications for cardiac pathophysiology. Circ. Res. 2007, 100, 1569–1578. [Google Scholar] [CrossRef]
- Carland, F.; Fujioka, S.; Nelson, T. The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 2010, 153, 741–756. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, H.; Fujioka, S.; Zhou, T.; Tian, H.; Tian, W.; Wang, X. Homeostasis of brassinosteroids regulated by DRL1, a putative acyltransferase in Arabidopsis. Mol. Plant 2013, 6, 546–558. [Google Scholar] [CrossRef] [PubMed]
- DeFalco, T.A.; Moeder, W.; Yoshioka, K. Opening the gates: Insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 2016, 21, 903–906. [Google Scholar] [CrossRef]
- Zelman, A.K.; Dawe, A.; Gehring, C.; Berkowitz, G.A. Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 2012, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Shi, Z.; Bao, Y.; Yan, J.; Yang, Z.; Yu, H.; Li, Y.; Gou, M.; Wang, S.; Zou, B.; et al. Calcium pumps and interacting BON1 protein modulate calcium signature, stomatal closure, and plant immunity. Plant Physiol. 2017, 175, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kumar, R.; Tripathi, A.K.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. Genome-wide investigation and expression analysis of Sodium/Calcium exchanger gene family in rice and Arabidopsis. Rice 2015, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Tian, W.; Kleist, T.; He, K.; Garcia, V.; Bai, F.; Hao, Y.; Luan, S.; Li, L. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014, 24, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, W.H. Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 2017, 39, 123–128. [Google Scholar] [CrossRef]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty years from transport to signaling networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef]
