The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction
Abstract
1. Introduction
2. Hulks and Deadpools of the Cytokinin Universe
3. Cytokinin Types
3.1. Cis or Trans—That’s What Matters
- Reduction of tRNA species bearing CK-like modifications and replacement with others, such as wybutosine.
- Preference for cis-zeatin over iP in tRNA.
- Modulation of CK perception to prefer trans-zeatin.
3.2. Dihydrozeatin
3.3. Aromatic Cytokinins
4. Cytokinin Biosynthesis
4.1. Isopentenyltransferase
4.2. Hydroxylation of Isopentenyladenine
4.3. Cis-Zeatin—Unde es?
- (1)
- Isoprenylation of adenine, either free or tRNA-bound, with (Z)-4-hydroxy-3-methylbut-2-enyl diphosphate.
- (2)
- cis-hydroxylation of iP, either free or tRNA-bound.
- (3)
- Isomerisation of tZ.
- (4)
- Dehydrogenation of dihydrozeatin.
5. Activation of Cytokinins
6. Deactivation of Cytokinins
6.1. Glycosylation
- I.
- They are active per se through:
- Binding to CK receptors—currently, N-glucosides were shown not to bind to CK receptors, but the reported specificities may not reflect situations in planta as could be illustrated by potent CKs DHZ or BAP, which are often reported with dissociation constants one-to-two orders of magnitude higher than tZ or iP (e.g., [13,58,68,70,190]).
- Binding to other receptors or binding proteins—CKs and their derivatives are known to bind various proteins, including cyclin-dependent kinases or CK binding protein [191].
- Alteration of CK metabolism, e.g., by inhibition of CKX or conjugation enzymes, which would prevent CK inactivation and thus increase the concentration of active CKs. This mode of action could work also in the case of the abovementioned BAP and DHZ to explain the discrepancy with the affinity of CK receptors and their biological activity.
- II.
- Hydrolysis to free bases, which in turn exert the activity; however, this would not explain the differential regulation of gene expression upon treatment with free base, N7- or N9-glucoside [187]
6.2. Cytokinin Oxidase/Dehydrogenase
7. Cytokinin Signalling
8. Cytokinin Transport
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miller, C.O.; Skoog, F.; Von Saltza, M.H.; Strong, F.M. Kinetin, a Cell Division Factor from Deoxyribonucleic Acid. J. Am. Chem. Soc. 1955, 77, 1392. [Google Scholar] [CrossRef]
- Shantz, E.M.; Steward, F.C. The Identification of Compound A from Coconut Milk as 1,3-Diphenylurea. J. Am. Chem. Soc. 1955, 77, 6351–6353. [Google Scholar] [CrossRef]
- Jacobs, W.P. Plant Hormones; Cambridge University Press: Cambridge, UK, 1979; ISBN 978-0-521-22062-0. [Google Scholar]
- Ge, L.; Yong, J.W.H.; Goh, N.K.; Chia, L.S.; Tan, S.N.; Ong, E.S. Identification of Kinetin and Kinetin Riboside in Coconut (Cocos Nucifera L.) Water Using a Combined Approach of Liquid Chromatography-Tandem Mass Spectrometry, High Performance Liquid Chromatography and Capillary Electrophoresis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005, 829, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Barciszewski, J.; Siboska, G.E.; Pedersen, B.O.; Clark, B.F.; Rattan, S.I. Evidence for the Presence of Kinetin in DNA and Cell Extracts. FEBS Lett. 1996, 393, 197–200. [Google Scholar] [CrossRef]
- Letham, D.S. Zeatin, a Factor Inducing Cell Division Isolated from Zea Mays. Life Sci. 1963, 2, 569–573. [Google Scholar] [CrossRef]
- Skoog, F.; Armstrong, D.J. Cytokinins. Ann. Rev. Plant Physiol. 1970, 21, 359–384. [Google Scholar] [CrossRef]
- Pongs, O.; Reinwald, E. Function of Y in Codon-Anticodon Interaction of TRNAPhe. Biochem. Biophys. Res. Commun. 1973, 50, 357–363. [Google Scholar] [CrossRef]
- Agris, P.F.; Vendeix, F.A.P.; Graham, W.D. TRNA’s Wobble Decoding of the Genome: 40 Years of Modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef]
- Persson, B.C.; Esberg, B.; Ólafsson, Ó.; Björk, G.R. Synthesis and Function of Isopentenyl Adenosine Derivatives in TRNA. Biochimie 1994, 76, 1152–1160. [Google Scholar] [CrossRef]
- Schweizer, U.; Bohleber, S.; Fradejas-Villar, N. The Modified Base Isopentenyladenosine and Its Derivatives in TRNA. RNA Biol. 2017, 14, 1197–1208. [Google Scholar] [CrossRef]
- Zürcher, E.; Müller, B. Cytokinin Synthesis, Signaling, and Function—Advances and New Insights. Int. Rev. Cell Mol. Biol. 2016, 324, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.V.; Osolodkin, D.I.; Schmülling, T.; Romanov, G.A. Plant Membrane Assays with Cytokinin Receptors Underpin the Unique Role of Free Cytokinin Bases as Biologically Active Ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Daudu, D.; Allion, E.; Liesecke, F.; Papon, N.; Courdavault, V.; Dugé de Bernonville, T.; Mélin, C.; Oudin, A.; Clastre, M.; Lanoue, A.; et al. CHASE-Containing Histidine Kinase Receptors in Apple Tree: From a Common Receptor Structure to Divergent Cytokinin Binding Properties and Specific Functions. Front. Plant Sci. 2017, 8, 1614. [Google Scholar] [CrossRef] [PubMed]
- Kamínek, M.; Březinov, A.; Gaudinová, A.; Motyka, V.; Vaňková, R.; Zažímalová, E. Purine Cytokinins: A Proposal of Abbreviations. Plant Growth Regul. 2000, 32, 253–256. [Google Scholar] [CrossRef]
- Mik, V.; Szüčová, L.; Spíchal, L.; Plíhal, O.; Nisler, J.; Zahajská, L.; Doležal, K.; Strnad, M. N9-Substituted N6-[(3-Methylbut-2-En-1-Yl)Amino]Purine Derivatives and Their Biological Activity in Selected Cytokinin Bioassays. Bioorg. Med. Chem. 2011, 19, 7244–7251. [Google Scholar] [CrossRef]
- Pokorná, E.; Hluska, T.; Galuszka, P.; Hallmark, H.T.; Dobrev, P.I.; Záveská Drábková, L.; Filipi, T.; Holubová, K.; Plíhal, O.; Rashotte, A.M.; et al. Cytokinin N-Glucosides: Occurrence, Metabolism and Biological Activities in Plants. Biomolecules 2021, 11, 24. [Google Scholar] [CrossRef]
- Galuszka, P.; Popelková, H.; Werner, T.; Frébortová, J.; Pospíšilová, H.; Mik, V.; Köllmer, I.; Schmülling, T.; Frébort, I. Biochemical Characterization of Cytokinin Oxidases/Dehydrogenases from Arabidopsis Thaliana Expressed in Nicotiana Tabacum L. J. Plant Growth Regul. 2007, 26, 255–267. [Google Scholar] [CrossRef]
- Zalabák, D.; Galuszka, P.; Mrízová, K.; Podlešáková, K.; Gu, R.; Frébortová, J. Biochemical Characterization of the Maize Cytokinin Dehydrogenase Family and Cytokinin Profiling in Developing Maize Plantlets in Relation to the Expression of Cytokinin Dehydrogenase Genes. Plant Physiol. Biochem. PPB Société Fr. Physiol. Végétale 2014, 74, 283–293. [Google Scholar] [CrossRef]
- Pertry, I.; Václavíková, K.; Depuydt, S.; Galuszka, P.; Spíchal, L.; Temmerman, W.; Stes, E.; Schmülling, T.; Kakimoto, T.; Van Montagu, M.C.E.; et al. Identification of Rhodococcus Fascians Cytokinins and Their Modus Operandi to Reshape the Plant. Proc. Natl. Acad. Sci. USA 2009, 106, 929–934. [Google Scholar] [CrossRef]
- Žižková, E.; Dobrev, P.I.; Muhovski, Y.; Hošek, P.; Hoyerová, K.; Haisel, D.; Procházková, D.; Lutts, S.; Motyka, V.; Hichri, I. Tomato (Solanum Lycopersicum L.) SlIPT3 and SlIPT4 Isopentenyltransferases Mediate Salt Stress Response in Tomato. BMC Plant Biol. 2015, 15, 85. [Google Scholar] [CrossRef]
- Jaworek, P.; Kopečný, D.; Zalabák, D.; Šebela, M.; Kouřil, Š.; Hluska, T.; Končitíková, R.; Podlešáková, K.; Tarkowski, P. Occurrence and Biosynthesis of Cytokinins in Poplar. Planta 2019, 250, 229–244. [Google Scholar] [CrossRef]
