Abstract
Interferon regulatory factor 5 (IRF5) has an important role in the inflammatory process, a fundamental component of coronary artery disease (CAD). Thus, the objective of this study was to evaluate the association of IRF5 polymorphisms with the development of premature CAD (pCAD) and cardiometabolic parameters. IRF5 polymorphisms (rs1874330, rs3778754, rs3757386, rs3757385, rs3807134, rs3807135, and rs6968563) were determined in 1116 pCAD patients and 1003 controls. Polymorphism distribution was similar in patients and controls; however, the haplotype analysis showed five haplotypes with a different distribution. TGCGTCT (OR (odds ratio) = 1.248, p = 0005) and TCTGCCT (OR = 10.73, p < 0.0001) were associated with a high risk, whereas TCCGTCT (OR = 0.155, p < 0.0001), CGCTTTT (OR = 0.108, p < 0.0001), and TCCGCCT (OR = 0.014, p < 0.0001) were associated with a low risk of pCAD. Associations with aspartate aminotransferase, hypertriglyceridemia, magnesium deficiency, triglycerides/HDL-C index, LDL-C, and adiponectin levels were observed in pCAD patients. In controls, associations with hypoalphalipoproteinemia, non-HDL-C, apolipoprotein B, hyperuricemia, TNF-α, IL-6, IL-15, valvular calcification, and subclinical hypothyroidism were observed. In summary, five haplotypes were associated with pCAD, two with high risk and three with low risk. Some IRF5 polymorphisms were associated with cardiometabolic parameters in pCAD patients and control.
1. Introduction
Coronary artery disease (CAD), a clinical manifestation of atherosclerosis, is one of the leading causes of death and morbidity worldwide [1]. The role of inflammation is well known in the progression of atherosclerosis. The infiltration of several types of cells, including T cells and macrophages, in the atherosclerotic plaque has been reported [2]. The production of cytokines, chemokines, and growth factors by these cells perpetuates the damage in the atherosclerotic lesion [3]. Interferon regulatory factor 5 (IRF5) plays a central role in inflammation, mediating the production of proinflammatory cytokines, such as IL-6, IL-12, IL-23, and TNF-α [4,5]. IRF5 is expressed in monocytes and macrophages and has an important role in defining the inflammatory macrophage phenotype [5]. It has been reported that IRF5 participates in the Akt2 activation, producing an increase of glycolysis and M1 macrophage polarization [6]. M1 macrophages can produce proinflammatory cytokines and are suggested to be involved in the development of atherosclerosis and affect the stabilization and impact of atherosclerotic plaques [7,8]. Seneviratne et al., using an animal model, established that IRF5 promotes the presence of proinflammatory CD11c+ macrophages within atherosclerotic lesions and controls the expansion of the necrotic core formation in atherosclerosis [9]. In murine models, it has been reported that the inhibition of IRF5 decreases myocardial infarction size, and its genetic deletion protects from inflammatory arthritis and insulin resistance [10,11,12]. The IRF5 gene is located in chromosome 7 in location 128,937,457–128,950,038 (ENSEMBL genome browser) and is highly polymorphic. IRF5 polymorphisms have been associated with the presence of coronary artery calcium in patients with systemic lupus erythematosus [13]. In another study, it was reported that IRF5 is expressed in cells of atherosclerotic tissue and that this expression is regulated by some IRF5 polymorphisms; however, these polymorphisms were not associated with CAD or related phenotypes [14]. Considering the important role of IRF5 in the atherosclerotic process suggested previously, the present study aimed to establish the association of IRF5 polymorphisms with the presence of pCAD and/or cardiometabolic parameters. Knowing genetic polymorphisms associated with the development of CAD can help select patients with high genetic risk or establish which therapy is the most appropriate for a specific patient. To our knowledge, our study is the first in which an association between IRF5 polymorphisms and the presence of CAD is established. After performing informatics analysis, seven polymorphisms with a minor allele frequency >5% and/or with probable functional effect were included in the study.