- He, Y.N.; Peng, J.S.; Cai, Y.; Liu, D.F.; Guan, Y.; Yi, H.Y.; Gong, J.M. Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in Arabidopsis. Sci. Rep. 2017, 7, 6417. [Google Scholar] [CrossRef]
- Li, J.Y.; Fu, Y.L.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.Z.; Zhang, Y.; Li, H.M.; Huang, J.; et al. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Kechid, M.; Desbrosses, G.; Rokhsi, W.; Varoquaux, F.; Djekoun, A.; Touraine, B. The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196. New Phytol. 2013, 198, 514–524. [Google Scholar] [CrossRef]
- Segonzac, C.; Boyer, J.C.; Ipotesi, E.; Szponarski, W.; Tillard, P.; Touraine, B.; Sommerer, N.; Rossignol, M.; Gibrat, R. Nitrate efflux at the root plasma membrane: Identification of an Arabidopsis excretion transporter. Plant Cell 2007, 19, 3760–3777. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef]
- Chantranupong, L.; Wolfson, R.L.; Sabatini, D.M. Nutrient-sensing mechanisms across evolution. Cell 2015, 161, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S.; Hellmann, H.A. Sugar sensing and signaling in plants. Front. Plant Sci. 2014, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Klemens, P.A.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef]
- Karve, A.; Xia, X.; Moore, B. Arabidopsis hexokinase-like1 and hexokinase1 form a critical node in mediating plant glucose and ethylene responses. Plant Physiol. 2012, 158, 1965–1975. [Google Scholar] [CrossRef]
- Klepek, Y.S.; Volke, M.; Konrad, K.R.; Wippel, K.; Hoth, S.; Hedrich, R.; Sauer, N. Arabidopsis thaliana POLYOL/MONOSACCHARIDE TRANSPORTERS 1 and 2: Fructose and xylitol/H+ symporters in pollen and young xylem cells. J. Exp. Bot. 2010, 61, 537–550. [Google Scholar] [CrossRef]
- Vandesteene, L.; Ramon, M.; Le Roy, K.; Van Dijck, P.; Rolland, F. A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol. Plant 2010, 3, 406–419. [Google Scholar] [CrossRef]