- Kasahara, H.; Takei, K.; Ueda, N.; Hishiyama, S.; Yamaya, T.; Kamiya, Y.; Yamaguchi, S.; Sakakibara, H. Distinct Isoprenoid Origins of Cis- and Trans.-Zeatin Biosyntheses in Arabidopsis. J. Biol. Chem. 2004, 279, 14049–14054. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Ueda, N.; Aoki, K.; Kuromori, T.; Hirayama, T.; Shinozaki, K.; Yamaya, T.; Sakakibara, H. AtIPT3 Is a Key Determinant of Nitrate-Dependent Cytokinin Biosynthesis in Arabidopsis. Plant Cell Physiol. 2004, 45, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional Analyses of LONELY GUY Cytokinin-Activating Enzymes Reveal the Importance of the Direct Activation Pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [PubMed]
- Cedzich, A.; Stransky, H.; Schulz, B.; Frommer, W.B. Characterization of Cytokinin and Adenine Transport in Arabidopsis Cell Cultures. Plant. Physiol. 2008, 148, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Gajdošová, S.; Spíchal, L.; Kamínek, M.; Hoyerová, K.; Novák, O.; Dobrev, P.I.; Galuszka, P.; Klíma, P.; Gaudinová, A.; Žižková, E.; et al. Distribution, Biological Activities, Metabolism, and the Conceivable Function of Cis-Zeatin-Type Cytokinins in Plants. J. Exp. Bot. 2011, 62, 2827–2840. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Brütting, C.; Meza-Canales, I.D.; Großkinsky, D.K.; Vaňková, R.; Baldwin, I.T.; Meldau, S. The Role of Cis-Zeatin-Type Cytokinins in Plant Growth Regulation and Mediating Responses to Environmental Interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.H.; Csonka, L.; David, H.; McLennan, B. Cytokinins in the Soluble RNA of Plant Tissues. Science 1967, 156, 69–71. [Google Scholar] [CrossRef]
- Mok, M.C.; Mok, D.W.S.; Armstrong, D.J. Differential Cytokinin Structure-Activity Relationships in Phaseolus. Plant Physiol. 1978, 61, 72–75. [Google Scholar] [CrossRef]
- Schmitz, R.Y.; Skoog, F.; Playtis, A.J.; Leonard, N.J. Cytokinins: Synthesis and Biological Activity of Geometric and Position Isomers of Zeatin. Plant Physiol. 1972, 50, 702–705. [Google Scholar] [CrossRef]
- Vreman, H.J.; Schmitz, R.Y.; Skoog, F.; Playtis, A.J.; Frihart, C.R.; Leonard, N.J. Synthesis of 2-Methylthio-Cis- and Trans.-Ribosylzeatin and Their Isolation from Pisum TRNA. Phytochemistry 1974, 13, 31–37. [Google Scholar] [CrossRef]
- Mauk, C.S.; Langille, A.R. Physiology of Tuberization in Solanum Tuberosum L.: Cis-Zeatin Riboside in the Potato Plant: Its Identification and Changes in Endogenous Levels as Influenced by Temperature and Photoperiod. Plant Physiol. 1978, 62, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Suttle, J.C.; Banowetz, G.M. Changes in Cis-Zeatin and Cis-Zeatin Riboside Levels and Biological Activity during Potato Tuber Dormancy. Physiol. Plant. 2000, 109, 68–74. [Google Scholar] [CrossRef]
- Lulai, E.C.; Suttle, J.C.; Olson, L.L.; Neubauer, J.D.; Campbell, L.G.; Campbell, M.A. Wounding Induces Changes in Cytokinin and Auxin Content in Potato Tuber, but Does Not Induce Formation of Gibberellins. J. Plant Physiol. 2016, 191, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Kolachevskaya, O.O.; Sergeeva, L.I.; Floková, K.; Getman, I.A.; Lomin, S.N.; Alekseeva, V.V.; Rukavtsova, E.B.; Buryanov, Y.I.; Romanov, G.A. Auxin Synthesis Gene Tms1 Driven by Tuber-Specific Promoter Alters Hormonal Status of Transgenic Potato Plants and Their Responses to Exogenous Phytohormones. Plant Cell Rep. 2017, 36, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Yokota, T.; Takahashi, N. Transfer RNA, a Possible Supplier of Free Cytokinins, Ribosyl-Cis-Zeatin and Ribosyl-2-Methylthiozeatin: Quantitative Comparison between Free and Transfer Cytokinins in Various Tissues of the Hop Plant. Plant Cell Physiol. 1982, 23, 479–488. [Google Scholar] [CrossRef]
- Kudo, T.; Makita, N.; Kojima, M.; Tokunaga, H.; Sakakibara, H. Cytokinin Activity of Cis-Zeatin and Phenotypic Alterations Induced by Overexpression of Putative Cis-Zeatin-O-Glucosyltransferase in Rice. Plant Physiol. 2012, 160, 319–331. [Google Scholar] [CrossRef]
- Takagi, M.; Yokota, T.; Murofushi, N.; Ota, Y.; Takahashi, N. Fluctuation of Endogenous Cytokinin Contents in Rice during Its Life Cycle-Quantification of Cytokinins by Selected Ion Monitoring Using Deuterium-Labelled Internal Standards. Agric. Biol. Chem. 1985, 49, 3271–3277. [Google Scholar] [CrossRef]
- Emery, R.J.N.; Ma, Q.; Atkins, C.A. The Forms and Sources of Cytokinins in Developing White Lupine Seeds and Fruits. Plant Physiol. 2000, 123, 1593–1604. [Google Scholar] [CrossRef]
- Emery, R.J.N.; Leport, L.; Barton, J.E.; Turner, N.C.; Atkins, C.A. Cis-Isomers of Cytokinins Predominate in Chickpea Seeds throughout Their Development. Plant Physiol. 1998, 117, 1515–1523. [Google Scholar] [CrossRef]
- Veach, Y.K.; Martin, R.C.; Mok, D.W.S.; Malbeck, J.; Vaňková, R.; Mok, M.C. O-Glucosylation of Cis-Zeatin in Maize. Characterization of Genes, Enzymes, and Endogenous Cytokinins. Plant Physiol. 2003, 131, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Vyroubalová, Š.; Václavíková, K.; Turečková, V.; Novák, O.; Šmehilová, M.; Hluska, T.; Ohnoutková, L.; Frébort, I.; Galuszka, P. Characterization of New Maize Genes Putatively Involved in Cytokinin Metabolism and Their Expression during Osmotic Stress in Relation to Cytokinin Levels. Plant Physiol. 2009, 151, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Stirk, W.A.; Gold, J.D.; Novák, O.; Strnad, M.; van Staden, J. Changes in Endogenous Cytokinins During Germination and Seedling Establishment of Tagetes Minuta L. Plant Growth Regul. 2005, 47, 1–7. [Google Scholar] [CrossRef]
- Stirk, W.A.; Novák, O.; Žižková, E.; Motyka, V.; Strnad, M.; van Staden, J. Comparison of Endogenous Cytokinins and Cytokinin Oxidase/Dehydrogenase Activity in Germinating and Thermoinhibited Tagetes Minuta Achenes. J. Plant Physiol. 2012, 169, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Quesnelle, P.E.; Emery, R.J.N. Cis-Cytokinins That Predominate in Pisum Sativum during Early Embryogenesis Will Accelerate Embryo Growth in Vitro. Can. J. Bot. 2007, 85, 91–103. [Google Scholar] [CrossRef]
- Goggin, D.E.; Emery, R.J.N.; Powles, S.B.; Steadman, K.J. Initial Characterisation of Low and High Seed Dormancy Populations of Lolium Rigidum Produced by Repeated Selection. J. Plant Physiol. 2010, 167, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Stirk, W.A.; Václavíková, K.; Novák, O.; Gajdošová, S.; Kotland, O.; Motyka, V.; Strnad, M.; van Staden, J. Involvement of Cis-Zeatin, Dihydrozeatin, and Aromatic Cytokinins in Germination and Seedling Establishment of Maize, Oats, and Lucerne. J. Plant Growth Regul. 2012, 31, 392–405. [Google Scholar] [CrossRef]
- Tarkowská, D.; Filek, M.; Biesaga-Kościelniak, J.; Marcińska, I.; Macháčková, I.; Krekule, J.; Strnad, M. Cytokinins in Shoot Apices of Brassica Napus Plants during Vernalization. Plant Sci. 2012, 187, 105–112. [Google Scholar] [CrossRef]
- Vondráková, Z.; Dobrev, P.I.; Pesek, B.; Fischerová, L.; Vágner, M.; Motyka, V. Profiles of Endogenous Phytohormones Over the Course of Norway Spruce Somatic Embryogenesis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Záveská Drábková, L.; Dobrev, P.I.; Motyka, V. Phytohormone Profiling across the Bryophytes. PLoS ONE 2015, 10, e0125411. [Google Scholar] [CrossRef]
