Pharmacological Manipulation of Early Zebrafish Skeletal Development Shows an Important Role for Smad9 in Control of Skeletal Progenitor Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry and Transgenic Lines
2.2. Pharmacological Treatment
2.3. Immunohistochemistry
2.4. RNAscope In Situ Hybridisation
2.5. Proliferation Assay
2.6. Alcian Blue Staining
2.7. Image Acquisition and Processing
2.8. Protein Sequence Alignment
2.9. Statistics
3. Results
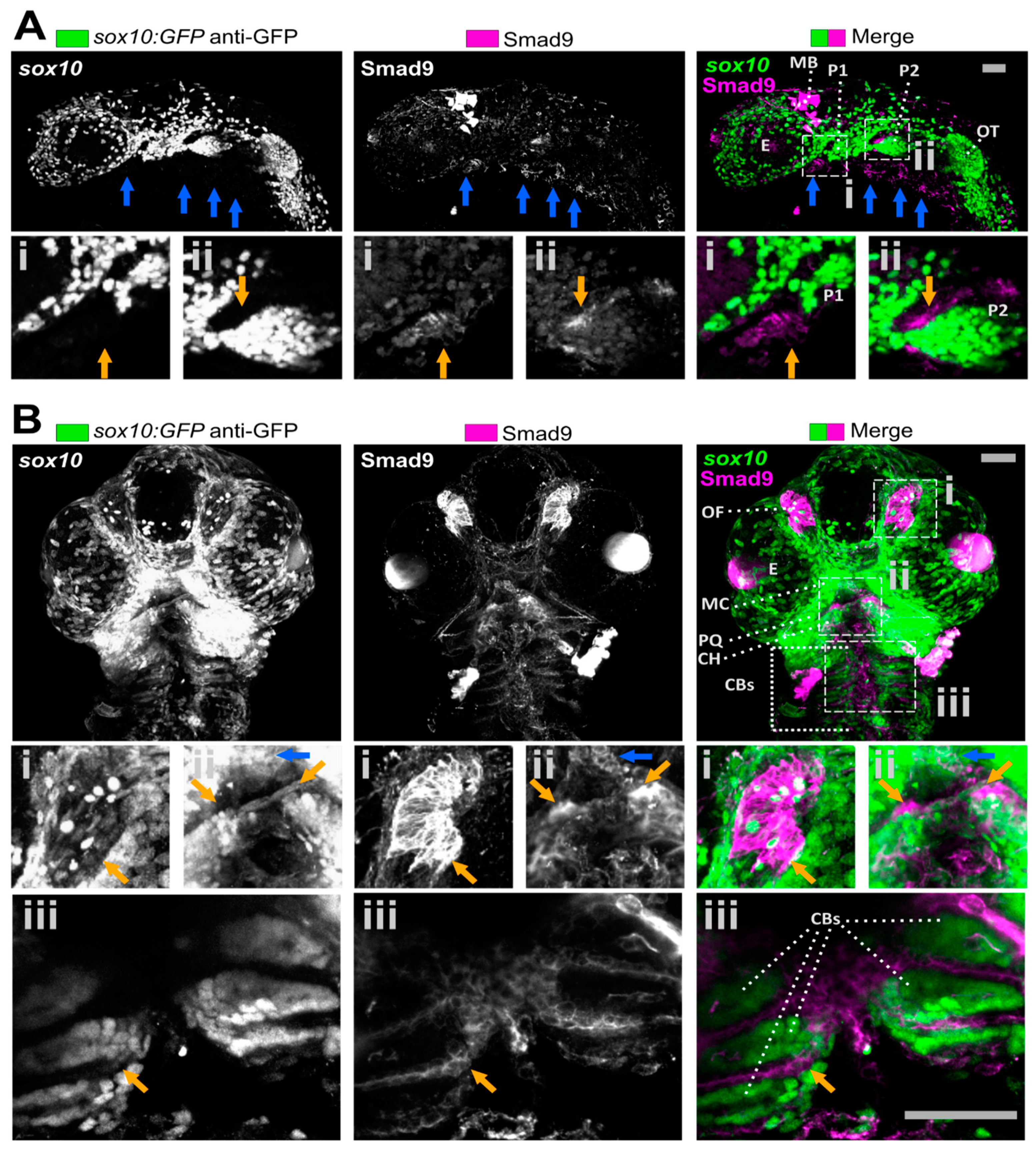
3.1. Smad9 Is Highly Expressed in Putative Osteochondral Progenitors during Embryonic Development
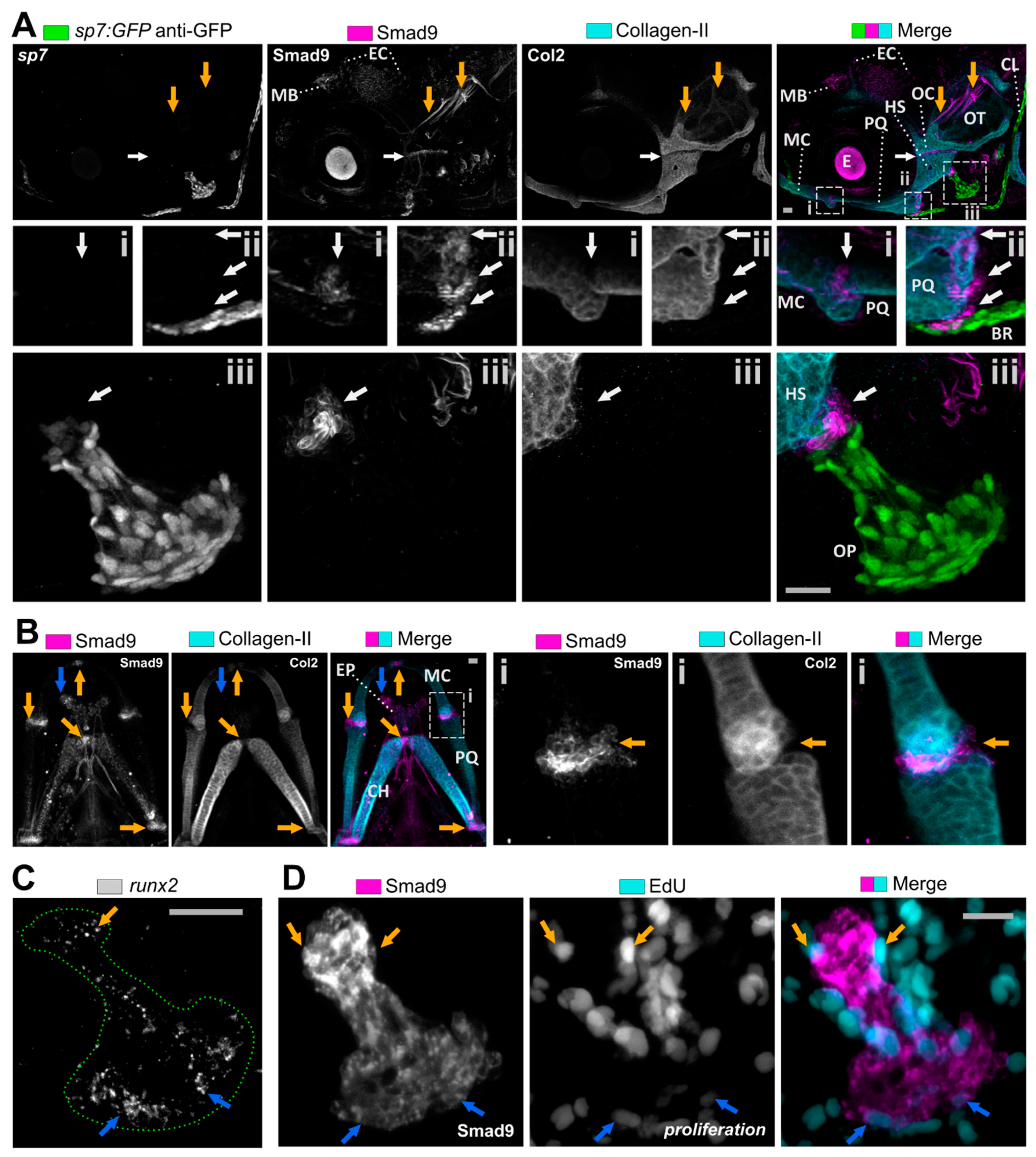
3.2. Smad9 Is Expressed within Joints and at Cartilage-Bone Interfaces during Early Jaw Development
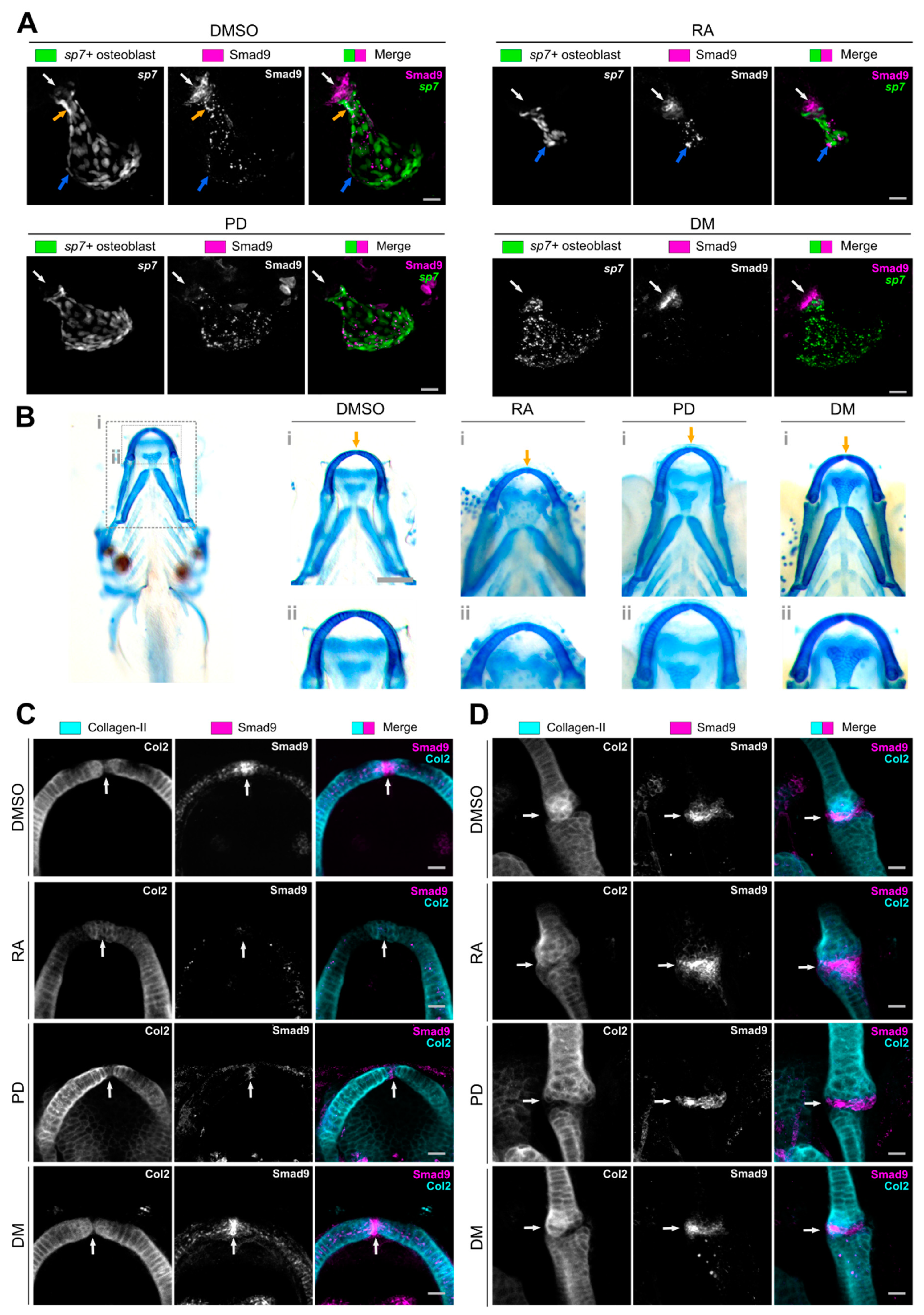
3.3. Pharmacological Manipulation Modulates Osteochondral Smad9 Expression at Skeletal Sites
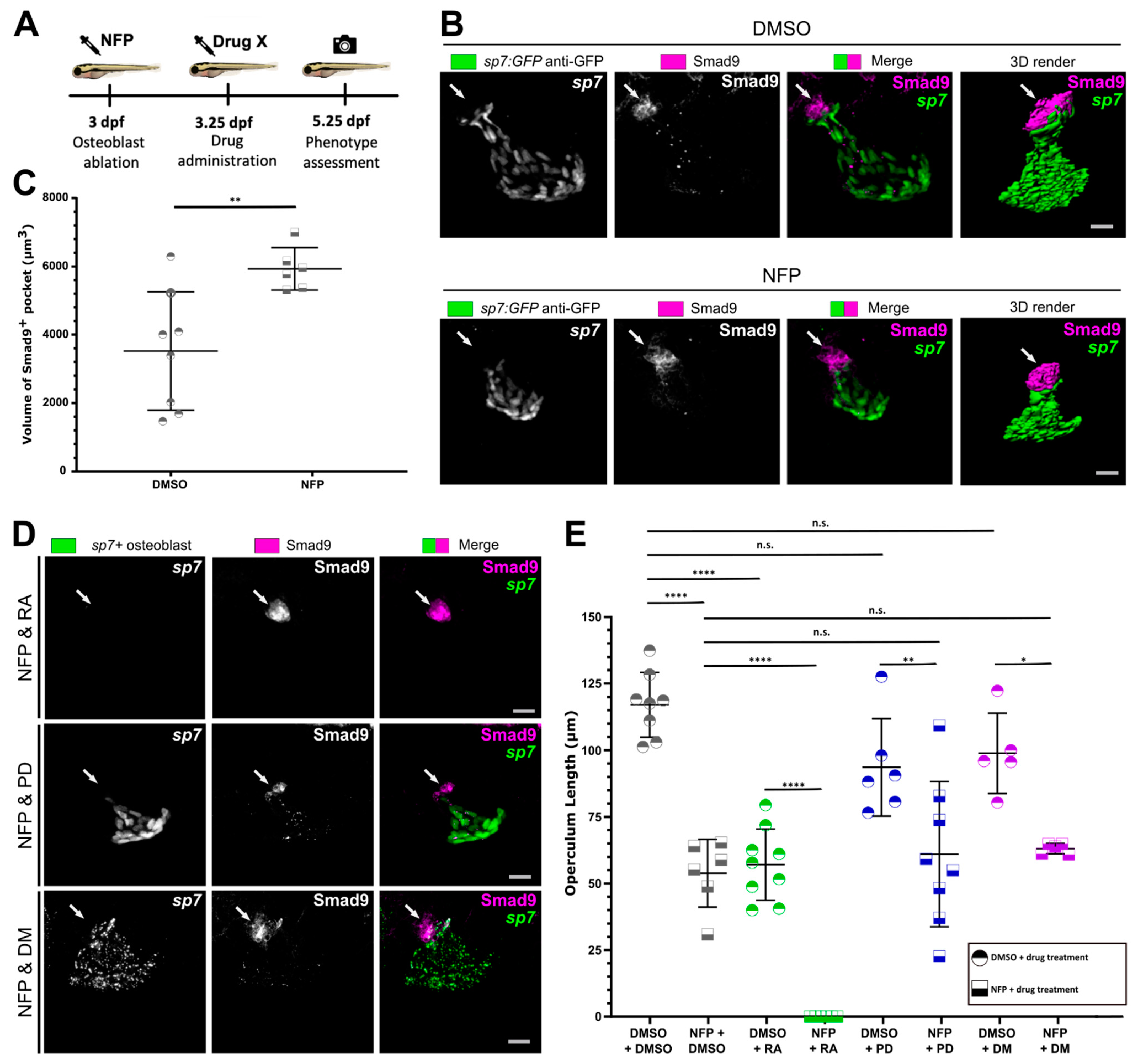
3.4. Osteoblast Ablation Induces Increased Expression of Smad9 during Larval De Novo Osteoblast Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 121–145. [Google Scholar] [CrossRef] [Green Version]
- Hernlund, E.; Svedbom, A.; Ivergård, M.; Compston, J.; Cooper, C.; Stenmark, J.; McCloskey, E.V.; Jonsson, B.; Kanis, J.A. Osteoporosis in the European Union: Medical management, epidemiology and economic burden. Arch. Osteoporos. 2013, 8, 1–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svedbom, A.; Hernlund, E.; Ivergård, M.; Compston, J.; Cooper, C.; Stenmark, J.; McCloskey, E.V.; Jönsson, B.; Kanis, J.A. Osteoporosis in the European Union: A compendium of country-specific reports. Arch. Osteoporos. 2013, 8, 137. [Google Scholar] [CrossRef] [Green Version]
- Metcalf, L.M.; Aspray, T.J.; McCloskey, E.V. The effects of parathyroid hormone peptides on the peripheral skeleton of postmenopausal women. A systematic review. Bone 2017, 99, 39–46. [Google Scholar] [CrossRef]