2. Materials and Methods
2.1. Ethics Statement
The study was approved by the Ethics Committee of the Instituto Nacional de Cardiología Ignacio Chávez (INCICH). All participants gave written informed consent, and the protocol followed the recommendations of the Declaration of Helsinki.
2.2. Subjects
The study included 1116 patients with pCAD and 1003 healthy controls, all of them belonging to the Genetics of Atherosclerotic Disease (GEA) Mexican Study. Premature CAD was defined as a history of myocardial infarction, revascularization surgery, angioplasty, and coronary stenosis >50% on angiography. Premature CAD was considered when the diagnosis was made before age 55 in men and before age 65 in women. The control group was recruited from the institute’s blood bank and by direct invitation. This group included healthy individuals with no family history of pCAD. Patients and controls were recruited regardless of the comorbidities they presented. Thus, comorbidities were not considered as an exclusion criterion in the study. In all individuals, a computed tomography (CT) of the chest and abdomen was performed. Total, subcutaneous, and visceral abdominal fat areas were quantified as described by Kvist et al. [15], and the coronary artery calcification (CAC) score using the Agatston method [16]. All individuals included in the control group presented a CAC score equal to zero. Demographic, clinical, and biochemical parameters and lifestyle characteristics were evaluated in all participants and defined as previously described [17,18,19,20,21,22].
To assess the possible influence of population stratification, a panel of 265 ancestry informative markers distinguishing mainly Amerindian, European, and African ancestries were determined in all individuals [23]. A similar global ancestry was observed in the study individuals with 54.0% of Amerindian, 35.8% of Caucasian, and 10.1% of African ancestry in controls and 55.8% of Amerindian, 34.3% of Caucasian, and 9.8% of African ancestry in pCAD patients [18].
2.3. Genetic Analysis
High-molecular-weight genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini kit (QIAGEN, Hilden, Germany). The possible functional effect of the IRF5 SNPs was evaluated using the SNP Function Prediction (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html, accessed on 15 March 2020), Splice Port: An Interactive Splice Site Analysis Tool (http://spliceport.cbcb.umd.edu/SplicingAnalyser.html, accessed on 15 March 2020), Human-Transcriptome Database for Alternative Splicing (http://www.h-invitational.jp/h-dbas/, accessed on 15 March 2020), HSF (http://www.umd.be/HSF/, accessed on 15 March 2020), ESE finder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi, accessed on 15 March 2020), and SNPs3D (http://www.snps3d.org/, accessed on 15 March 2020) bioinformatics tools. After this functional analysis, we selected for the study seven IRF5 polymorphisms (rs1874330, rs3778754, rs3757386, rs3757385, rs3807134, rs3807135, and rs6968563) that were determined using 5′ exonuclease TaqMan genotyping assays. The polymorphisms were genotyped on an ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). To corroborate the adequate assignment of the genotypes in the TaqMan assays, 10% of the samples were randomly selected and repeated. These samples were 100% concordant in two independent assays.
2.4. Statistical Analysis
Data are expressed as frequencies, median (interquartile range), or mean ± standard deviation, as appropriate. Either Mann–Whitney U or Student’s t-test was used for continuous variable comparisons, while the chi-square test was employed for categorical variable comparisons. Alleles and genotype frequencies were determined by direct counting. Hardy–Weinberg’s equilibrium was determined by the chi-square test. The association of the polymorphisms with pCAD and with cardiometabolic parameters was evaluated using logistic regression analysis under different inheritance models (additive, codominant 1, codominant 2, dominant, heterozygote, and recessive). The different models were adjusted for confounding variables as appropriate. Haploview version 4.1 (https://www.broadinstitute.org/haploview/haploview, accessed on 15 October 2020) (Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, USA) was used to establish linkage disequilibrium (LD, D’) and construction of haplotypes.