- Žižková, E.; Kubeš, M.; Dobrev, P.I.; Přibyl, P.; Šimura, J.; Zahajská, L.; Záveská Drábková, L.; Novák, O.; Motyka, V. Control of Cytokinin and Auxin Homeostasis in Cyanobacteria and Algae. Ann. Bot. 2017, 119, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.N.; Knowles, S.; Hayward, A.; Thorn, R.G.; Saville, B.J.; Emery, R.J.N. Detection of Phytohormones in Temperate Forest Fungi Predicts Consistent Abscisic Acid Production and a Common Pathway for Cytokinin Biosynthesis. Mycologia 2015, 107, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP Isopentenyltransferases and TRNA Isopentenyltransferases in Cytokinin Biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [PubMed]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the Cytosolic Cytokinin Oxidase/Dehydrogenase (CKX7) from Arabidopsis Causes Specific Changes in Root Growth and Xylem Differentiation. Plant J. Cell Mol. Biol. 2014, 78, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of Plant Growth by Cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Martin, R.C.; Mok, M.C.; Habben, J.E.; Mok, D.W.S. A Maize Cytokinin Gene Encoding an O-Glucosyltransferase Specific to Cis-Zeatin. Proc. Natl. Acad. Sci. USA 2001, 98, 5922–5926. [Google Scholar] [CrossRef]
- Spíchal, L.; Rakova, N.Y.; Riefler, M.; Mizuno, T.; Romanov, G.A.; Strnad, M.; Schmülling, T. Two Cytokinin Receptors of Arabidopsis Thaliana, CRE1/AHK4 and AHK3, Differ in Their Ligand Specificity in a Bacterial Assay. Plant Cell Physiol. 2004, 45, 1299–1305. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Kojima, M.; Yamaya, T.; Sakakibara, H. Molecular Characterization of Cytokinin-Responsive Histidine Kinases in Maize. Differential Ligand Preferences and Response to Cis-Zeatin. Plant Physiol. 2004, 134, 1654–1661. [Google Scholar] [CrossRef]
- Park, E.J.; Kim, T.-H. Fine-Tuning of Gene Expression by TRNA-Derived Fragments during Abiotic Stress Signal Transduction. Int. J. Mol. Sci. 2018, 19, 518. [Google Scholar] [CrossRef]
- Kamínek, M. Letter: Evolution of TRNA and Origin of the Two Positional Isomers of Zeatin. J. Theor. Biol. 1974, 48, 489–492. [Google Scholar] [CrossRef]
- Kamínek, M. Tracking the Story of Cytokinin Research. J. Plant Growth Regul. 2015, 34, 723–739. [Google Scholar] [CrossRef]
- Podlešáková, K.; Fardoux, J.; Patrel, D.; Bonaldi, K.; Novák, O.; Strnad, M.; Giraud, E.; Spíchal, L.; Nouwen, N. Rhizobial Synthesized Cytokinins Contribute to but Are Not Essential for the Symbiotic Interaction between Photosynthetic Bradyrhizobia and Aeschynomene Legumes. Mol. Plant Microbe Interact. MPMI 2013, 26, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, J.; Galuszka, P.; Tudzynski, P. Functional Characterization of the First Filamentous Fungal TRNA-Isopentenyltransferase and Its Role in the Virulence of Claviceps Purpurea. New Phytol. 2016, 211, 980–992. [Google Scholar] [CrossRef] [PubMed]
- Koshimizu, K.; Kusaki, T.; Mitsui, T.; Matsubara, S. Isolation of a Cytokinin, (−)-Dihydrozeatin, from Immature Seeds of Lupinus Luteus. Tetrahedron Lett. 1967, 8, 1317–1320. [Google Scholar] [CrossRef]
- Koshimizu, K.; Matsubara, S.; Kusaki, T.; Mitsui, T. Isolation of a New Cytokinin from Immature Yellow Lupin Seeds. Agric. Biol. Chem. 1967, 31, 795–801. [Google Scholar] [CrossRef]
- Choi, J.; Lee, J.; Kim, K.; Cho, M.; Ryu, H.; An, G.; Hwang, I. Functional Identification of OsHk6 as a Homotypic Cytokinin Receptor in Rice with Preferential Affinity for IP. Plant Cell Physiol. 2012, 53, 1334–1343. [Google Scholar] [CrossRef]
- Kuderová, A.; Gallová, L.; Kuricová, K.; Nejedlá, E.; Čurdová, A.; Micenková, L.; Plíhal, O.; Šmajs, D.; Spíchal, L.; Hejátko, J. Identification of AHK2- and AHK3-like Cytokinin Receptors in Brassica Napus Reveals Two Subfamilies of AHK2 Orthologues. J. Exp. Bot. 2015, 66, 339–353. [Google Scholar] [CrossRef]
- Lomin, S.N.; Yonekura-Sakakibara, K.; Romanov, G.A.; Sakakibara, H. Ligand-Binding Properties and Subcellular Localization of Maize Cytokinin Receptors. J. Exp. Bot. 2011, 62, 5149–5159. [Google Scholar] [CrossRef]
- Romanov, G.A.; Lomin, S.N.; Schmülling, T. Biochemical Characteristics and Ligand-Binding Properties of Arabidopsis Cytokinin Receptor AHK3 Compared to CRE1/AHK4 as Revealed by a Direct Binding Assay. J. Exp. Bot. 2006, 57, 4051–4058. [Google Scholar] [CrossRef]
- Gaudinová, A.; Dobrev, P.I.; Šolcová, B.; Novák, O.; Strnad, M.; Friedecký, D.; Motyka, V. The Involvement of Cytokinin Oxidase/Dehydrogenase and Zeatin Reductase in Regulation of Cytokinin Levels in Pea (Pisum Sativum L.) Leaves. J. Plant Growth Regul. 2005, 24, 188–200. [Google Scholar] [CrossRef]
- Martin, R.C.; Mok, M.C.; Shaw, G.; Mok, D.W.S. An Enzyme Mediating the Conversion of Zeatin to Dihydrozeatin in Phaseolus Embryos. Plant Physiol. 1989, 90, 1630–1635. [Google Scholar] [CrossRef]
- Sondheimer, E.; Tzou, D.-S. The Metabolism of Hormones during Seed Germination and Dormancy II. The Metabolism of 8-14C-Zeatin in Bean Axes. Plant Physiol. 1971, 47, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Letham, D.S.; Jameson, P.E.; Zhang, R.; Parker, C.W.; Bandenoch-Jones, J.; Noodén, L.D. Cytokinin Biochemistry in Relation to Leaf Senescence: IV. Cytokinin Metabolism in Soybean Explants. Plant Physiol. 1988, 88, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Podlešáková, K.; Zalabák, D.; Čudejková, M.; Plíhal, O.; Szüčová, L.; Doležal, K.; Spíchal, L.; Strnad, M.; Galuszka, P. Novel Cytokinin Derivatives Do Not Show Negative Effects on Root Growth and Proliferation in Submicromolar Range. PLoS ONE 2012, 7, e39293. [Google Scholar] [CrossRef]
- Arnau, J.A.; Tadeo, F.R.; Guerri, J.; Primo-Millo, E. Cytokinins in Peach: Endogenous Levels during Early Fruit Development. Plant Physiol. Biochem. 1999, 37, 741–750. [Google Scholar] [CrossRef]
- Strnad, M. The Aromatic Cytokinins. Physiol. Plant. 1997, 101, 674–688. [Google Scholar] [CrossRef]
- Tarkowská, D.; Dolezal, K.; Tarkowski, P.; Astot, C.; Holub, J.; Fuksová, K.; Schmülling, T.; Sandberg, G.; Strnad, M. Identification of New Aromatic Cytokinins in Arabidopsis Thaliana and Populus x Canadensis Leaves by LC-(+)ESI-MS and Capillary Liquid Chromatography/Frit-Fast Atom Bombardment Mass Spectrometry. Physiol. Plant. 2003, 117, 579–590. [Google Scholar] [CrossRef]
- Miller, C.O.; Skoog, F.; Okumura, F.S.; Von Saltza, M.H.; Strong, F.M. Structure and Synthesis of Kinetin. J. Am. Chem. Soc. 1955, 77, 2662–2663. [Google Scholar] [CrossRef]
- Haidoune, M.; Mornet, R.; Laloue, M. Synthesis of 6-(3-Methylpyrrol-1-Yl)-9-β-d-Ribofuranosyl Purine, a Novel Metabolite of Zeatin Riboside. Tetrahedron Lett. 1990, 31, 1419–1422. [Google Scholar] [CrossRef]