- Figueroa, C.M.; Lunn, J.E. A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Sakr, S.; Wang, M.; Dedaldechamp, F.; Perez-Garcia, M.D.; Oge, L.; Hamama, L.; Atanassova, R. The sugar-signaling hub: Overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef]
- Tengholm, A.; Gylfe, E. cAMP signalling in insulin and glucagon secretion. Diabetes Obes. Metab. 2017, 19 (Suppl. 1), 42–53. [Google Scholar] [CrossRef] [PubMed]
- Brinkert, K.; De Causmaecker, S.; Krieger-Liszkay, A.; Fantuzzi, A.; Rutherford, A.W. Bicarbonate-induced redox tuning in Photosystem II for regulation and protection. Proc. Natl. Acad. Sci. USA 2016, 113, 12144–12149. [Google Scholar] [CrossRef] [PubMed]
- Negi, J.; Matsuda, O.; Nagasawa, T.; Oba, Y.; Takahashi, H.; Kawai-Yamada, M.; Uchimiya, H.; Hashimoto, M.; Iba, K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 2008, 452, 483–486. [Google Scholar] [CrossRef]
- Geiger, D.; Maierhofer, T.; Al-Rasheid, K.A.; Scherzer, S.; Mumm, P.; Liese, A.; Ache, P.; Wellmann, C.; Marten, I.; Grill, E.; et al. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 2011, 4, ra32. [Google Scholar] [CrossRef] [PubMed]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Jang, I.S.; Yeo, E.J.; Park, J.A.; Ahn, J.S.; Park, J.S.; Cho, K.A.; Juhnn, Y.S.; Park, S.C. Altered cAMP signaling induced by lysophosphatidic acid in senescent human diploid fibroblasts. Biochem. Biophys. Res. Commun. 2003, 302, 778–784. [Google Scholar] [CrossRef]
- Traverso, V.; Bush, R.A.; Sieving, P.A.; Deretic, D. Retinal cAMP levels during the progression of retinal degeneration in rhodopsin P23H and S334ter transgenic rats. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1655–1661. [Google Scholar]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Zentgraf, U.; Doll, J. Arabidopsis WRKY53, a node of multi-layer regulation in the network of senescence. Plants (Basel) 2019, 8, 578. [Google Scholar] [CrossRef]