3. Results
3.1. Demographic, Clinical, Biochemical, and Lifestyle Characteristics
The analysis included 2119 individuals, 1116 with pCAD and 1003 healthy controls (CAC score equal to zero). Demographic, clinical, biochemical, and lifestyle characteristics in the studied groups are shown in Table 1.

Table 1.
Demographic, clinical, biochemical, and lifestyle characteristics in the study groups.
Compared with controls, body mass index (BMI), triglycerides, triglycerides/HDL-C index, aspartate aminotransferase (AST), uric acid, and interleukin 6 were higher in patients with pCAD. In the same way, the pCAD patients showed a high frequency of obesity, hypertension, type 2 diabetes mellitus, hypoalphalipoproteinemia, hypertriglyceridemia, triglycerides/HDL-C index, hypoadiponectinemia, hyperuricemia, and magnesium deficiency when compared with the healthy controls. In contrast, the patients showed lower levels of LDL cholesterol, apolipoprotein B, and non-HDL cholesterol. This decrease may be due to the statin treatment that the patients received (Table 2).

Table 2.
Prevalence of coronary risk factors in the study groups.
High non-high-density lipoprotein cholesterol (non-HDL-C) was defined when its values were >160 mg/dL. Increased TNF-α, IL-6, and IL-15 were defined as follow: TNF-α > 75th percentile (0.97 pg/mL in women and 2.13 pg/mL in men); IL-6 > 75th percentile (1.30 pg/mL in women and 1.78 pg/mL in men); IL-15 > 75th percentile (2.02 pg/mL in women and 2.41 pg/mL in men). These cutoff points were obtained from a GEA Mexican study sample of 131 men and 185 women without obesity and with normal values of blood pressure, fasting glucose, and lipids.
3.2. Association of IRF5 Polymorphisms with pCAD
The seven polymorphisms evaluated independently were not associated with the risk of premature pCAD (data not shown).
3.3. Distribution of IRF5 Haplotypes in pCAD Patients and Healthy Controls
After the linkage disequilibrium analysis, eight haplotypes were formed. Table 3 shows the distribution of haplotypes in pCAD patients and healthy controls. TGCGTCT (OR (odds ratio) = 1.248, 95% CI: 1.102–1.413, p = 0005) and TCTGCCT (OR = 10.73, 95% CI: 5.416–21.26, p < 0.0001) were associated with a high risk, whereas TCCGTCT (OR = 0.155, 95% CI: 0.089–0.269, p < 0.0001), CGCTTTT (OR = 0.108, 95% CI: 0.051–0.226, p < 0.0001), and TCCGCCT (OR = 0.014, 95% CI: 0.002–0.102, p < 0.0001) were associated with a low risk of pCAD when compared with healthy controls.

Table 3.
IRF5 haplotype frequencies and the presence of pCAD.
3.4. Association of IRF5 Polymorphisms with Cardiometabolic Parameters
The associations of IRF5 polymorphisms with cardiometabolic parameters were evaluated independently in pCAD patients and healthy controls. In healthy controls, rs3757385 (OR = 0.63, 95% CI: 0.46–0.85, pdominant = 0.002) and rs3807135 (OR = 0.67, 95% CI: 0.49–0.92, pcodominant 1 = 0.013) were associated with a low risk of hypoalphalipoproteinemia, rs3807134 with elevated non-HDL-cholesterol (OR = 1.78, 95% CI: 1.12–2.82, pheterozygote = 0.015), rs6968563 with elevated apolipoprotein B (OR = 2.04, 95% CI: 1.07–3.88, pheterozygote = 0.030), rs3807134 with a high risk of hyperuricemia (OR = 2.07, 95% CI: 1.24–3.43, pcodominant 1 = 0.005), and rs3778754 with elevated levels of TNF-α (OR = 1.51, 95% CI: 1.08–2.11, precessive = 0.017). In the same way, rs1874330 (OR = 0.72, 95% CI: 0.53–0.98, precessive = 0.038) and rs3807135 (OR = 0.69, 95% CI: 0.49–0.96, precessive = 0.030) were associated with a low risk to have elevated IL-6 levels, rs1874330 (OR = 0.71, 95% CI: 0.53–0.96, precessive = 0.023) with a low risk to have elevated IL-15 levels, rs3778754 (OR = 1.565, 95% CI: 1.12–2.16, padditive = 0.009) with a high risk of valvular calcification, and rs3757385 and rs3807135 with a high risk of subclinical hypothyroidism (OR = 1.73, 95% CI: 1.20–2.51, precessive = 0.004) (Figure 1). The models were adjusted by age, sex, and body mass index.