- Hluska, T.; Dobrev, P.I.; Tarkowská, D.; Frébortová, J.; Zalabák, D.; Kopečný, D.; Plíhal, O.; Kokáš, F.; Briozzo, P.; Zatloukal, M.; et al. Cytokinin Metabolism in Maize: Novel Evidence of Cytokinin Abundance, Interconversions and Formation of a New Trans.-Zeatin Metabolic Product with a Weak Anticytokinin Activity. Plant Sci. 2016, 247, 127–137. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Benfield, A.H.; Wollenberg, R.D.; Westphal, K.; Wimmer, R.; Nielsen, M.R.; Nielsen, K.F.; Carere, J.; Covarelli, L.; Beccari, G.; et al. The Cereal Pathogen Fusarium Pseudograminearum Produces a New Class of Active Cytokinins during Infection. Mol. Plant Pathol. 2018, 19, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Laloue, M.; Fox, J.E. Cytokinin Oxidase from Wheat: Partial Purification and General Properties. Plant Physiol. 1989, 90, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Mok, M.C.; Martin, R.C.; Dobrev, P.I.; Vanková, R.; Ho, P.S.; Yonekura-Sakakibara, K.; Sakakibara, H.; Mok, D.W.S. Topolins and Hydroxylated Thidiazuron Derivatives Are Substrates of Cytokinin O-Glucosyltransferase with Position Specificity Related to Receptor Recognition. Plant Physiol. 2005, 137, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.W.H.; Ge, L.; Ng, Y.F.; Tan, S.N. The Chemical Composition and Biological Properties of Coconut (Cocos Nucifera L.) Water. Molecules 2009, 14, 5144–5164. [Google Scholar] [CrossRef] [PubMed]
- Stirk, W.A.; Novák, O.; Václavíková, K.; Tarkowski, P.; Strnad, M.; van Staden, J. Spatial and Temporal Changes in Endogenous Cytokinins in Developing Pea Roots. Planta 2008, 227, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Doležal, K.; Åstot, C.; Hanuš, J.; Holub, J.; Peters, W.; Beck, E.; Strnad, M.; Sandberg, G. Identification of Aromatic Cytokinins in Suspension Cultured Photoautotrophic Cells of Chenopodium Rubrum by Capillary Liquid Chromatography/Frit-Fast Atom Bombarded Mass Spectrometry. Plant Growth Regul. 2002, 36, 181–189. [Google Scholar] [CrossRef]
- Edlund, E.; Novák, O.; Karady, M.; Ljung, K.; Jansson, S. Contrasting Patterns of Cytokinins between Years in Senescing Aspen Leaves. Plant Cell Environ. 2017, 40, 622–634. [Google Scholar] [CrossRef]
- Horgan, R.; Hewett, E.W.; Purse, J.G.; Wareing, P.F. A New Cytokinin from Populus Robusta. Tetrahedron Lett. 1973, 14, 2827–2828. [Google Scholar] [CrossRef]
- Chaves das Neves, H.J.; Pais, M.S.S. Identification of a Spathe Regreening Factor in Zantedeschia Aethiopica. Biochem. Biophys. Res. Commun. 1980, 95, 1387–1392. [Google Scholar] [CrossRef]
- Ge, L.; Yong, J.W.H.; Tan, S.N.; Yang, X.H.; Ong, E.S. Analysis of Positional Isomers of Hydroxylated Aromatic Cytokinins by Micellar Electrokinetic Chromatography. Electrophoresis 2005, 26, 1768–1777. [Google Scholar] [CrossRef]
- De Meutter, J.; Tytgat, T.; Witters, E.; Gheysen, G.; Van Onckelen, H.; Gheysen, G. Identification of Cytokinins Produced by the Plant Parasitic Nematodes Heterodera Schachtii and Meloidogyne Incognita. Mol. Plant Pathol. 2003, 4, 271–277. [Google Scholar] [CrossRef]
- Murfin, K.E.; Dillman, A.R.; Foster, J.M.; Bulgheresi, S.; Slatko, B.E.; Sternberg, P.W.; Goodrich-Blair, H. Nematode-Bacterium Symbioses—Cooperation and Conflict Revealed in the “Omics” Age. Biol. Bull. 2012, 223, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Juříková, S. Cytokininy v Řasách. Bachelor’s Thesis, Universita Palackého v Olomouci, Olomouc, Czech Republic, 2016. [Google Scholar]
- Kieber, J.J.; Schaller, G.E. Cytokinin Signaling in Plant Development. Dev. Camb. Engl. 2018, 145. [Google Scholar] [CrossRef]
- Rohmer, M. The Discovery of a Mevalonate-Independent Pathway for Isoprenoid Biosynthesis in Bacteria, Algae and Higher Plants. Nat. Prod. Rep. 1999, 16, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Kuzuyama, T.; Seto, H. Diversity of the Biosynthesis of the Isoprene Units. Nat. Prod. Rep. 2003, 20, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Kamada-Nobusada, T.; Sakakibara, H. Molecular Basis for Cytokinin Biosynthesis. Phytochemistry 2009, 70, 444–449. [Google Scholar] [CrossRef]
- Laule, O.; Fürholz, A.; Chang, H.-S.; Zhu, T.; Wang, X.; Heifetz, P.B.; Gruissem, W.; Lange, M. Crosstalk between Cytosolic and Plastidial Pathways of Isoprenoid Biosynthesis in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6866–6871. [Google Scholar] [CrossRef]
- Sakakibara, H.; Kasahara, H.; Ueda, N.; Kojima, M.; Takei, K.; Hishiyama, S.; Asami, T.; Okada, K.; Kamiya, Y.; Yamaya, T.; et al. Agrobacterium Tumefaciens Increases Cytokinin Production in Plastids by Modifying the Biosynthetic Pathway in the Host Plant. Proc. Natl. Acad. Sci. USA 2005, 102, 9972–9977. [Google Scholar] [CrossRef]
- Abe, I.; Tanaka, H.; Abe, T.; Noguchi, H. Enzymatic Formation of Unnatural Cytokinin Analogs by Adenylate Isopentenyltransferase from Mulberry. Biochem. Biophys. Res. Commun. 2007, 355, 795–800. [Google Scholar] [CrossRef]
- Chu, H.-M.; Ko, T.-P.; Wang, A.H.-J. Crystal Structure and Substrate Specificity of Plant Adenylate Isopentenyltransferase from Humulus Lupulus: Distinctive Binding Affinity for Purine and Pyrimidine Nucleotides. Nucleic Acids Res. 2010, 38, 1738–1748. [Google Scholar] [CrossRef][Green Version]
- Chu, H.-M.; Chen, F.-Y.; Ko, T.-P.; Wang, A.H.-J. Binding and Catalysis of Humulus Lupulus Adenylate Isopentenyltransferase for the Synthesis of Isopentenylated Diadenosine Polyphosphates. FEBS Lett. 2010, 584, 4083–4088. [Google Scholar] [CrossRef] [PubMed]
- Skoog, F.; Hamzi, H.Q.; Szweykowska, A.M.; Leonard, N.J.; Carraway, K.L.; Fujii, T.; Helgeson, J.P.; Loeppky, R.N. Cytokinins: Structure/Activity Relationships. Phytochemistry 1967, 6, 1169–1192. [Google Scholar] [CrossRef]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of Cytokinin Biosynthetic Isopentenyltransferase Genes in Arabidopsis: Tissue Specificity and Regulation by Auxin, Cytokinin, and Nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Sakakibara, H.; Kojima, M.; Yamamoto, Y.; Nagasaki, H.; Inukai, Y.; Sato, Y.; Matsuoka, M. Ectopic Expression of KNOTTED1-like Homeobox Protein Induces Expression of Cytokinin Biosynthesis Genes in Rice. Plant Physiol. 2006, 142, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Wu, S.; Kong, F.; Zhou, C.; Yang, Q.; Sun, Y.; Wang, B. Identification and Characterization of an Isopentenyltransferase (IPT) Gene in Soybean (Glycine Max L.). Plant Sci. 2006, 170, 542–550. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Identification, Expression, and Comparative Genomic Analysis of the IPT and CKX Gene Families in Chinese Cabbage (Brassica Rapa ssp. Pekinensis). BMC Genom. 2013, 14, 594. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, W.; Li, X.; Jiang, H.; Wu, P.; Xia, K.; Yang, Y.; Wu, G. Knockdown of LjIPT3 Influences Nodule Development in Lotus Japonicus. Plant Cell Physiol. 2014, 55, 183–193. [Google Scholar] [CrossRef]
- Song, J.; Jiang, L.; Jameson, P.E. Expression Patterns of Brassica Napus Genes Implicate IPT, CKX, Sucrose Transporter, Cell Wall Invertase, and Amino Acid Permease Gene Family Members in Leaf, Flower, Silique, and Seed Development. J. Exp. Bot. 2015, 66, 5067–5082. [Google Scholar] [CrossRef]
- Li, M.; Wei, Q.; Xiao, Y.; Peng, F. The Effect of Auxin and Strigolactone on ATP/ADP Isopentenyltransferase Expression and the Regulation of Apical Dominance in Peach. Plant Cell Rep. 2018, 37, 1693–1705. [Google Scholar] [CrossRef]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin Controls Local Cytokinin Biosynthesis in the Nodal Stem in Apical Dominance. Plant J. Cell Mol. Biol. 2006, 45, 1028–1036. [Google Scholar] [CrossRef]