- Watanabe, A.; Yoneyama, S.; Nakajima, M.; Sato, N.; Takao-Kawabata, R.; Isogai, Y.; Sakurai-Tanikawa, A.; Higuchi, K.; Shimoi, A.; Yamatoya, H.; et al. Osteosarcoma in Sprague-Dawley rats after long-term treatment with teriparatide (human parathyroid hormone (1–34)). J. Toxicol. Sci. 2012, 37, 617–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregson, C.L.; Steel, S.A.; O’Rourke, K.P.; Allan, K.; Ayuk, J.; Bhalla, A.; Clunie, G.; Crabtree, N.; Fogelman, I.; Goodby, A.; et al. “Sink or swim”: An evaluation of the clinical characteristics of individuals with high bone mass. Osteoporos. Int. 2012, 23, 643–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, M.R.; Grauer, A.; Boonen, S.; Bolognese, M.A.; Brown, J.P.; Diez-Perez, A.; Langdahl, B.L.; Reginster, J.-Y.; Zanchetta, J.R.; Wasserman, S.M.; et al. Romosozumab in postmenopausal women with low bone mineral density. N. Engl. J. Med. 2014, 370, 412–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balemans, W. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum. Mol. Genet. 2001, 10, 537–543. [Google Scholar] [CrossRef] [Green Version]
- Gregson, C.L.; Bergen, D.J.M.; Leo, P.; Sessions, R.B.; Wheeler, L.; Hartley, A.; Youlten, S.; Croucher, P.; McInerney-Leo, A.M.; Fraser, W.; et al. A Rare Mutation in SMAD9 Associated with High Bone Mass Identifies the SMAD-Dependent BMP Signaling Pathway as a Potential Anabolic Target for Osteoporosis. J. Bone Miner. Res. 2020, 35, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Nakao, A.; Imamura, T.; Souchelnytskyi, S.; Kawabata, M.; Ishisaki, A.; Oeda, E.; Tamaki, K.; Hanai, J.-I.; Heldin, C.-C.; Miyazono, K.; et al. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997, 16, 5353–5362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukamoto, S.; Mizuta, T.; Fujimoto, M.; Ohte, S.; Osawa, K.; Miyamoto, A.; Yoneyama, K.; Murata, E.; Machiya, A.; Jimi, E.; et al. Smad9 is a new type of transcriptional regulator in bone morphogenetic protein signaling. Sci. Rep. 2014, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.B.; Hong, C.C.; Sachidanandan, C.; Babitt, J.L.; Deng, D.Y.; Hoyng, S.A.; Lin, H.Y.; Bloch, K.D.; Peterson, R.T. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008, 4, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askary, A.; Smeeton, J.; Paul, S.; Schindler, S.; Braasch, I.; Ellis, N.A.; Postlethwait, J.; Miller, C.T.; Crump, J.G. Ancient origin of lubricated joints in bony vertebrates. eLife 2016, 5, e16415. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Miller, C.T.; Moens, C.B. Specification and morphogenesis of the zebrafish larval head skeleton. Dev. Biol. 2001, 233, 239–257. [Google Scholar] [CrossRef] [Green Version]
- Eames, B.F.; DeLaurier, A.; Ullmann, B.; Huycke, T.R.; Nichols, J.T.; Dowd, J.; McFadden, M.; Sasaki, M.M.; Kimmel, C.B. FishFace: Interactive atlas of zebrafish craniofacial development at cellular resolution. BMC Dev. Biol. 2013, 13, 23. [Google Scholar] [CrossRef] [Green Version]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. In Nature Reviews Drug Discovery; Nature Publishing Group: Berlin, Germany, 2015; Volume 14, pp. 721–731. [Google Scholar]
- Lleras-Forero, L.; Winkler, C.; Schulte-Merker, S. Zebrafish and Medaka as Models for Biomedical Research of Bone Diseases; Developmental Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2020; Volume 457, pp. 191–205. [Google Scholar]
- Singh, S.P.; Holdway, J.E.; Poss, K.D. Regeneration of Amputated Zebrafish Fin Rays from De Novo Osteoblasts. Dev. Cell 2012, 22, 879–886. [Google Scholar] [CrossRef] [Green Version]
- Mantoku, A.; Chatani, M.; Aono, K.; Inohaya, K.; Kudo, A. Osteoblast and osteoclast behaviors in the turnover of attachment bones during medaka tooth replacement. Dev. Biol. 2016, 409, 370–381. [Google Scholar] [CrossRef] [Green Version]
- Witten, P.E.; Huysseune, A. A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol. Rev. 2009, 84, 315–346. [Google Scholar] [CrossRef]
- Hammond, C.L.; Schulte-Merker, S. Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development 2009, 136, 3991–4000. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast–osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef]
