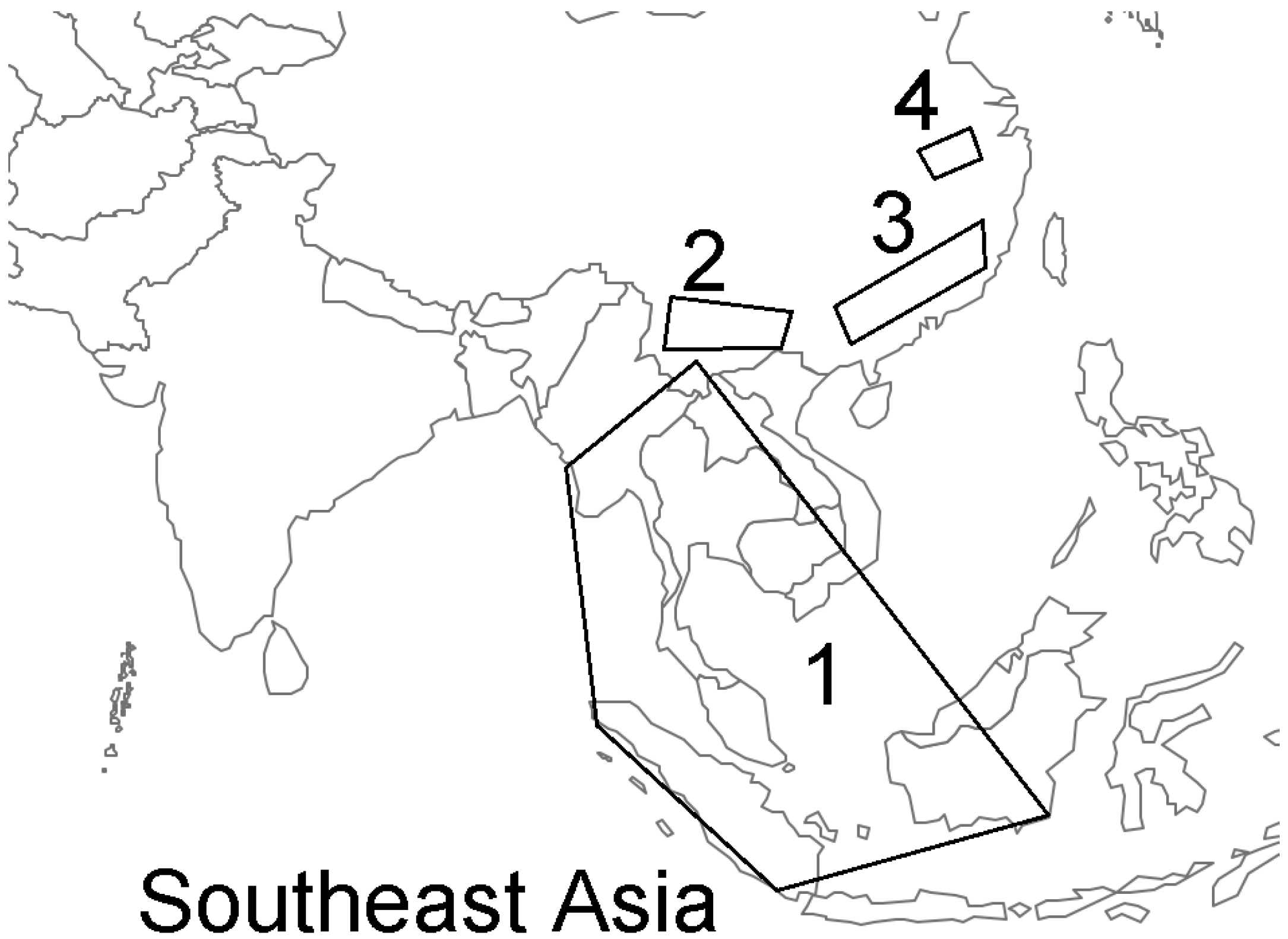
Urgent Need for Field Surveys of Coronaviruses in Southeast Asia to Understand the SARS-CoV-2 Phylogeny and Risk Assessment for Future Outbreaks †
Funding
Conflicts of Interest
References
- Olival, K.J.; Cryan, P.M.; Amman, B.R.; Baric, R.S.; Blehert, D.S.; Brook, C.E.; Calisher, C.H.; Castle, K.T.; Coleman, J.T.; Daszak, P.; et al. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: A case study of bats. PLoS Pathog. 2020, 16, e1008758. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203.e3. [Google Scholar] [CrossRef]
- Wong, A.C.P.; Li, X.; Lau, S.K.P.; Woo, P.C.Y. Global Epidemiology of Bat Coronaviruses. Viruses 2019, 11, 174. [Google Scholar] [CrossRef]
- Han, Y.; Du, J.; Su, H.; Zhang, J.; Zhu, G.; Zhang, S.; Wu, Z.; Jin, Q. Identification of Diverse Bat Alphacoronaviruses and Betacoronaviruses in China Provides New Insights into the Evolution and Origin of Coronavirus-Related Diseases. Front. Microbiol. 2019, 10, 1900. [Google Scholar] [CrossRef]
- Han, G.Z. Pangolins Harbor SARS-CoV-2-Related Coronaviruses. Trends Microbiol. 2020, 28, 515–517. [Google Scholar] [CrossRef]
- Lam, T.T.; Jia, N.; Zhang, Y.W.; Shum, M.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jiang, J.Z.; Wan, X.F.; Hua, Y.; Li, L.; Zhou, J.; Wang, X.; Hou, F.; Chen, J.; Zou, J.; et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020, 16, e1008421. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.J.; Li, N.; Guo, Y.; Li, X.; Shen, X.; et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Afrasiabi, A.; Alinejad-Rokny, H.; Lovell, N.; Xu, Z.; Ebrahimi, D. Insight into the origin of 5’UTR and source of CpG reduction in SARS-CoV-2 genome. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fischer, H.; Tschachler, E.; Eckhart, L. Pangolins Lack IFIH1/MDA5, a Cytoplasmic RNA Sensor That Initiates Innate Immune Defense Upon Coronavirus Infection. Front Immunol. 2020, 11, 939. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, D.; Halbwax, M.L.; Makaga, L.; Whytock, R.; Ndindiwe Malata, L.L.; Bombenda Mouele, W.; Momboua, B.R.; Koumba Pambo, A.F.; White, L.J.T. Pangolins and bats living together in underground burrows in Lopé National Park, Gabon. Afr. J. Ecol. 2020. [Google Scholar] [CrossRef]
- Hassanin, A.; Grandcolas, P.; Veron, G. Covid-19: Natural or anthropic origin? Mammalia 2020, 85, 1–7. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef]