- Landrein, B.; Formosa-Jordan, P.; Malivert, A.; Schuster, C.; Melnyk, C.W.; Yang, W.; Turnbull, C.G.N.; Meyerowitz, E.M.; Locke, J.C.W.; Jönsson, H. Nitrate Modulates Stem Cell Dynamics in Arabidopsis Shoot Meristems through Cytokinins. Proc. Natl. Acad. Sci. USA 2018, 115, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, K.; Nie, H.; Zeng, Q.; Wu, B.; Qian, J.; Fang, Z. Rice Nitrate Transporter OsNPF7.2 Positively Regulates Tiller Number and Grain Yield. Rice 2018, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Galichet, A.; Hoyerová, K.; Kamínek, M.; Gruissem, W. Farnesylation Directs AtIPT3 Subcellular Localization and Modulates Cytokinin Biosynthesis in Arabidopsis. Plant Physiol. 2008, 146, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Miura, G.A.; Miller, C.O. 6-(γ,γ-Dimethylallylamino)Purine As A Precursor of Zeatin. Plant Physiol. 1969, 44, 372–376. [Google Scholar] [CrossRef]
- Miura, G.A.; Hall, R.H. Trans-Ribosylzeatin: Its Biosynthesis in Zea Mays Endosperm and the Mycorrhizal Fungus, Rhizopogon Roseolus. Plant Physiol. 1973, 51, 563–569. [Google Scholar] [CrossRef]
- Chen, C.M.; Leisner, S.M. Modification of Cytokinins by Cauliflower Microsomal Enzymes. Plant Physiol. 1984, 75, 442–446. [Google Scholar] [CrossRef]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 Encode Cytokinin Hydroxylases That Catalyze the Biosynthesis of Trans.-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]
- Kiba, T.; Takei, K.; Kojima, M.; Sakakibara, H. Side-Chain Modification of Cytokinins Controls Shoot Growth in Arabidopsis. Dev. Cell 2013, 27, 452–461. [Google Scholar] [CrossRef]
- Chang, L.; Ramireddy, E.; Schmülling, T. Cytokinin as a Positional Cue Regulating Lateral Root Spacing in Arabidopsis. J. Exp. Bot. 2015, 66, 4759–4768. [Google Scholar] [CrossRef]
- Persson, B.C.; Björk, G.R. Isolation of the Gene (MiaE) Encoding the Hydroxylase Involved in the Synthesis of 2-Methylthio-Cis-Ribozeatin in TRNA of Salmonella Typhimurium and Characterization of Mutants. J. Bacteriol. 1993, 175, 7776–7785. [Google Scholar] [CrossRef]
- Kaminska, K.H.; Baraniak, U.; Boniecki, M.; Nowaczyk, K.; Czerwoniec, A.; Bujnicki, J.M. Structural Bioinformatics Analysis of Enzymes Involved in the Biosynthesis Pathway of the Hypermodified Nucleoside Ms2Io6A37 in TRNA. Proteins Struct. Funct. Bioinform. 2008, 70, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Bassil, N.V.; Mok, D.W.S.; Mok, M.C. Partial Purification of a Cis-Trans.-Isomerase of Zeatin from Immature Seed of Phaseolus Vulgaris L. Plant Physiol. 1993, 102, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, E.; Ogura, T.; Takei, K.; Kojima, M.; Kitahata, N.; Sakakibara, H.; Asami, T.; Shimada, Y. Uniconazole, a Cytochrome P450 Inhibitor, Inhibits Trans.-Zeatin Biosynthesis in Arabidopsis. Phytochemistry 2013, 87, 30–38. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, L.; Fu, Q.; Xu, Z.-F. Identification and Expression Analysis of Cytokinin Metabolic Genes IPTs, CYP735A and CKXs in the Biofuel Plant Jatropha Curcas. PeerJ 2018, 6, e4812. [Google Scholar] [CrossRef] [PubMed]
- Eisermann, I.; Motyka, V.; Kümmel, S.; Dobrev, P.I.; Hübner, K.; Deising, H.B.; Wirsel, S.G.R. CgIPT1 Is Required for Synthesis of Cis-Zeatin Cytokinins and Contributes to Stress Tolerance and Virulence in Colletotrichum Graminicola. Fungal Genet. Biol. 2020, 143, 103436. [Google Scholar] [CrossRef] [PubMed]
- Hluska, T.; Šebela, M.; Lenobel, R.; Frébort, I.; Galuszka, P. Purification of Maize Nucleotide Pyrophosphatase/Phosphodiesterase Casts Doubt on the Existence of Zeatin Cis–Trans. Isomerase in Plants. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Kuroha, T.; Kato, H.; Asami, T.; Yoshida, S.; Kamada, H.; Satoh, S. A Trans.-Zeatin Riboside in Root Xylem Sap Negatively Regulates Adventitious Root Formation on Cucumber Hypocotyls. J. Exp. Bot. 2002, 53, 2193–2200. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S. Metabolism of Zeatin Riboside in a Hormone Autonomous Genetic Tumour Line of Tobacco. Plant Growth Regul. 1997, 23, 159–166. [Google Scholar] [CrossRef]
- Trdá, L.; Barešová, M.; Šašek, V.; Nováková, M.; Zahajská, L.; Dobrev, P.I.; Motyka, V.; Burketová, L. Cytokinin Metabolism of Pathogenic Fungus Leptosphaeria Maculans Involves Isopentenyltransferase, Adenosine Kinase and Cytokinin Oxidase/Dehydrogenase. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Šmehilová, M.; Galuszka, P.; Bilyeu, K.D.; Jaworek, P.; Kowalska, M.; Šebela, M.; Sedlárová, M.; English, J.T.; Frébort, I. Subcellular Localization and Biochemical Comparison of Cytosolic and Secreted Cytokinin Dehydrogenase Enzymes from Maize. J. Exp. Bot. 2009, 60, 2701–2712. [Google Scholar] [CrossRef]
- Václavíková, K.; Hluska, T.; Floková, K.; Slováková, K.; Galuszka, P.; Tarkowski, P. The Cytokinin Metabolism in Maize. In Proceedings of the 3rd ACPD International Symposium, Prague, Czech Republic, 10–14 July 2009. [Google Scholar]
- Hluska, T. On the Hunt for Zeatin Cis-Trans. Isomerase. Bachelor’s Thesis, Palacký University, Olomouc, Czech Republic, 2010. [Google Scholar]
- Chen, C. Cytokinin Biosynthesis and Interconversion. Physiol. Plant. 1997, 101, 665–673. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, J.; Vrabka, J.; Oeser, B.; Novák, O.; Galuszka, P.; Tudzynski, P. De Novo Biosynthesis of Cytokinins in the Biotrophic Fungus Claviceps Purpurea. Environ. Microbiol. 2015, 17, 2935–2951. [Google Scholar] [CrossRef] [PubMed]
- Samanovic, M.I.; Tu, S.; Novák, O.; Iyer, L.M.; McAllister, F.E.; Aravind, L.; Gygi, S.P.; Hubbard, S.R.; Strnad, M.; Darwin, K.H. Proteasomal Control of Cytokinin Synthesis Protects Mycobacterium Tuberculosis against Nitric Oxide. Mol. Cell 2015, 57, 984–994. [Google Scholar] [CrossRef]
- Seo, H.; Kim, S.; Sagong, H.-Y.; Son, H.F.; Jin, K.S.; Kim, I.-K.; Kim, K.-J. Structural Basis for Cytokinin Production by LOG from Corynebacterium Glutamicum. Sci. Rep. 2016, 6, 31390. [Google Scholar] [CrossRef]
- Seo, H.; Kim, K.-J. Structural Basis for a Novel Type of Cytokinin-Activating Protein. Sci. Rep. 2017, 7, 45985. [Google Scholar] [CrossRef]
- Seo, H.; Kim, K.-J. Structural Insight into Molecular Mechanism of Cytokinin Activating Protein from Pseudomonas Aeruginosa PAO1. Environ. Microbiol. 2018, 20, 3214–3223. [Google Scholar] [CrossRef]