- Yu, P.B.; Deng, D.Y.; Lai, C.S.; Hong, C.C.; Cuny, G.D.; Bouxsein, M.L.; Hond, D.W.; McManus, P.M.; Katagiri, T.; Sachidanandan, C.; et al. BMP type I receptor inhibition reduces heterotopic ossification. Nat. Med. 2008, 14, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Wang, C.; Tang, Q.; Yang, F.; Xu, Y. Possible mechanisms of prednisolone-induced osteoporosis in zebrafish larva. Biomed. Pharmacother. 2018, 101, 981–987. [Google Scholar] [CrossRef]
- Pasqualetti, S.; Congiu, T.; Banfi, G.; Mariotti, M. Alendronate rescued osteoporotic phenotype in a model of glucocorticoid-induced osteoporosis in adult zebrafish scale. Int. J. Exp. Pathol. 2015, 96, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Geurtzen, K.; Vernet, A.; Freidin, A.; Rauner, M.; Hofbauer, L.C.; Schneider, J.E.; Brand, M.; Knopf, F. Immune Suppressive and Bone Inhibitory Effects of Prednisolone in Growing and Regenerating Zebrafish Tissues. J. Bone Miner. Res. 2017, 32, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Bergen, D.J.M.; Kague, E.; Hammond, C.L. Zebrafish as an emerging model for osteoporosis: A primary testing platform for screening new osteo-active compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vrieze, E.; Zethof, J.; Schulte-Merker, S.; Flik, G.; Metz, J.R. Identification of novel osteogenic compounds by an ex-vivo sp7:luciferase zebrafish scale assay. Bone 2015, 74, 106–113. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef] [Green Version]
- DeLaurier, A.; Frank Eames, B.; Blanco-Sánchez, B.; Peng, G.; He, X.; Swartz, M.E.; Ullmann, B.; Westerfield, M.; Kimmel, C.B. Zebrafish sp7:EGFP: A transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 2010, 48, 505–511. [Google Scholar] [CrossRef]
- Alexander, C.; Zuniga, E.; Blitz, I.L.; Wada, N.; le Pabic, P.; Javidan, Y.; Zhang, T.; Cho, K.W.; Crump, J.G.; Schilling, T.F. Combinatorial roles for BMPs and endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development 2011, 138, 5135–5146. [Google Scholar] [CrossRef] [Green Version]