- Fischer-Kilbienski, I.; Miao, Y.; Roitsch, T.; Zschiesche, W.; Humbeck, K.; Krupinska, K. Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol. Biol. 2010, 73, 379–390. [Google Scholar] [CrossRef]
- Kimura, M.; Yamamoto, Y.Y.; Seki, M.; Sakurai, T.; Sato, M.; Abe, T.; Yoshida, S.; Manabe, K.; Shinozaki, K.; Matsui, M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003, 77, 226–233. [Google Scholar]
- Uematsu, K.; Nakajima, M.; Yamaguchi, I.; Yoneyama, K.; Fukui, Y. Role of cAMP in gibberellin promotion of seed germination in Orobanche minor smith. J. Plant Growth Regul. 2007, 26, 245–254. [Google Scholar] [CrossRef]
- Mukherjee, S.; Jansen, V.; Jikeli, J.F.; Hamzeh, H.; Alvarez, L.; Dombrowski, M.; Balbach, M.; Strunker, T.; Seifert, R.; Kaupp, U.B.; et al. A novel biosensor to study cAMP dynamics in cilia and flagella. eLife 2016, 5, e14052. [Google Scholar] [CrossRef]
- Serezani, C.H.; Ballinger, M.N.; Aronoff, D.M.; Peters-Golden, M. Cyclic AMP: Master regulator of innate immune cell function. Am. J. Respir. Cell Mol. Biol. 2008, 39, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Raker, V.K.; Becker, C.; Steinbrink, K. The cAMP pathway as therapeutic target in autoimmune and inflammatory diseases. Front. Immunol. 2016, 7, 123. [Google Scholar] [CrossRef]
- Botman, D.; O’Toole, T.G.; Goedhart, J.; Bruggeman, F.J.; Van Heerden, J.H.; Teusink, B. A new FRET biosensor enlightens cAMP signalling in budding yeast. bioRxiv 2019, 831354. [Google Scholar] [CrossRef]
- Mika, D.; Richter, W.; Conti, M. A CaMKII/PDE4D negative feedback regulates cAMP signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 2023–2028. [Google Scholar] [CrossRef]