- Mayaka, J.B.; Huang, Q.; Xiao, Y.; Zhong, Q.; Ni, J.; Shen, Y. The Lonely Guy (LOG) Homologue SiRe_0427 from the Thermophilic Archaeon Sulfolobus Islandicus REY15A Is a Phosphoribohydrolase Representing a Novel Group. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Nishii, K.; Wright, F.; Chen, Y.-Y.; Möller, M. Tangled History of a Multigene Family: The Evolution of ISOPENTENYLTRANSFERASE Genes. PLoS ONE 2018, 13, e0201198. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Zhang, D.; Schäffer, D.E.; Iyer, L.M.; Aravind, L. Comparative Genomic Analyses Reveal a Vast, Novel Network of Nucleotide-Centric Systems in Biological Conflicts, Immunity and Signaling. Nucleic Acids Res. 2015, 43, 10633–10654. [Google Scholar] [CrossRef]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis Lonely Guy (LOG) Multiple Mutants Reveal a Central Role of the LOG-Dependent Pathway in Cytokinin Activation. Plant J. 2012, 69, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Creason, A.L.; Vandeputte, O.M.; Savory, E.A.; Davis, E.W.; Putnam, M.L.; Hu, E.; Swader-Hines, D.; Mol, A.; Baucher, M.; Prinsen, E.; et al. Analysis of Genome Sequences from Plant Pathogenic Rhodococcus Reveals Genetic Novelties in Virulence Loci. PLoS ONE 2014, 9, e101996. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, E.-M.; Münsterkötter, M.; Proctor, R.H.; Brown, D.W.; Sharon, A.; Idan, Y.; Oren-Young, L.; Sieber, C.M.; Novák, O.; Pěnčík, A.; et al. Comparative “Omics” of the Fusarium Fujikuroi Species Complex Highlights Differences in Genetic Potential and Metabolite Synthesis. Genome Biol. Evol. 2016, 8, 3574–3599. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of Cytokinin Biosynthesis, Compartmentalization and Translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef]
- Osugi, A.; Kojima, M.; Takebayashi, Y.; Ueda, N.; Kiba, T.; Sakakibara, H. Systemic Transport of Trans.-Zeatin and Its Precursor Have Differing Roles in Arabidopsis Shoots. Nat. Plants 2017, 3, 17112. [Google Scholar] [CrossRef]
- Kind, S.; Hinsch, J.; Vrabka, J.; Hradilová, M.; Majeská-Čudejková, M.; Tudzynski, P.; Galuszka, P. Manipulation of Cytokinin Level in the Ergot Fungus Claviceps Purpurea Emphasizes Its Contribution to Virulence. Curr. Genet. 2018, 64, 1303–1319. [Google Scholar] [CrossRef]
- Pertry, I.; Václavíková, K.; Gemrotová, M.; Spíchal, L.; Galuszka, P.; Depuydt, S.; Temmerman, W.; Stes, E.; De Keyser, A.; Riefler, M.; et al. Rhodococcus Fascians Impacts Plant Development through the Dynamic Fas-Mediated Production of a Cytokinin Mix. Mol. Plant Microbe Interact. 2010, 23, 1164–1174. [Google Scholar] [CrossRef]
- Schoor, S.; Farrow, S.; Blaschke, H.; Lee, S.; Perry, G.; von Schwartzenberg, K.; Emery, N.; Moffatt, B. Adenosine Kinase Contributes to Cytokinin Interconversion in Arabidopsis. Plant Physiol. 2011, 157, 659–672. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Lin, X.; Hong, X.; Zhu, Y.; Li, W.; He, W.; An, F.; Guo, H. Adenine Phosphoribosyl Transferase 1 Is a Key Enzyme Catalyzing Cytokinin Conversion from Nucleobases to Nucleotides in Arabidopsis. Mol. Plant 2013, 6, 1661–1672. [Google Scholar] [CrossRef]
- Kwade, Z.; Swiatek, A.; Azmi, A.; Goossens, A.; Inzé, D.; Van Onckelen, H.; Roef, L. Identification of Four Adenosine Kinase Isoforms in Tobacco By-2 Cells and Their Putative Role in the Cell Cycle-Regulated Cytokinin Metabolism. J. Biol. Chem. 2005, 280, 17512–17519. [Google Scholar] [CrossRef]
- Bromley, J.R.; Warnes, B.J.; Newell, C.A.; Thomson, J.C.P.; James, C.M.; Turnbull, C.G.N.; Hanke, D.E. A Purine Nucleoside Phosphorylase in Solanum Tuberosum L. (Potato) with Specificity for Cytokinins Contributes to the Duration of Tuber Endodormancy. Biochem. J. 2014, 458, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Kristopeit, S.M. Metabolism of Cytokinin: Deribosylation of Cytokinin Ribonucleoside by Adenosine Nucleosidase from Wheat Germ Cells. Plant Physiol. 1981, 68, 1020–1023. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Kristopeit, S.M. Metabolism of Cytokinin: Dephosphorylation of cytokinin ribonucleotide by 5’-nucleotidases from wheat germ cytosol. Plant Physiol. 1981, 67, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Kakimoto, T. Identification of Plant Cytokinin Biosynthetic Enzymes as Dimethylallyl Diphosphate: ATP/ADP Isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Sakano, Y.; Okada, Y.; Matsunaga, A.; Suwama, T.; Kaneko, T.; Ito, K.; Noguchi, H.; Abe, I. Molecular Cloning, Expression, and Characterization of Adenylate Isopentenyltransferase from Hop (Humulus Lupulus L.). Phytochemistry 2004, 65, 2439–2446. [Google Scholar] [CrossRef]
- Parker, C.W.; Letham, D.S.; Gollnow, B.I.; Summons, R.E.; Duke, C.C.; Macleod, J.K. Regulators of Cell Division in Plant Tissues: XXV. Metabolism of Zeatin by Lupin Seedlings. Planta 1978, 142, 239–251. [Google Scholar] [CrossRef]
- Palni, L.M.; Palmer, M.V.; Letham, D.S. The Stability and Biological Activity of Cytokinin Metabolites in Soybean Callus Tissue. Planta 1984, 160, 242–249. [Google Scholar] [CrossRef]
- Badenoch-Jones, J.; Rolfe, B.G.; Letham, D.S. Phytohormones, Rhizobium Mutants, and Nodulation in Legumes V. Cytokinin Metabolism in Effective and Ineffective Pea Root Nodules. Plant Physiol. 1984, 74, 239–246. [Google Scholar] [CrossRef]
- Entsch, B.; Letham, D.S.; Parker, C.W.; Summons, R.E.; Gollnow, B.I. Metabolites of Cytokinins. In Proceedings of the Plant Growth Substances 1979; Skoog, F., Ed.; Springer: Berlin/Heidelberg, Germany, 1980; pp. 109–118. [Google Scholar]
- Letham, D.S.; Summons, R.E.; Parker, C.W.; MacLeod, J.K. Regulators of Cell Division in Plant Tissues: XXVII. Identification of an Amino-Acid Conjugate of 6-Benzylaminopurine Formed in Phaseolus Vulgaris Seedlings. Planta 1979, 146, 71–74. [Google Scholar] [CrossRef]
- Elliott, D.C.; Thompson, M.J. The Identity of the Major Metabolite of Benzylaminopurine in Soybean Cultures, and the Inhibition of Its Formation by Aminophylline. Plant Sci. Lett. 1982, 28, 29–38. [Google Scholar] [CrossRef]
- Entsch, B.; Parker, C.W.; Letham, D.S. An Enzyme from Lupin Seeds Forming Alanine Derivatives of Cytokinins. Phytochemistry 1983, 22, 375–381. [Google Scholar] [CrossRef]
- Nomura, T.; Tanaka, Y.; Abe, H.; Uchiyama, M. Cytokinin Activity of Discadenine: A Spore Germination Inhibitor of Dictyostelium Discoideum. Phytochemistry 1977, 16, 1819–1820. [Google Scholar] [CrossRef]
- Mik, V.; Mičková, Z.; Doležal, K.; Frébort, I.; Pospíšil, T. Activity of (+)-Discadenine as a Plant Cytokinin. J. Nat. Prod. 2017. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.M.; Kisiala, A.B.; Li, S.; Stock, N.L.; Brunetti, C.R.; Huber, R.J.; Emery, R.J.N. Cytokinin Detection during the Dictyostelium Discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination. Biomolecules 2019, 9, 702. [Google Scholar] [CrossRef]
- Adl, S.M.; Simpson, A.G.B.; Lane, C.E.; Lukeš, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The Revised Classification of Eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–514. [Google Scholar] [CrossRef]
- Turner, J.E.; Mok, D.W.S.; Mok, M.C.; Shaw, G. Isolation and Partial Purification of an Enzyme Catalyzing the Formation of O-Xylosylzeatin in Phaseolus Vulgaris Embryos. Proc. Natl. Acad. Sci. USA 1987, 84, 3714–3717. [Google Scholar] [CrossRef]
- Taylor, J.S.; Koshioka, M.; Pharis, R.P.; Sweet, G.B. Changes in Cytokinins and Gibberellin-Like Substances in Pinus Radiata Buds during Lateral Shoot Initiation and the Characterization of Ribosyl Zeatin and a Novel Ribosyl Zeatin Glycoside. Plant Physiol. 1984, 74, 626–631. [Google Scholar] [CrossRef]
- Zhang, H.; Horgan, K.J.; Stewart Reynolds, P.H.; Norris, G.E.; Jameson, P.E. Novel Cytokinins: The Predominant Forms in Mature Buds of Pinus Radiata. Physiol. Plant. 2001, 112, 127–134. [Google Scholar] [CrossRef]
- Kobayashi, H.; Morisaki, N.; Tago, Y.; Hashimoto, Y.; Iwasaki, S.; Kawachi, E.; Nagata, R.; Shudo, K. Identification of a Major Cytokinin in Coconut Milk. Experientia 1995, 51, 1081–1084. [Google Scholar] [CrossRef]