- Dutton, J.R.; Antonellis, A.; Carney, T.J.; Rodrigues, F.S.L.M.; Pavan, W.J.; Ward, A.; Kelsh, R.N. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev. Biol. 2008, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Moro, E.; Ozhan-Kizil, G.; Mongera, A.; Beis, D.; Wierzbicki, C.; Young, R.M.; Bournele, D.; Domenichini, A.; Valdivia, L.E.; Lum, L.; et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 2012, 366, 327–340. [Google Scholar] [CrossRef]
- Dee, C.T.; Nagaraju, R.T.; Athanasiadis, E.I.; Gray, C.; del Ama, L.F.; Johnston, S.A.; Secombes, C.J.; Cvejic, A.; Hurlstone, A.F.L. CD4-Transgenic Zebrafish Reveal Tissue-Resident Th2- and Regulatory T Cell–like Populations and Diverse Mononuclear Phagocytes. J. Immunol. 2016, 197, 3520–3530. [Google Scholar] [CrossRef]
- Spoorendonk, K.M.; Peterson-Maduro, J.; Renn, J.; Trowe, T.; Kranenbarg, S.; Winkler, C.; Schulte-Merker, S. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development 2008, 135, 3765–3774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, L.; Wang, L.; Yang, Z.; Li, P.; Geng, D.; Xu, Y. Prednisolone induces osteoporosis-like phenotypes via focal adhesion signaling pathway in zebrafish larvae. Biol. Open. 2018, 15, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassmann, T.; Frings, O.; Sonnhammer, E.L.L. Kalign2: High-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009, 37, 858–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, T.; Azuma, Y.; Fukuyama, R.; Hattori, Y.; Yoshida, C.; Koida, M.; Ogita, K.; Komori, T. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J. Cell Biol. 2004, 166, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Bergström, I.; Isaksson, H.; Koskela, A.; Tuukkanen, J.; Ohlsson, C.; Andersson, G.; Windahl, S. Prednisolone treatment reduces the osteogenic effects of loading in mice. Bone 2018, 112, 10–18. [Google Scholar] [CrossRef]
- De Vrieze, E.; Van Kessel, M.A.H.J.; Peters, H.M.; Spanings, F.A.T.; Flik, G.; Metz, J.R. Prednisolone induces osteoporosis-like phenotype in regenerating zebrafish scales. Osteoporos. Int. 2014, 25, 567–578. [Google Scholar] [CrossRef]