- Wenzel, J. Origins of SARS-CoV-1 and SARS-CoV-2 are often poorly explored in leading publications. Cladistics 2020, 36, 374–379. [Google Scholar] [CrossRef]
- Mavian, C.; Pond, S.K.; Marini, S.; Magalis, B.R.; Vandamme, A.M.; Dellicour, S.; Scarpino, S.V.; Houldcroft, C.; Villabona-Arenas, J.; Paisie, T.K.; et al. Sampling bias and incorrect rooting make phylogenetic network tracing of SARS-COV-2 infections unreliable. Proc. Natl. Acad. Sci. USA 2020, 117, 12522–12523. [Google Scholar] [CrossRef]
- Garamszegi, L.Z.; Møller, A.P. Effects of sample size and intraspecific variation in phylogenetic comparative studies: A meta-analytic review. Biol. Rev. Camb. Philos. Soc. 2010, 85, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Seyran, M.; Pizzol, D.; Adadi, P.; El-Aziz, T.M.A.; Hassan, S.S.; Soares, A.; Kandimalla, R.; Lundstrom, K.; Tambuwala, M.; Aljabali, A.A.A.; et al. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 2020, 93, 1204–1206. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Shao, Y.; Tian, L.; Flasch, D.A.; Mulder, H.L.; Edmonson, M.N.; Liu, Y.; Chen, X.; Newman, S.; Nakitandwe, J.; et al. Analysis of error profiles in deep next-generation sequencing data. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Segreto, R.; Deigin, Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin: SARS-COV-2 chimeric structure and furin cleavage site might be the result of genetic manipulation. Bioessays 2020, 43, 2000240. [Google Scholar] [CrossRef]
- Singla, M.; Ahmad, S.; Gupta, C.; Sethi, T. De-novo assembly of RaTG13 Genome Reveals Inconsistencies further Obscuring SARS-CoV-2 Origins. Preprints 2020. [Google Scholar] [CrossRef]
- Hassanin, A. The SARS-CoV-2-like virus found in captive pangolins from Guangdong should be better sequenced. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mickleburgh, S.; Waylen, K.; Race, P. Bats as bushmeat: A global review. Oryx 2009, 43, 217–234. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014, 101, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hughes, T.; Lee, M.H.; Field, H.; Rovie-Ryan, J.J.; Sitam, F.T.; Sipangkui, S.; Nathan, S.K.; Ramirez, D.; Kumar, S.V.; et al. No Evidence of Coronaviruses or Other Potentially Zoonotic Viruses in Sunda pangolins (Manis javanica) Entering the Wildlife Trade via Malaysia. Ecohealth 2020, 17, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.R.; Schoeder, C.T.; Brown, J.A.; Smart, C.D.; Moth, C.; Wikswo, J.P.; Capra, J.A.; Meiler, J.; Chen, W.; Madhur, M.S. Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species. FASEB J. 2020, 34, 15946–15960. [Google Scholar] [CrossRef] [PubMed]