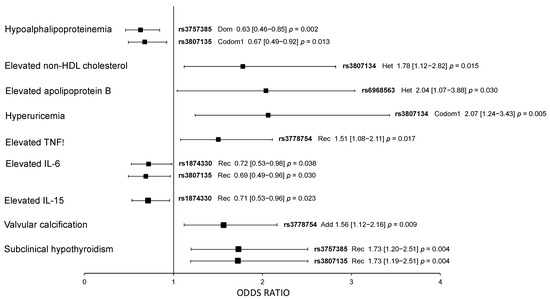
Figure 1.
Association of IRF5 polymorphisms with cardiometabolic parameters in healthy controls. All models were adjusted by age, sex, and BMI. Dom: dominant model; Rec: recessive model; Codom1: codominant 1 model; Het: heterozygote model; Add: additive model. OR (95% CI) p values.
In the patient group, four polymorphisms (rs1874330, rs3778754, rs3757385, and rs3807135) were associated with low AST levels, high LDL-C levels, and a low risk of magnesium deficiency. Three of them (rs3778754, rs3757385, and rs3807135) were associated with a low risk of hypertriglyceridemia, and four (rs3757386, rs3757385, rs3807134, and rs3807135) were associated with elevated triglycerides/HDL-cholesterol index. Finally, two polymorphisms (rs1874330 and rs3757385) were associated with a high risk of having low adiponectin levels (Figure 2). The models were adjusted by age, sex, and body mass index. A summary of results is shown in Figure 3.
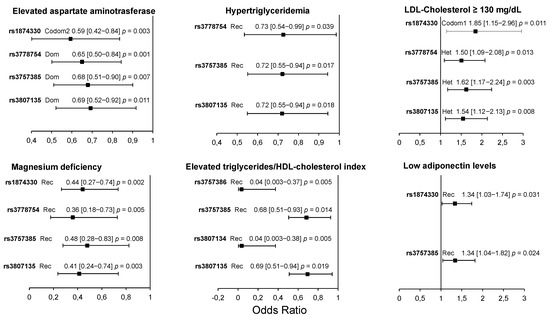
Figure 2.
Association of IRF5 polymorphisms with cardiometabolic parameters in pCAD patients. All models were adjusted by age, sex, and BMI. Dom: dominant model; Rec: recessive model; Codom1: codominant 1 model; Codom2: codominant 2 model; Het: heterozygote model. OR (95% CI) p values.
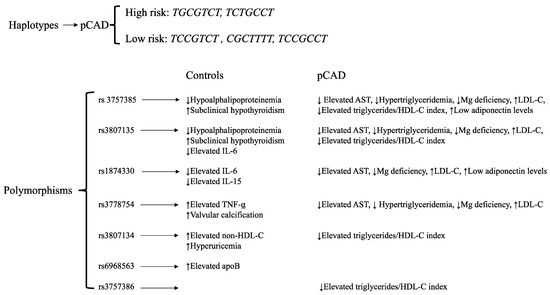
Figure 3.
Summary results. AST: aspartate aminotransferase.