- Letham, D.S.; Zhang, R. Cytokinin Translocation and Metabolism in Lupin Species II. New Nucleotide Metabolites of Cytokinins. Plant Sci. 1989, 64, 161–165. [Google Scholar] [CrossRef]
- Brzobohatý, B.; Moore, I.; Kristoffersen, P.; Bako, L.; Campos, N.; Schell, J.; Palme, K. Release of Active Cytokinin by a β-Glucosidase Localized to the Maize Root Meristem. Science 1993, 262, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, Biosynthesis, and Translocation. Ann. Rev. Plant. Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Letham, D.S.; Palni, L.M.S. The Biosynthesis and Metabolism of Cytokinins. Ann. Rev. Plant Physiol. 1983, 34, 163–197. [Google Scholar] [CrossRef]
- Letham, D.S.; Palni, L.M.S.; Tao, G.-Q.; Gollnow, B.I.; Bates, C.M. Regulators of Cell Division in Plant Tissues XXIX. The Activities of Cytokinin Glucosides and Alanine Conjugates in Cytokinin Bioassays. J. Plant Growth Regul. 1983, 2, 103–115. [Google Scholar] [CrossRef]
- Fusseder, A.; Ziegler, P. Metabolism and Compartmentation of Dihydrozeatin Exogenously Supplied to Photoautotrophic Suspension Cultures of Chenopodium Rubrum. Planta 1988, 173, 104–109. [Google Scholar] [CrossRef]
- Jiskrová, E.; Novák, O.; Pospíšilová, H.; Holubová, K.; Karády, M.; Galuszka, P.; Robert, S.; Frébort, I. Extra- and Intracellular Distribution of Cytokinins in the Leaves of Monocots and Dicots. New Biotechnol. 2016, 33, 735–742. [Google Scholar] [CrossRef]
- Benková, E.; Witters, E.; Van Dongen, W.; Kolář, J.; Motyka, V.; Brzobohatý, B.; Van Onckelen, H.A.; Macháčková, I. Cytokinins in Tobacco and Wheat Chloroplasts. Occurrence and Changes Due to Light/Dark Treatment. Plant Physiol. 1999, 121, 245–252. [Google Scholar] [CrossRef]
- Polanská, L.; Vičánková, A.; Nováková, M.; Malbeck, J.; Dobrev, P.I.; Brzobohatý, B.; Vaňková, R.; Macháčková, I. Altered Cytokinin Metabolism Affects Cytokinin, Auxin, and Abscisic Acid Contents in Leaves and Chloroplasts, and Chloroplast Ultrastructure in Transgenic Tobacco. J. Exp. Bot. 2007, 58, 637–649. [Google Scholar] [CrossRef]
- Kiran, N.S.; Benková, E.; Reková, A.; Dubová, J.; Malbeck, J.; Palme, K.; Brzobohatý, B. Retargeting a Maize β-Glucosidase to the Vacuole—Evidence from Intact Plants That Zeatin-O-Glucoside Is Stored in the Vacuole. Phytochemistry 2012, 79, 67–77. [Google Scholar] [CrossRef]
- Fox, J.E.; Cornette, J.; Deleuze, G.; Dyson, W.; Giersak, C.; Niu, P.; Zapata, J.; McChesney, J. The Formation, Isolation, and Biological Activity of a Cytokinin 7-Glucoside. Plant Physiol. 1973, 52, 627–632. [Google Scholar] [CrossRef]
- Parker, C.W.; Letham, D.S. Regulators of Cell Division in Plant Tissues. XVI Metabolism of Zeatin by Radish Cotyledons and Hypocotyls. Planta 1973, 114, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Hallmark, H.T.; Černý, M.; Brzobohatý, B.; Rashotte, A.M. Trans-Zeatin-N-Glucosides Have Biological Activity in Arabidopsis Thaliana. PLoS ONE 2020, 15, e0232762. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Onckelen, H.V.; Schmülling, T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Pospíšilová, H.; Jiskrová, E.; Vojta, P.; Mrízová, K.; Kokáš, F.; Čudejková, M.M.; Bergougnoux, V.; Plíhal, O.; Klimešová, J.; Novák, O.; et al. Transgenic Barley Overexpressing a Cytokinin Dehydrogenase Gene Shows Greater Tolerance to Drought Stress. New Biotechnol. 2016, 33, 692–705. [Google Scholar] [CrossRef]
- Lomin, S.N.; Myakushina, Y.A.; Kolachevskaya, O.O.; Getman, I.A.; Arkhipov, D.V.; Savelieva, E.M.; Osolodkin, D.I.; Romanov, G.A. Cytokinin Perception in Potato: New Features of Canonical Players. J. Exp. Bot. 2018, 69, 3839–3853. [Google Scholar] [CrossRef]
- Simerský, R.; Chamrád, I.; Kania, J.; Strnad, M.; Šebela, M.; Lenobel, R. Chemical Proteomic Analysis of 6-Benzylaminopurine Molecular Partners in Wheat Grains. Plant Cell Rep. 2017, 36, 1561–1570. [Google Scholar] [CrossRef]
- Bajguz, A.; Piotrowska, A. Conjugates of Auxin and Cytokinin. Phytochemistry 2009, 70, 957–969. [Google Scholar] [CrossRef]
- Gawer, M.; Laloue, M.; Terrine, C.; Guern, J. Metabolism and Biological Significance of Benzyladenine-7-Glucoside. Plant Sci. Lett. 1977, 8, 267–274. [Google Scholar] [CrossRef]
- Faiss, M.; Strnad, M.; Redig, P.; Doležal, K.; Hanuš, J.; Van Onckelen, H.A.; Schmülling, T. Chemically Induced Expression of the RolC-Encoded β-Glucosidase in Transgenic Tobacco Plants and Analysis of Cytokinin Metabolism: RolC Does Not Hydrolyze Endogenous Cytokinin Glucosides in Planta. Plant J. 1996, 10, 33–46. [Google Scholar] [CrossRef]
- Zhang, R.; Letham, D.S. Cytokinin Biochemistry in Relation to Leaf Senescence. III. The Senescence-Retarding Activity and Metabolism of 9-Substituted 6-Benzylaminopurines in Soybean Leaves. J. Plant Growth Regul. 1989, 8, 181–197. [Google Scholar] [CrossRef]
- Hošek, P.; Hoyerová, K.; Kiran, N.S.; Dobrev, P.I.; Zahajská, L.; Filepová, R.; Motyka, V.; Müller, K.; Kamínek, M. Distinct Metabolism of N-Glucosides of Isopentenyladenine and Trans.-Zeatin Determines Cytokinin Metabolic Spectrum in Arabidopsis. New Phytol. 2020, 225, 2423–2438. [Google Scholar] [CrossRef] [PubMed]
- Entsch, B.; Parker, C.W.; Letham, D.S.; Summons, R.E. Preparation and Characterization, Using High-Performance Liquid Chromatography, of an Enzyme Forming Glucosides of Cytokinins. Biochim. Biophys. Acta BBA Enzymol. 1979, 570, 124–139. [Google Scholar] [CrossRef]
- Dixon, S.C.; Martin, R.C.; Mok, M.C.; Shaw, G.; Mok, D.W.S. Zeatin Glycosylation Enzymes in Phaseolus Isolation of O-Glucosyltransferase from P. Lunatus and Comparison to O-Xylosyltransferase from P. Vulgaris. Plant Physiol. 1989, 90, 1316–1321. [Google Scholar] [CrossRef]
- Martin, R.C.; Mok, M.C.; Mok, D.W.S. A Gene Encoding the Cytokinin Enzyme Zeatin O-Xylosyltransferase of Phaseolus Vulgaris. Plant Physiol. 1999, 120, 553–558. [Google Scholar] [CrossRef]
- Martin, R.C.; Mok, M.C.; Mok, D.W.S. Isolation of a Cytokinin Gene, ZOG1, Encoding Zeatin O-Glucosyltransferase from Phaseolus Lunatus. Proc. Natl. Acad. Sci. USA 1999, 96, 284–289. [Google Scholar] [CrossRef]
- Hou, B.-K.; Lim, E.-K.; Higgins, G.S.; Bowles, D.J. N-Glucosylation of Cytokinins by Glycosyltransferases of Arabidopsis Thaliana. J. Biol. Chem. 2004, 279, 47822–47832. [Google Scholar] [CrossRef]
- Gandia-Herrero, F.; Lorenz, A.; Larson, T.; Graham, I.A.; Bowles, D.J.; Rylott, E.L.; Bruce, N.C. Detoxification of the Explosive 2,4,6-Trinitrotoluene in Arabidopsis: Discovery of Bifunctional O- and C-Glucosyltransferases. Plant J. 2008, 56, 963–974. [Google Scholar] [CrossRef]
- Poppenberger, B.; Fujioka, S.; Soeno, K.; George, G.L.; Vaistij, F.E.; Hiranuma, S.; Seto, H.; Takatsuto, S.; Adam, G.; Yoshida, S.; et al. The UGT73C5 of Arabidopsis Thaliana Glucosylates Brassinosteroids. Proc. Natl. Acad. Sci. USA 2005, 102, 15253–15258. [Google Scholar] [CrossRef]
- Pineda Rodó, A.; Brugière, N.; Vaňková, R.; Malbeck, J.; Olson, J.M.; Haines, S.C.; Martin, R.C.; Habben, J.E.; Mok, D.W.S.; Mok, M.C. Over-Expression of a Zeatin O-Glucosylation Gene in Maize Leads to Growth Retardation and Tasselseed Formation. J. Exp. Bot. 2008, 59, 2673–2686. [Google Scholar] [CrossRef]