- Schmidt, J.R.; Geurtzen, K.; von Bergen, M.; Schubert, K.; Knopf, F. Glucocorticoid Treatment Leads to Aberrant Ion and Macromolecular Transport in Regenerating Zebrafish Fins. Front. Endocrinol. 2019, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Laue, K.; Pogoda, H.M.; Daniel, P.B.; Van Haeringen, A.; Alanay, Y.; Von Ameln, S.; Rachwalski, M.; Morgan, T.; Gray, M.J.; Breuning, M.H.; et al. Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am. J. Hum. Genet. 2011, 89, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Jeradi, S.; Hammerschmidt, M. Retinoic acid-induced premature osteoblast-to-preosteocyte transitioning has multiple effects on calvarial development. Development 2016, 143, 1205–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laue, K.; Jänicke, M.; Plaster, N.; Sonntag, C.; Hammerschmidt, M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development 2008, 135, 3775–3787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunt, L.H.; Begg, K.; Kague, E.; Cross, S.; Hammond, C.L. Wnt signalling controls the response to mechanical loading during zebrafish joint development. Development 2017, 144, 2798–2809. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, N.; Ye, L.; Kobayashi, T.; Mochida, Y.; Yamauchi, M.; Kronenberg, H.M.; Feng, J.Q.; Mishina, Y. BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development 2008, 135, 3801–3811. [Google Scholar] [CrossRef] [Green Version]
- Bergemann, D.; Massoz, L.; Bourdouxhe, J.; Carril Pardo, C.A.; Voz, M.L.; Peers, B.; Manfroid, I. Nifurpirinol: A more potent and reliable substrate compared to metronidazole for nitroreductase-mediated cell ablations. Wound Repair Regen. 2018, 26, 238–244. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, D.; Ihida-Stansbury, K.; Jones, P.L.; Martin, J.F. Defective pulmonary vascular remodeling in Smad8 mutant mice. Hum. Mol. Genet. 2009, 18, 2791–2801. [Google Scholar] [CrossRef] [Green Version]