4. Discussion
Atherosclerosis is a chronic and progressive disease that begins early in life and is characterized by a long subclinical phase, which progresses, producing coronary artery disease. Unfortunately, atherosclerosis is frequently diagnosed in advanced stages and generally after a sudden and sometimes fatal cardiovascular event. Genetic background is involved in both the triggering and progression of atherosclerosis [24]. Thus, we analyzed the distribution of seven polymorphisms of the IRF5 gene in patients with pCAD and healthy controls. The distribution of the polymorphisms was similar in the study groups; however, different haplotypes were associated with pCAD. TGCGTCT and TCTGCCT were associated with a high risk, and TCCGTCT, CGCTTTT, and TCCGCCT with a low risk of pCAD. In the same way, some polymorphisms were associated with cardiometabolic parameters.
IRF5 is an important regulator of the production of pro-inflammatory cytokines, such as TNF-alpha, IL-6, IL-12, and IL-23 [4,5]. Given these effects, IRF5 is an important regulator of inflammation. In the same way, IRF5 has been reported to modulate the genotype and function of macrophages, affecting the formation and stability of atherosclerotic plaque [9]. Mälarstig et al. reported that IRF5 is expressed in cells in atherosclerotic plaques and that some polymorphisms located in the IRF5 gene modified this expression [14]. In this study, the authors determined 10 polymorphisms, none of which were associated with CAD. Of those polymorphisms, in our study only one of them was included (rs3757385). The selection of the polymorphisms for the Mälarstig study was made considering those polymorphisms previously associated with some autoimmune diseases and with expression levels. In our case, the polymorphisms were selected using informatics tools. Similar to the Mälarstig study, we did not detect associations of the polymorphisms studied with pCAD. However, when the haplotype analysis was made, some haplotypes were associated with pCAD. It has been suggested that the use of haplotypes could be a better tool to capture more relevant information in a specific region compared with the analysis of independent polymorphisms [25]. The combination of polymorphisms in a haplotype could have a greater effect on the genotype of interest, and this effect would be greater than when analyzing the polymorphisms independently [26].
Association of the polymorphisms with cardiometabolic parameters was made independently in pCAD patients and healthy controls. In patients, the association of the polymorphisms with AST, LDL-C, magnesium deficiency, hypertriglyceridemia, triglycerides/HDL-C index, and adiponectin levels is worth mentioning. It is important to note that the rs3757385 polymorphism was associated with six cardiometabolic variables in this group of patients. In the study of Mälarstig et al., this polymorphism was not associated with CAD; however, it was significantly associated with IRF5 mRNA expression levels in carotid plaques [14]. This polymorphism is located in the promoter region of the gene and, according to the informatics analysis, is a tag SNP. The change in this position produces binding sites for BCL6 (G allele), STAT (G allele), and YY1 (T allele) transcription factors. This polymorphism has been associated with interstitial lung disease associated with systemic sclerosis [27,28] and unexplained recurrent pregnancy loss [29]. On the other hand, in controls, the polymorphisms were associated with hypoalphalipoproteinemia, non-HDL cholesterol, apolipoprotein B, hyperuricemia, TNF-α, IL-6, IL-15, valvular calcification, and subclinical hypothyroidism. As can be seen, the associations detected were different in both study groups, which is to be expected because these two groups are different. It is noteworthy that the control group does not have a family history of cardiovascular disease. Sindhu et al. reported that adipose tissue IRF5 gene expression was associated with cardiometabolic parameters in diabetic obese patients. A positive correlation was reported for LDL-C, HDL-C, triglycerides, TNF-α, and IL-6 [30], variables associated with IRF5 polymorphisms in our study. In an animal model of IRF5-deficient systemic lupus erythematosus, an increase in atherosclerosis was observed. This was accompanied by metabolic disturbances, such as insulin resistance, hyperglycemia, hyperlipidemia, increased adiposity, and hepatic steatosis [31].
The detection of polymorphisms associated with the development of CAD using either genome-wide association studies or candidate gene studies can be very important for the detection of individuals at high risk of developing CAD even before they present symptoms. Early detection can help define better and more targeted treatments. In the same way, in the context of precision medicine, knowledge of these polymorphisms can contribute to better individual therapy with beneficial results for affected patients.