- Frutos, R.; Serra-Cobo, J.; Chen, T.; Devaux, C.A. COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans. Infect. Genet. Evol. 2020, 84, 104493. [Google Scholar] [CrossRef]
- Wu, L.; Chen, Q.; Liu, K.; Wang, J.; Han, P.; Zhang, Y.; Hu, Y.; Meng, Y.; Pan, X.; Qiao, C.; et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020, 6, 1–12. [Google Scholar] [CrossRef]
- Seyran, M.; Takayama, K.; Uversky, V.N.; Lundstrom, K.; Palù, G.; Sherchan, S.P.; Attrish, D.; Rezaei, N.; Aljabali, A.A.; Ghosh, S.; et al. The Structural Basis of Accelerated Host Cell Entry by SARS-CoV-2. FEBS J. 2020. [Google Scholar] [CrossRef]
- Freuling, C.M.; Breithaupt, A.; Müller, T.; Sehl, J.; Balkema-Buschmann, A.; Rissmann, M.; Klein, A.; Wylezich, C.; Höper, D.; Wernike, K.; et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg. Infect. Dis. 2020, 26, 2982–2985. [Google Scholar] [CrossRef]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2021, 11, 100192. [Google Scholar] [CrossRef]
- Hobbs, E.C.; Reid, T.J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Morozov, I.; Trujillo, J.D.; Gaudreault, N.N.; Bold, D.; Carossino, M.; Artiaga, B.L.; Indran, S.V.; Kwon, T.; Balaraman, V.; et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 2278–2288. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220-20. [Google Scholar] [CrossRef] [PubMed]
- Pollock, D.D.; Castoe, T.A.; Perry, B.W.; Lytras, S.; Wade, K.J.; Robertson, D.L.; Holmes, E.C.; Boni, M.F.; Kosakovsky Pond, S.L.; Parry, R.; et al. Viral CpG Deficiency Provides No Evidence That Dogs Were Intermediate Hosts for SARS-CoV-2. Mol. Biol. Evol. 2020, 37, 2706–2710. [Google Scholar] [CrossRef] [PubMed]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Uversky, V.N.; Elrashdy, F.; Aljadawi, A.; Redwan, E.M. Household pets and SARS-CoV2 transmissibility in the light of the ACE2 intrinsic disorder status. J. Biomol. Struct. Dyn. 2020, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yu, D.D.; Ma, Y.H.; Yao, Y.L.; Luo, R.H.; Feng, X.L.; Cai, H.R.; Han, J.B.; Wang, X.H.; Li, M.H.; et al. COVID-19-like symptoms observed in Chinese tree shrews infected with SARS-CoV-2. Zool. Res. 2020, 41, 517–526. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2020, 371, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Musso, N.; Costantino, A.; La Spina, S.; Finocchiaro, A.; Andronico, F.; Stracquadanio, S.; Liotta, L.; Visalli, R.; Emmanuele, G. New SARS-CoV-2 Infection Detected in an Italian Pet Cat by RT-qPCR from Deep Pharyngeal Swab. Pathogens 2020, 9, 746. [Google Scholar] [CrossRef]