- Abarca-Buis, R.F.; Bustamante, M.; Cuervo, R.; Aguilar-Fernández-de-Lara, D.; Chimal-Monroy, J. Smad8 is expressed in the anterior necrotic zone: Evidence for a role of bone morphogenetic proteins/SMAD signaling in the activation of a molecular cascade that culminates in cell death. Dev. Growth Differ. 2011, 3, 780–792. [Google Scholar] [CrossRef]
- Kemp, J.P.; Morris, J.A.; Medina-Gomez, C.; Forgetta, V.; Warrington, N.M.; Youlten, S.E.; Zheng, J.; Gregson, C.L.; Grundberg, E.; Trajanoska, K.; et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 2017, 49, 1468. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Trajanoska, K.; Rivadeneira, F. The genetic architecture of osteoporosis and fracture risk. Bone 2019, 126, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Tobias, J.H.; Duncan, E.L.; Kague, E.; Hammond, C.L.; Gregson, C.L.; Bassett, J.H.D.; Williams, G.R.; Min, J.L.; Gaunt, T.R.; Karasik, D.; et al. Opportunities and challenges in functional genomics research in osteoporosis: Report from a workshop held by the Causes Working Group of the Osteoporosis and Bone Research Academy of the Royal Osteoporosis Society on October 5th 2020. Front. Endocrinol. 2020, 11, 1127. [Google Scholar]
- Brunkow, M.E.; Gardner, J.C.; Van Ness, J.; Paeper, B.W.; Kovacevich, B.R.; Proll, S.; Skonier, J.E.; Zhao, L.; Sabo, P.J.; Fu, Y.-H.; et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet. 2001, 68, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Cosman, F.; Crittenden, D.B.; Adachi, J.D.; Binkley, N.; Czerwinski, E.; Ferrari, S.; Hofbauer, L.C.; Lau, E.; Lewiecki, E.M.; Miyauchi, A.; et al. Romosozumab treatment in postmenopausal women with osteoporosis. N. Engl. J. Med. 2016, 375, 1532–1543. [Google Scholar] [CrossRef]
- Carney, T.J.; Feitosa, N.M.; Sonntag, C.; Slanchev, K.; Kluger, J.; Kiyozumi, D.; Gebauer, J.M.; Talbot, J.C.; Kimmel, C.B.; Sekiguchi, K.; et al. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet. 2010, 6, e1000907. [Google Scholar] [CrossRef] [Green Version]