Our study has several strengths, among which the inclusion of a large number of patients and controls well characterized from the demographic, clinical, and biochemical points of view stands out. This allowed us to analyze the association of polymorphisms with pCAD and with cardiometabolic variables. In the same way, it allowed us to adjust the inheritance models for confounding factors that are directly associated with the disease.
Among the limitations, we can mention that the possible functional effect was only evaluated with bioinformatics tools, and an experimental design was not included. Additionally, in our study, it was not possible to measure the levels of expression of IRF5. Finally, the results were not replicated in an independent group of individuals with and without pCAD.
5. Conclusions
In summary, five haplotypes were associated with pCAD, two with a high risk (TGCGTCT and TCTGCCT) and three with a low risk (TCCGTCT, CGCTTTT, and TCCGCCT). Some IRF5 polymorphisms were associated with cardiometabolic parameters in pCAD patients and healthy controls (Figure 3).
Author Contributions
Conceptualization, R.P.-S. and G.V.-A.; methodology, G.C.-S. and J.M.F.; formal analysis, R.P.-S., G.C.-S. and J.M.F.; resources, G.V.-A. and R.P.-S.; supervision, G.V.-A.; project administration, G.V.-A.; funding acquisition, G.V.-A.; writing—original draft preparation, R.P.-S. and G.V.-A.; writing—review and editing, G.V.-A. and R.P.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by a grant from the Consejo Nacional de Ciencia y Tecnología, México (Project No. FC-1958).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the Instituto Nacional de Cardiología Ignacio Chávez (protocol 19-1104, 18 February 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Informed: The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors are grateful to the study participants. Institutional Review Board approval was obtained for all sample collections.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Globalburdenofcoronaryheartdisease. Geneva: World Health Organization. 2004. Available online: http://www.who.int/cardiovascular_diseases/en/cvd_atlas_13_coronaryHD.pdf (accessed on 10 January 2007).
- Libby, P.; Buring, J.; Badimon, L.; Hansson, G.; Deanfield, J.; Bittencourt, M.; Tokgözoğlu, L.; Lewis, E. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56. [Google Scholar] [CrossRef]
- Gencer, S.; Evans, B.; van der Vorst, E.; Döring, Y.; Weber, C. Inflammatory Chemokines in Atherosclerosis. Cells 2021, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, A.; Yanai, H.; Kondo, S.; Duncan, G.; Negishi, H.; Mizutani, T.; Kano, S.; Honda, K.; Ohba, Y.; Mak, T.; et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 2005, 434, 243–249. [Google Scholar] [CrossRef]
- Krausgruber, T.; Blazek, K.; Smallie, T.; Alzabin, S.; Lockstone, H.; Sahgal, N.; Hussell, T.; Feldmann, M.; Udalova, I. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 2011, 13, 231–238. [Google Scholar] [CrossRef]
- Hedl, M.; Yan, J.; Abraham, C. IRF5 and IRF5 Disease-Risk Variants Increase Glycolysis and Human M1 Macrophage Polarization by Regulating Proximal Signaling and Akt2 Activation. Cell Rep. 2016, 16, 2442–2455. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Biswas, S.; Chittezhath, M.; Shalova, I.; Lim, J. Macrophage polarization and plasticity in health and disease. Immunol. Res. 2012, 53, 11–24. [Google Scholar] [CrossRef]