- Nikawa, J.; Cameron, S.; Toda, T.; Ferguson, K.M.; Wigler, M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987, 1, 931–937. [Google Scholar] [CrossRef][Green Version]
- Edmunds, L.N., Jr. Clocks, cell cycles, cancer, and aging. Role of the adenylate cyclase-cyclic AMP-phosphodiesterase axis in signal transduction between circadian oscillator and cell division cycle. Ann. N. Y. Acad. Sci. 1994, 719, 77–96. [Google Scholar] [CrossRef]
- Domańska, A.; Godlewski, M.; Kobylińska, A. Effect of cAMP level on thallus growth and gametangia development in macroalga Chara Vulgaris (Charales, Charophyta). Acta Biol. Crac. Ser. Bot. 2013, 55, 90–98. [Google Scholar] [CrossRef]
- Gordillo, F.J.; Segovia, M.; Lopez-Figueroa, F. Cyclic AMP levels in several macroalgae and their relation to light quantity and quality. J. Plant Physiol. 2004, 161, 211–217. [Google Scholar] [CrossRef][Green Version]
- Ehsan, H.; Reichheld, J.P.; Roef, L.; Witters, E.; Lardon, F.; Van Bockstaele, D.; Van Montagu, M.; Inze, D.; Van Onckelen, H. Effect of indomethacin on cell cycle dependent cyclic AMP fluxes in tobacco BY-2 cells. FEBS Lett. 1998, 422, 165–169. [Google Scholar] [CrossRef]
- Sampathkumar, A. Mechanical feedback-loop regulation of morphogenesis in plants. Development 2020, 147, dev177964. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, A.; Furumoto, T.; Ishida, S.; Takahashi, Y. AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 2007, 143, 1152–1162. [Google Scholar] [CrossRef]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, L.A.; Nikolaeva, D.A.; Fedotkina, T.V.; Govardovskii, V.I.; Firsov, M.L. Elevated cAMP improves signal-to-noise ratio in amphibian rod photoreceptors. J. Gen. Physiol. 2017, 149, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, S.V.; Steegborn, C.; Levin, L.R.; Buck, J. Pharmacological modulation of the CO2/HCO3-/pH-, calcium-, and ATP-sensing soluble adenylyl cyclase. Pharmacol. Ther. 2018, 190, 173–186. [Google Scholar] [CrossRef]