- Zaeck, L.M.; Scheibner, D.; Sehl, J.; Müller, M.; Hoffmann, D.; Beer, M.; Abdelwhab, E.M.; Mettenleiter, T.C.; Breithaupt, A.; Finke, S.; et al. 3D reconstruction of SARS-CoV-2 infection in ferrets emphasizes focal infection pattern in the upper respiratory tract. bioRxiv 2020. [Google Scholar] [CrossRef]
- Relman, D.A. Opinion: To stop the next pandemic, we need to unravel the origins of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 29246–29248. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seyran, M.; Hassan, S.S.; Uversky, V.N.; Pal Choudhury, P.; Uhal, B.D.; Lundstrom, K.; Attrish, D.; Rezaei, N.; Aljabali, A.A.A.; Ghosh, S.; et al. Urgent Need for Field Surveys of Coronaviruses in Southeast Asia to Understand the SARS-CoV-2 Phylogeny and Risk Assessment for Future Outbreaks. Biomolecules 2021, 11, 398. https://doi.org/10.3390/biom11030398
Seyran M, Hassan SS, Uversky VN, Pal Choudhury P, Uhal BD, Lundstrom K, Attrish D, Rezaei N, Aljabali AAA, Ghosh S, et al. Urgent Need for Field Surveys of Coronaviruses in Southeast Asia to Understand the SARS-CoV-2 Phylogeny and Risk Assessment for Future Outbreaks. Biomolecules. 2021; 11(3):398. https://doi.org/10.3390/biom11030398
Chicago/Turabian StyleSeyran, Murat, Sk. Sarif Hassan, Vladimir N. Uversky, Pabitra Pal Choudhury, Bruce D. Uhal, Kenneth Lundstrom, Diksha Attrish, Nima Rezaei, Alaa A. A. Aljabali, Shinjini Ghosh, and et al. 2021. "Urgent Need for Field Surveys of Coronaviruses in Southeast Asia to Understand the SARS-CoV-2 Phylogeny and Risk Assessment for Future Outbreaks" Biomolecules 11, no. 3: 398. https://doi.org/10.3390/biom11030398
APA StyleSeyran, M., Hassan, S. S., Uversky, V. N., Pal Choudhury, P., Uhal, B. D., Lundstrom, K., Attrish, D., Rezaei, N., Aljabali, A. A. A., Ghosh, S., Pizzol, D., Adadi, P., El-Aziz, T. M. A., Kandimalla, R., Tambuwala, M. M., Lal, A., Azad, G. K., Sherchan, S. P., Baetas-da-Cruz, W., ... Brufsky, A. M. (2021). Urgent Need for Field Surveys of Coronaviruses in Southeast Asia to Understand the SARS-CoV-2 Phylogeny and Risk Assessment for Future Outbreaks. Biomolecules, 11(3), 398. https://doi.org/10.3390/biom11030398