- Seneviratne, A.; Edsfeldt, A.; Cole, J.; Kassiteridi, C.; Swart, M.; Park, I.; Green, P.; Khoyratty, T.; Saliba, D.; Goddard, M.; et al. Interferon Regulatory Factor 5 Controls Necrotic Core Formation in Atherosclerotic Lesions by Impairing Efferocytosis. Circulation 2017, 136, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Dalmas, E.; Toubal, A.; Alzaid, F.; Blazek, K.; Eames, H.; Lebozec, K.; Pini, M.; Hainault, I.; Montastier, E.; Denis, R.; et al. IRF5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nat. Med. 2015, 21, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.; Byrne, A.; Blazek, K.; Saliba, D.; Pease, J.; Perocheau, D.; Feldmann, M.; Udalova, I. IRF5 controls both acute and chronic inflammation. Proc. Natl. Acad. Sci. USA 2015, 112, 11001–11006. [Google Scholar] [CrossRef]
- Courties, G.; Heidt, T.; Sebas, M.; Iwamoto, Y.; Jeon, D.; Truelove, J.; Tricot, B.; Wojtkiewicz, G.; Dutta, P.; Sager, H.; et al. In vivo silencing of the transcription factor IRF5 reprograms the macrophage phenotype and improves infarct healing. J. Am. Coll. Cardiol. 2014, 63, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Solus, J.; Oeser, A.; Li, C.; Raggi, P.; Smith, J.; Stein, C. Genetic Variation and Coronary Atherosclerosis in Patients with Systemic Lupus Erythematosus. Lupus 2014, 23, 876–880. [Google Scholar] [CrossRef]
- Mälarstig, A.; Sigurdsson, S.; Eriksson, P.; Paulsson-Berne, G.; Hedin, U.; Wallentin, L.; Siegbahn, A.; Hamsten, A.; Syvänen, A. Variants of the Interferon Regulatory Factor 5 Gene Regulate Expression of IRF5 mRNA in Atherosclerotic Tissue but Are Not Associated with Myocardial Infarction. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 975–982. [Google Scholar] [CrossRef]
- Kvist, H.; Chowdhury, B.; Grangård, U.; Tylén, U.; Sjöström, L. Total and visceral adipose-tissue volumes derived from measurements with computed tomography in adult men and women: Predictive equations. Am. J. Clin. Nutr. 1988, 48, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Mautner, G.C.; Mautner, S.L.; Froehlich, J.; Feuerstein, I.M.; Proschan, M.A.; Roberts, W.C.; Doppman, J.L. Coronary artery calcification: Assessment with electron beam CT and histomorphometric correlation. Radiology 1994, 192, 619–623. [Google Scholar] [CrossRef]
- Posadas-Sánchez, R.; López-Uribe, Á.R.; Posadas-Romero, C.; Pérez-Hernández, N.; Rodríguez-Pérez, J.M.; Ocampo-Arcos, W.A.; Fragoso, J.M.; Cardoso-Saldaña, G.; Vargas-Alarcón, G. Association of the I148M/PNPLA3 (rs738409) polymorphism with premature coronary artery disease, fatty liver, and insulin resistance in type 2 diabetic patients and healthy controls. The GEA study. Immunobiology 2017, 222, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Posadas-Sánchez, R.; Pérez-Hernández, N.; Angeles-Martínez, J.; López-Bautista, F.; Villarreal-Molina, T.; Rodríguez-Pérez, J.M.; Fragoso, J.M.; Posadas-Romero, C.; Vargas-Alarcón, G. Interleukin 35 Polymorphisms Are Associated with Decreased Risk of Premature Coronary Artery Disease, Metabolic Parameters, and IL-35 Levels: The Genetics of Atherosclerotic Disease (GEA) Study. Mediat. Inflamm. 2017, 2017, 6012795. [Google Scholar] [CrossRef]