- Lymperopoulos, P.; Msanne, J.; Rabara, R. Phytochrome and phytohormones: Working in tandem for plant growth and development. Front. Plant Sci. 2018, 9, 1037. [Google Scholar] [CrossRef]
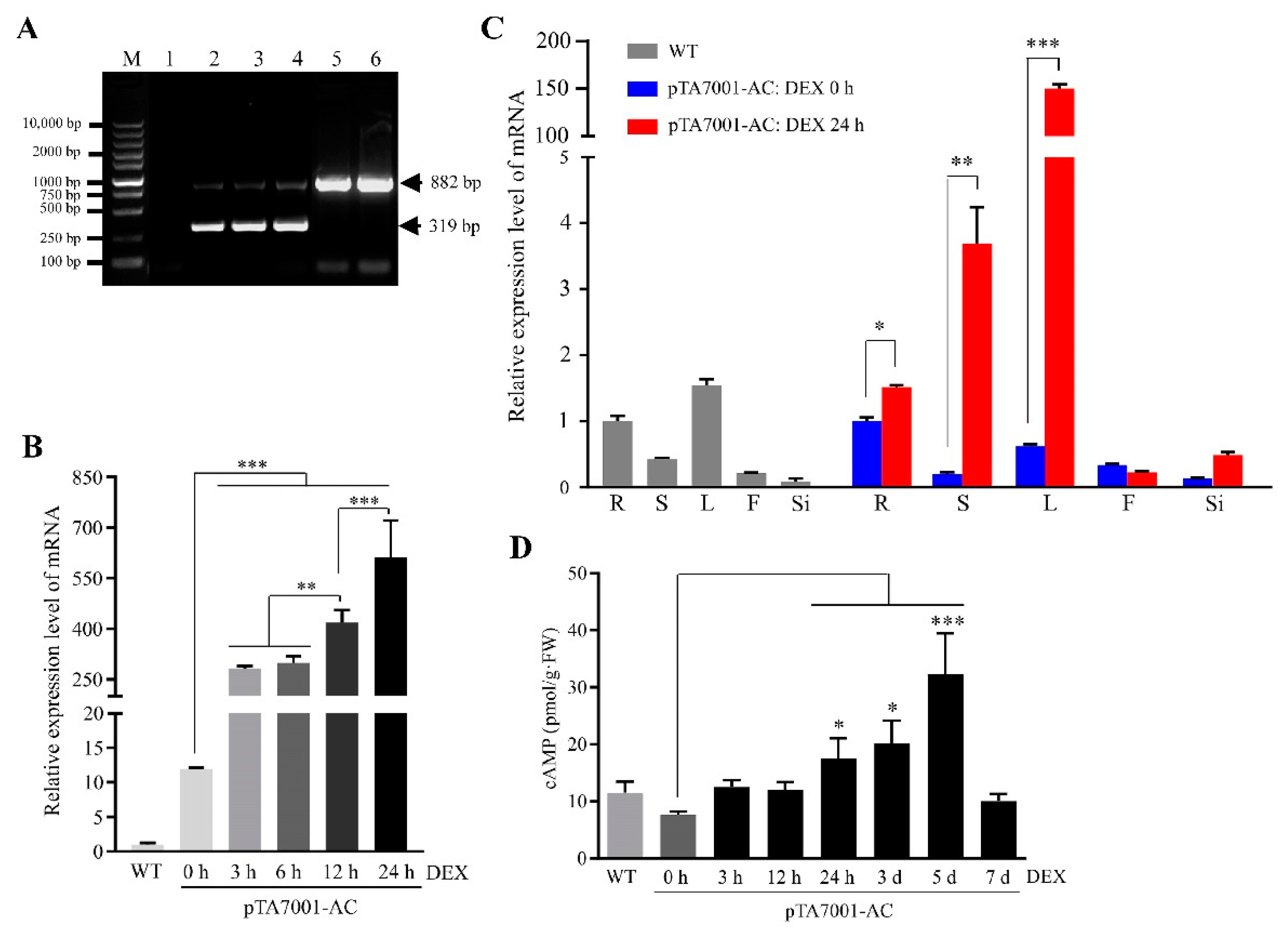
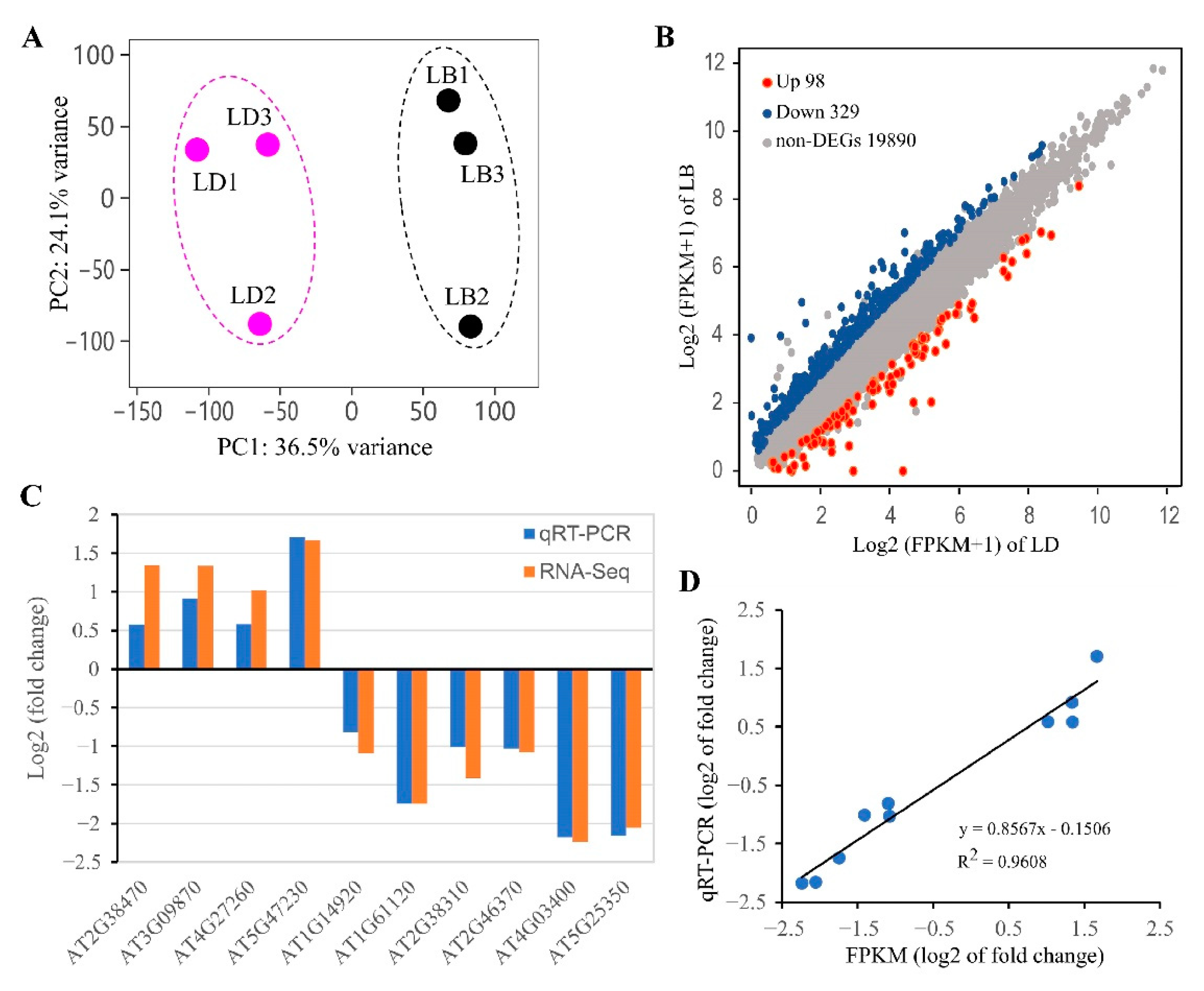
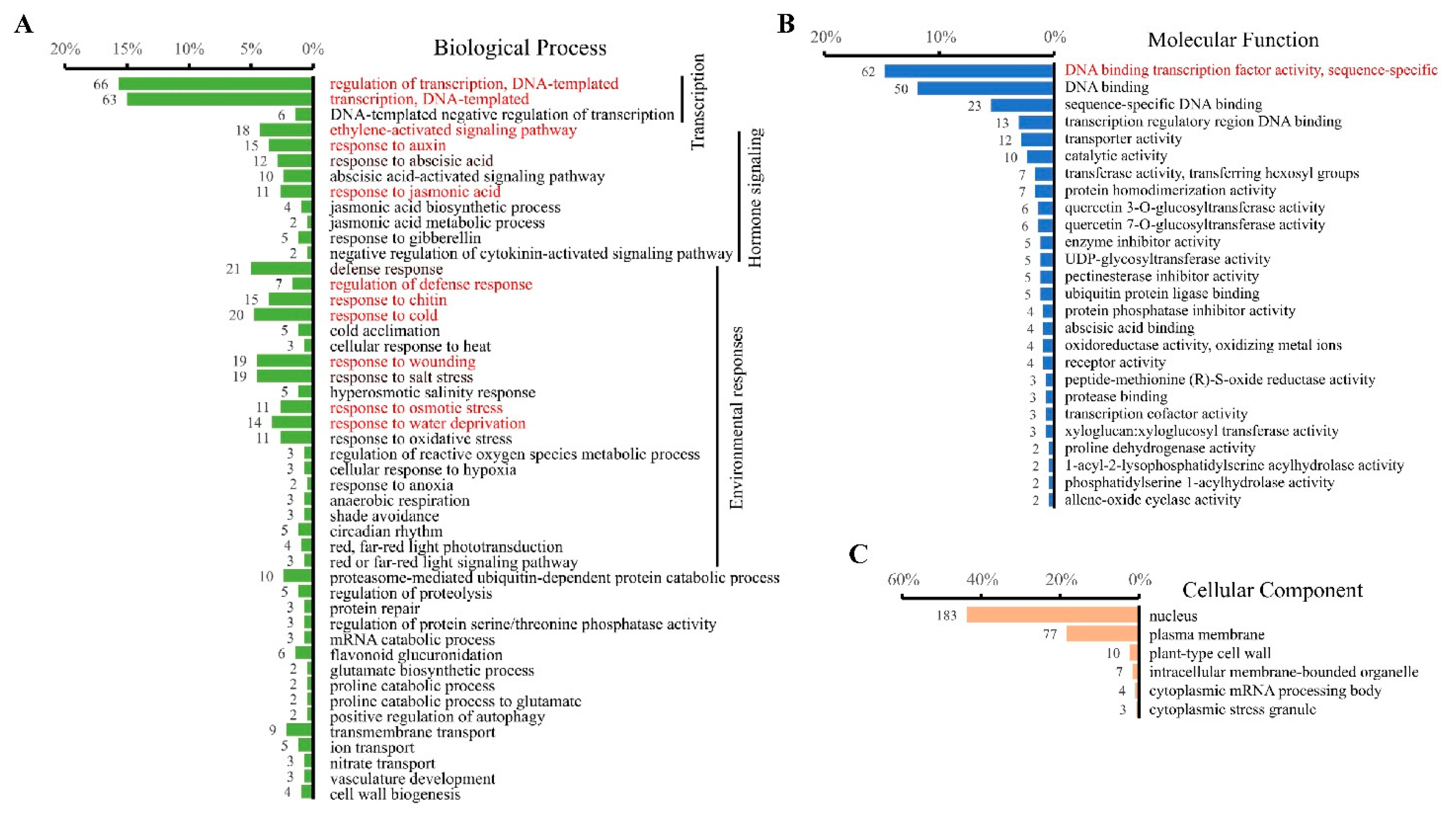
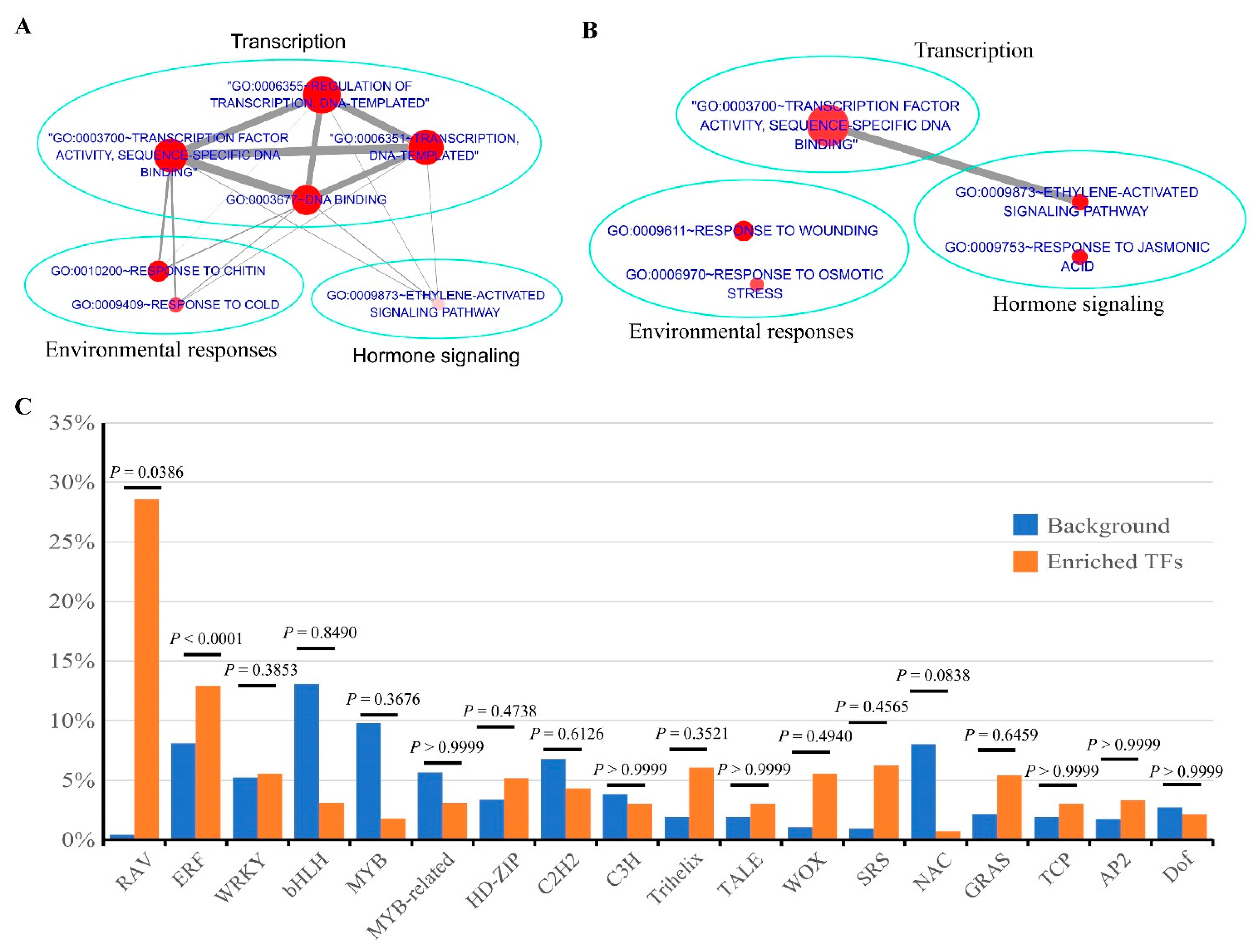
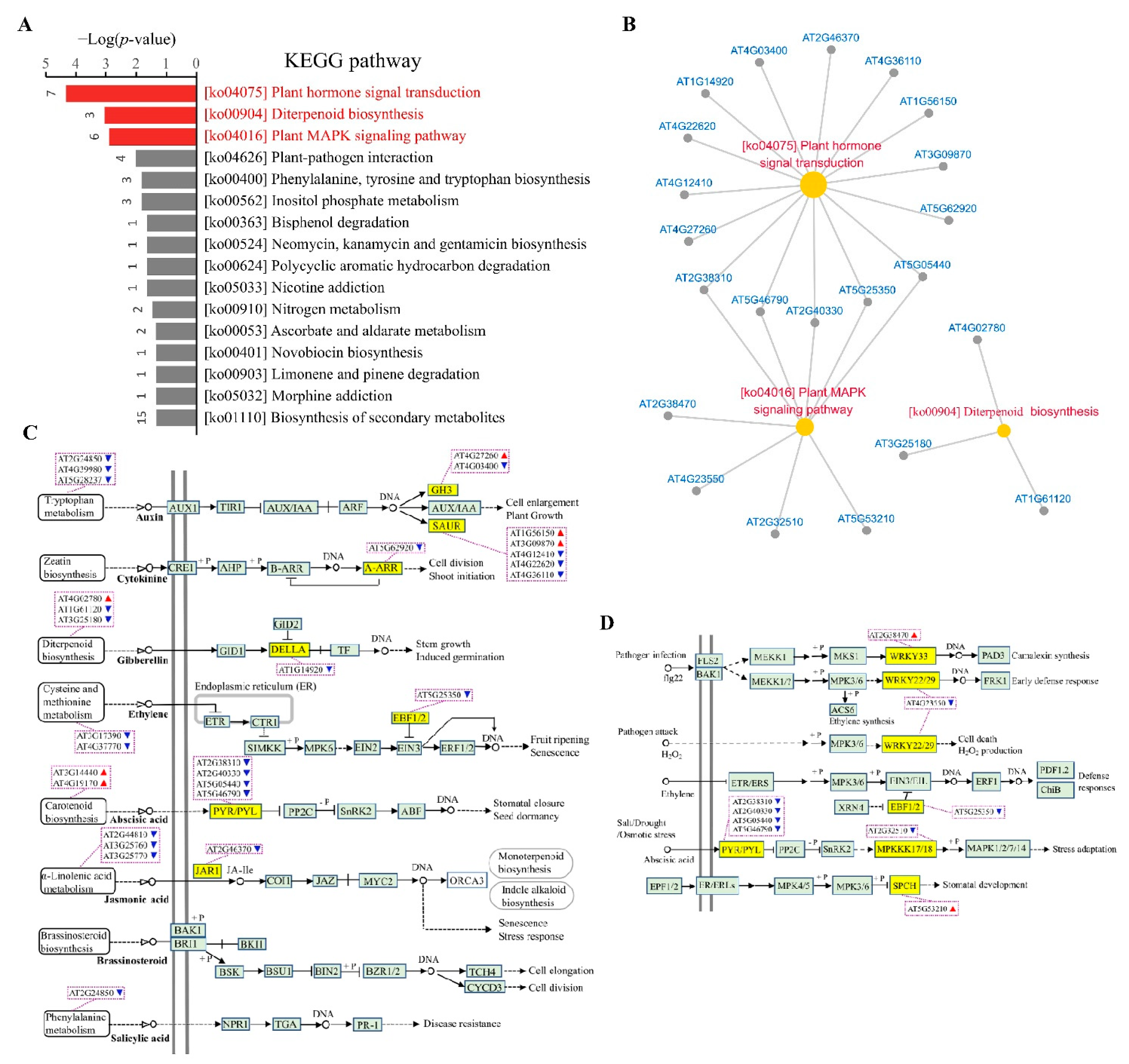
| Biological Processes | CRGs | |
|---|---|---|
| Phytohormone biosynthesis and homeostasis | PIN6 (AT1G77110), PID (AT2G34650), JAR1 (AT2G46370), DND1 (AT2G44810), AOC1 (AT3G25760), AOC2 (AT3G25770), GA1 (AT4G02780), CPS (AT4G02780), ACS8 (AT4G37770), LOG7 (AT5G06300), SMT3 (AT1G76090), CYP702A3 (AT4G15310), HXXXD-type acyl-transferase family protein (AT2G40230) | |
| mRNA degradation | SOV (AT1G77680), CAF1-5 (AT1G61470), TSN2 (AT5G61780), BRN2 (AT1G03457) | |
| Protein synthesis | LOS1 (AT1G56070) | |
| Proteasomal degradation | PUB29 (AT3G18710), PUB45 (AT1G27910), RING/U-box superfamily protein (AT5G47610), SGR9 (AT5G02750), SNIPER1 (AT1G14200), EBF2 (AT5G25350), PP2-A14 (AT5G52120), KMD2 (AT1G15670), KMD4 (AT3G59940), F-box/kelch-repeat protein (AT1G14330), HUP6 (AT3G27220), BTB/POZ domain-containing protein (AT2G30600) | |
| Ion transport and cell signaling | ||
| Ca2+ | AtCNGC2 (AT5G15410), CAX7 (AT5G17860), CSC1-like proteins (AT1G10090; AT1G62320) | |
| K+ | AtKUP7 | |
| Nitrate | NPF5.12 (AT1G72140), NPF7.2/NRT1.8 (AT4G21680), NRT2.6 (AT3G45060), NPF2.7/NAXT1(AT3G45650), CLC-b (AT3G27170) | |
| Sugar | SWEET16 (AT3G16690), HKL1 (AT1G50460), PMT6/PLT6 (AT4G36670) | |
| Lipid | MIOX2 (AT2G19800), ITPK3 (AT4G08170), SFH14 (AT5G56160) | |
| Light | BG1 (AT5G12050), ERD7 (AT2G17840), KNAT4 (AT5G11060), DFL2 (AT4G03400), PKS1 (AT2G02950), PKS2 (AT1G14280), BRN2 (AT1G03457), TEM1 (AT1G25560), BBX30 (AT4G15248) | |