- Lowery, J.W.; Rosen, V. Bone morphogenetic protein–based therapeutic approaches. Cold Spring Harb. Perspect. Biol. 2018, 10, a022327. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.R.; Lai, Y.H.; Tsai, J.J.; Hsiao, C.D. Live fluorescent staining platform for drug-screening and mechanism-analysis in zebrafish for bone mineralization. Molecules 2017, 22, 2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shintani, N.; Hunziker, E.B. Differential effects of dexamethasone on the chondrogenesis of mesenchymal stromal cells: Influence of microenvironment, tissue origin and growth factor. Eur. Cells Mater. 2011, 22, 302–319. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.S. Glucocorticoid-Induced Osteoporosis and Osteonecrosis. Endocrinol. Metab. Clin. N. Am. 2012, 41, 595. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonald, G.L.K.; Wang, M.; Hammond, C.L.; Bergen, D.J.M. Pharmacological Manipulation of Early Zebrafish Skeletal Development Shows an Important Role for Smad9 in Control of Skeletal Progenitor Populations. Biomolecules 2021, 11, 277. https://doi.org/10.3390/biom11020277
McDonald GLK, Wang M, Hammond CL, Bergen DJM. Pharmacological Manipulation of Early Zebrafish Skeletal Development Shows an Important Role for Smad9 in Control of Skeletal Progenitor Populations. Biomolecules. 2021; 11(2):277. https://doi.org/10.3390/biom11020277
Chicago/Turabian StyleMcDonald, Georgina L. K., Mengdi Wang, Chrissy L. Hammond, and Dylan J. M. Bergen. 2021. "Pharmacological Manipulation of Early Zebrafish Skeletal Development Shows an Important Role for Smad9 in Control of Skeletal Progenitor Populations" Biomolecules 11, no. 2: 277. https://doi.org/10.3390/biom11020277
APA StyleMcDonald, G. L. K., Wang, M., Hammond, C. L., & Bergen, D. J. M. (2021). Pharmacological Manipulation of Early Zebrafish Skeletal Development Shows an Important Role for Smad9 in Control of Skeletal Progenitor Populations. Biomolecules, 11(2), 277. https://doi.org/10.3390/biom11020277