- Posadas-Sánchez, R.; Posadas-Romero, C.; Cardoso-Saldaña, G.; Vargas-Alarcón, G.; Villarreal-Molina, M.T.M.T.; Pérez-Hernández, N.; Rodríguez-Pérez, J.M.J.M.; Medina-Urrutia, A.; Jorge-Galarza, E.; Juárez-Rojas, J.G.J.G.; et al. Serum magnesium is inversely associated with coronary artery calcification in the Genetics of Atherosclerotic Disease (GEA) study. Nutr. J. 2016, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Posadas-Romero, C.; Jorge-Galarza, E.; Posadas-Sánchez, R.; Acuña-Valerio, J.; Juárez-Rojas, J.G.J.G.; Kimura-Hayama, E.; Medina-Urrutia, A.; Cardoso-Saldaña, G.C.G.C. Fatty liver largely explains associations of subclinical hypothyroidism with insulin resistance, metabolic syndrome, and subclinical coronary atherosclerosis. Eur. J. Endocrinol. 2014, 171, 319–325. [Google Scholar] [CrossRef]
- Cardoso-Saldaña, G.; Fragoso, J.; Lale-Farjat, S.; Torres-Tamayo, M.; Posadas-Romero, C.; Vargas-Alarcón, G.; Posadas-Sánchez, R. The rs10455872-G allele of the LPA gene is associated with high lipoprotein(a) levels and increased aortic valve calcium in a Mexican adult population. Genet. Mol. Biol. 2019, 42, 519–525. [Google Scholar] [CrossRef]
- Baecke, J.A.; Burema, J.; Frijters, J.E. A short questionnaire for the measurement of habitual physical activity in epidemiological studies. Am. J. Clin. Nutr. 1982, 36, 936–942. [Google Scholar] [CrossRef]
- Silva-Zolezzi, I.; Hidalgo-Miranda, A.; Estrada-Gil, J.; Fernandez-Lopez, J.; Uribe-Figueroa, L.; Contreras, A.; Balam-Ortiz, E.; del Bosque-Plata, L.; Velazquez-Fernandez, D.; Lara, C.; et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. USA 2009, 106, 8611–8616. [Google Scholar] [CrossRef]
- Labos, C.; Thanassoulis, G. Genetic Risk Prediction for Primary and Secondary Prevention of Atherosclerotic Cardiovascular Disease: An Update. Curr. Cardiol. Rep. 2018, 20, 36. [Google Scholar] [CrossRef]
- Daly, M.; Rioux, J.; Schaffner, S.; Hudson, T.; Lander, E. High-resolution haplotype structure in the human genome. Nat. Genet. 2001, 29, 229–232. [Google Scholar] [CrossRef]
- Drysdale, C.; McGraw, D.; Stack, C.; Stephens, J.; Judson, R.; Nandabalan, K.; Arnold, K.; Ruano, G.; Liggett, S. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. USA 2000, 97, 10483–10488. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yi, L.; Guo, X.; Liu, M.; Li, H.; Zou, H.; Gu, Y.; Tu, W.; Guo, G.; Yang, L.; et al. Association of the IRF5 SNP rs2004640 with systemic sclerosis in Han Chinese. Int. J. Immunopathol. Pharmacol. 2014, 27, 635–638. [Google Scholar] [CrossRef]
- Dieude, P.; Dawidowicz, K.; Guedj, M.; Legrain, Y.; Wipff, J.; Hachulla, E.; Diot, E.; Sibilia, J.; Mouthon, L.; Cabane, J.; et al. Phenotype-haplotype correlation of IRF5 in systemic sclerosis: Role of 2 haplotypes in disease severity. J. Rheumatol. 2010, 37, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Amiri Jahromi, R.; Nasiri, M.; Jahromi, A. Interferon Regulatory Factor 5 Gene Polymorphisms in Iranian Women with Unexplained Recurrent Pregnancy Loss. Immunol. Invest. 2017, 46, 97–107. [Google Scholar] [CrossRef]
- Sindhu, S.; Kochumon, S.; Thomas, R.; Bennakhi, A.; Al-Mulla, F.; Ahmad, R. Enhanced Adipose Expression of Interferon Regulatory Factor (IRF)-5 Associates with the Signatures of Metabolic Inflammation in Diabetic Obese Patients. Cells 2020, 9, 730. [Google Scholar] [CrossRef]
- Watkins, A.; Yasuda, K.; Wilson, G.; Aprahamian, T.; Xie, Y.; Maganto-Garcia, E.; Shukla, P.; Oberlander, L.; Laskow, B.; Menn-Josephy, H.; et al. IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus-associated atherosclerosis. J. Immunol. 2015, 194, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).