| CO2/HCO3−/pH sensing | βCA3 (AT1G23730), SLAH3 (AT5G24030) | |
| Cell cycle | CDKG1 (AT5G63370) | |
| Secondary metabolism | AtGES (AT1G61120), CYP82G1 (AT3G25180), TAT3 (AT2G24850), DHS1 (AT4G39980), Tryptophan synthase (AT5G28237) | |
| Cell wall assembly and remodeling | XTH16 (AT3G23730), XTH23 (AT4G25810), XTH33 (AT1G10550), CSLA3 (AT1G23480), LRX2 (AT1G62440), AtGH9C2 (AT1G64390), KOR2 (AT1G65610), EXPA12 (AT3G15370), EXPA15 (AT2G03090), GALS1 (AT2G33570), FLA13 (AT5G44130), Exostosin family protein (AT4G32790), PME20 (AT2G47550), PMEI7 (AT4G25260), PMEI11 (AT3G47380), PMEI13/MMI9.1 (AT5G62360), MMI9.18 (AT5G62350), PER62 (AT5G39580), MAP70.5 (AT4G17220), EARLI1-like lipid transfer protein 3 (AT4G12500), Cell wall integrity/stress response component-like protein (AT4G39840), OFUT39 (AT5G65470), EXO70D1 (AT1G72470) | |
| Mitochondria-associated | PDC1 (AT4G33070), QCR7-2 (AT5G25450), PCMP-E34 (AT1G28690), PCMP-H43 (AT3G12770), POX1 (AT3G30775), POX2 (AT5G38710), DFR1 (AT5G17460), APC1 (AT5G61810), PUMP4 (AT4G24570) | |
| Chloroplasts-associated | MSL3 (AT1G58200) | |
| GO Term | Up-Regulated CRGs | Down-Regulated CRGs |
|---|---|---|
| Transcription Factors (TF family) * | ||
| GO:0003700~transcription factor activity andGO:0006355~regulation of transcription | AT1G19210 (ERF), AT1G74930 (ERF), AT4G25470 (ERF), AT4G25490 (ERF), AT5G21960 (ERF), AT5G47230 (ERF), AT5G51190 (ERF), AT5G51990 (ERF), AT5G61600 (ERF), AT2G38470 (WRKY), AT2G46400 (WRKY), AT4G23810 (WRKY), AT5G03150 (C2H2), AT5G67450 (C2H2), AT2G40140 (C3H), AT5G53210 (bHLH), AT5G11060 (TALE), AT1G46480 (WOX), AT3G10590 (MYB-related), AT5G66350 (SRS), AT2G17040 (NAC), AT3G21150 (C2C2 $), AT4G14465 (AT-hook $) | AT1G22810 (ERF), AT1G43160 (ERF), AT1G72360 (ERF), AT4G06746 (ERF), AT4G32800 (ERF), AT5G25810 (ERF), AT5G53290 (ERF), AT5G64750 (ERF),AT5G67190 (ERF), AT1G12540 (bHLH), AT1G18400 (bHLH), AT1G62975 (bHLH), AT1G73830 (bHLH), AT2G22770 (bHLH), AT4G29930 (bHLH), AT1G18710 (MYB), AT1G57560 (MYB), AT4G05100 (MYB), AT5G56840 (MYB-related), AT5G58900 (MYB-related), AT1G10480 (C2H2), AT2G41940 (C2H2), AT3G53600 (C2H2), AT5G44260 (C3H), AT4G16780 (HD-ZIP), AT4G17460 (HD-ZIP), AT4G37790 (HD-ZIP), AT3G01080 (WRKY), AT4G23550 (WRKY), AT2G38250 (Trihelix), AT5G01380 (Trihelix), AT1G25560 (RAV), AT3G25730 (RAV)AT1G14920 (GRAS), AT3G50650 (GRAS), AT3G02150 (TCP), AT2G28550 (AP2), AT1G69570 (Dof), AT5G49700 (AT-hook $) |
| Regulation of Transcription (gene description) | ||
| GO:0006355~regulation of transcription | AT1G61470 (CCR4-ASSOCIATED FACTOR 1E, CAF1E), AT2G40130 (SMAX1-LIKE 8, SMXL8), AT3G04930 (DNA-binding storekeeper protein-related transcriptional regulator), AT5G36740 (Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger), AT5G57180 (CHLOROPLAST IMPORT APPARATUS 2, CIA2) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, R.; Guo, Y.; Peng, S.; Liu, J.; Li, P.; Jia, W.; Zhao, J. Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis. Biomolecules 2021, 11, 688. https://doi.org/10.3390/biom11050688
Xu R, Guo Y, Peng S, Liu J, Li P, Jia W, Zhao J. Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis. Biomolecules. 2021; 11(5):688. https://doi.org/10.3390/biom11050688
Chicago/Turabian StyleXu, Ruqiang, Yanhui Guo, Song Peng, Jinrui Liu, Panyu Li, Wenjing Jia, and Junheng Zhao. 2021. "Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis" Biomolecules 11, no. 5: 688. https://doi.org/10.3390/biom11050688
APA StyleXu, R., Guo, Y., Peng, S., Liu, J., Li, P., Jia, W., & Zhao, J. (2021). Molecular Targets and Biological Functions of cAMP Signaling in Arabidopsis. Biomolecules, 11(5), 688. https://doi.org/10.3390/biom11050688