- Cucinotta, M.; Manrique, S.; Cuesta, C.; Benková, E.; Novák, O.; Colombo, L. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 Regulate Cytokinin Homeostasis to Determine Ovule Number in Arabidopsis. J. Exp. Bot. 2018, 69, 5169–5176. [Google Scholar] [CrossRef]
- Shang, X.-L.; Xie, R.-R.; Tian, H.; Wang, Q.-L.; Guo, F.-Q. Putative Zeatin O-Glucosyltransferase OscZOG1 Regulates Root and Shoot Development and Formation of Agronomic Traits in Rice. J. Integr. Plant Biol. 2016, 58, 627–641. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of Leaf Senescence by Autoregulated Production of Cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- Frébort, I.; Kowalska, M.; Hluska, T.; Frébortová, J.; Galuszka, P. Evolution of Cytokinin Biosynthesis and Degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Manns, I.B. y Structure and Function of Cytokinin Oxidase/Dehydrogenase Genes of Maize, Rice, Arabidopsis and Other Species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef]
- Pačes, V.; Werstiuk, E.; Hall, R.H. Conversion of N6-(Δ2-Isopentenyl)Adenosine to Adenosine by Enzyme Activity in Tobacco Tissue. Plant Physiol. 1971, 48, 775–778. [Google Scholar] [CrossRef]
- Houba-Hérin, N.; Pethe, C.; D’Alayer, J.; Laloue, M. Cytokinin Oxidase from Zea Mays: Purification, CDNA Cloning and Expression in Moss Protoplasts. Plant J. 1999, 17, 615–626. [Google Scholar] [CrossRef]
- Morris, R.O.; Bilyeu, K.D.; Laskey, J.G.; Cheikh, N.N. Isolation of a Gene Encoding a Glycosylated Cytokinin Oxidase from Maize. Biochem. Biophys. Res. Commun. 1999, 255, 328–333. [Google Scholar] [CrossRef]
- Galuszka, P.; Frébort, I.; Šebela, M.; Sauer, P.; Jacobsen, S.; Peč, P. Cytokinin Oxidase or Dehydrogenase? Mechanism of Cytokinin Degradation in Cereals. Eur. J. Biochem. 2001, 268, 450–461. [Google Scholar] [CrossRef]
- Frébortová, J.; Novák, O.; Frébort, I.; Jorda, R. Degradation of Cytokinins by Maize Cytokinin Dehydrogenase Is Mediated by Free Radicals Generated by Enzymatic Oxidation of Natural Benzoxazinones. Plant J. 2010, 61, 467–481. [Google Scholar] [CrossRef]
- Niemann, M.C.E.; Weber, H.; Hluska, T.; Leonte, G.; Anderson, S.M.; Novák, O.; Senes, A.; Werner, T. The Cytokinin Oxidase/Dehydrogenase CKX1 Is a Membrane-Bound Protein Requiring Homooligomerization in the Endoplasmic Reticulum for Its Cellular Activity. Plant Physiol. 2018, 176, 2024–2039. [Google Scholar] [CrossRef]
- Frébortová, J.; Galuszka, P.; Werner, T.; Schmülling, T.; Frébort, I. Functional Expression and Purification of Cytokinin Dehydrogenase from Arabidopsis Thaliana (AtCKX2) in Saccharomyces Cerevisiae. Biol. Plant. 2007, 51, 673–682. [Google Scholar] [CrossRef]
- Frébortová, J.; Greplová, M.; Seidl, M.F.; Heyl, A.; Frébort, I. Biochemical Characterization of Putative Adenylate Dimethylallyltransferase and Cytokinin Dehydrogenase from Nostoc sp. PCC 7120. PLoS ONE 2015, 10, e0138468. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, M.; Galuszka, P.; Frébortová, J.; Šebela, M.; Béres, T.; Hluska, T.; Šmehilová, M.; Bilyeu, K.D.; Frébort, I. Vacuolar and Cytosolic Cytokinin Dehydrogenases of Arabidopsis Thaliana: Heterologous Expression, Purification and Properties. Phytochemistry 2010, 71, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Chatfield, J.M.; Armstrong, D.J. Regulation of Cytokinin Oxidase Activity in Callus Tissues of Phaseolus Vulgaris L. Cv Great Northern. Plant Physiol. 1986, 80, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Kamínek, M.; Armstrong, D.J. Genotypic Variation in Cytokinin Oxidase from Phaseolus Callus Cultures. Plant Physiol. 1990, 93, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Brugière, N.; Jiao, S.; Hantke, S.; Zinselmeier, C.; Roessler, J.A.; Niu, X.; Jones, R.J.; Habben, J.E. Cytokinin Oxidase Gene Expression in Maize Is Localized to the Vasculature, and Is Induced by Cytokinins, Abscisic Acid, and Abiotic Stress. Plant Physiol. 2003, 132, 1228–1240. [Google Scholar] [CrossRef]
- Motyka, V.; Vaňková, R.; Čapková, V.; Petrášek, J.; Kamínek, M.; Schmülling, T. Cytokinin-Induced Upregulation of Cytokinin Oxidase Activity in Tobacco Includes Changes in Enzyme Glycosylation and Secretion. Physiol. Plant. 2003, 117, 11–21. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.-H.; Schaller, G.E.; Kieber, J.J. Characterization of Genes Involved in Cytokinin Signaling and Metabolism from Rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef]
- Panda, B.B.; Sekhar, S.; Dash, S.K.; Behera, L.; Shaw, B.P. Biochemical and Molecular Characterisation of Exogenous Cytokinin Application on Grain Filling in Rice. BMC Plant Biol. 2018, 18, 89. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin Oxidase Regulates Rice Grain Production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield in Arabidopsis Thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, Y.-L.; Gao, L.-F.; Zhao, G.-Y.; Zhou, R.-H.; Zhang, B.-S.; Jia, J.-Z. TaCKX6-D1, the Ortholog of Rice OsCKX2, Is Associated with Grain Weight in Hexaploid Wheat. New Phytol. 2012, 195, 574–584. [Google Scholar] [CrossRef]
- Yeh, S.-Y.; Chen, H.-W.; Ng, C.-Y.; Lin, C.-Y.; Tseng, T.-H.; Li, W.-H.; Ku, M.S.B. Down-Regulation of Cytokinin Oxidase 2 Expression Increases Tiller Number and Improves Rice Yield. Rice 2015, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, G.; Gao, J.; Zhang, S.; Zhang, R.; Li, W.; Chen, M.; Liu, M.; Xia, X.; Risacher, T.; et al. Enhancement of Grain Number per Spike by RNA Interference of Cytokinin Oxidase 2 Gene in Bread Wheat. Hereditas 2018, 155, 33. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Yang, X.; Gao, A.; Li, X.; Wu, X.; Li, L. Isolation and Characterization of Two Putative Cytokinin Oxidase Genes Related to Grain Number per Spike Phenotype in Wheat. Mol. Biol. Rep. 2011, 38, 2337–2347. [Google Scholar] [CrossRef]
- Mrízová, K.; Jiskrová, E.; Vyroubalová, Š.; Novák, O.; Ohnoutková, L.; Pospíšilová, H.; Frébort, I.; Harwood, W.A.; Galuszka, P. Overexpression of Cytokinin Dehydrogenase Genes in Barley (Hordeum Vulgare Cv. Golden Promise) Fundamentally Affects Morphology and Fertility. PLoS ONE 2013, 8, e79029. [Google Scholar] [CrossRef]
- Werner, T.; Nehnevajova, E.; Köllmer, I.; Novák, O.; Strnad, M.; Krämer, U.; Schmülling, T. Root-Specific Reduction of Cytokinin Causes Enhanced Root Growth, Drought Tolerance, and Leaf Mineral Enrichment in Arabidopsis and Tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef]
- Ramireddy, E.; Hosseini, S.A.; Eggert, K.; Gillandt, S.; Gnad, H.; von Wirén, N.; Schmülling, T. Root Engineering in Barley: Increasing Cytokinin Degradation Produces a Larger Root System, Mineral Enrichment in the Shoot and Improved Drought Tolerance. Plant Physiol. 2018, 177, 1078–1095. [Google Scholar] [CrossRef]
- Gao, S.; Fang, J.; Xu, F.; Wang, W.; Sun, X.; Chu, J.; Cai, B.; Feng, Y.; Chu, C. CYTOKININ OXIDASE/DEHYDROGENASE4 Integrates Cytokinin and Auxin Signaling to Control Rice Crown Root Formation. Plant Physiol. 2014, 165, 1035–1046. [Google Scholar] [CrossRef]
- Li, W.; Zhai, L.; Strauss, S.H.; Yer, H.; Merewitz, E.; Chen, J.; Wang, X.; Zhuang, W.; Fang, C.; Chen, Y.; et al. Transgenic Reduction of Cytokinin Levels in Roots Inhibits Root-Sprouting in Populus. Plant Physiol. 2019, 180, 1788–1792. [Google Scholar] [CrossRef]
- Gao, S.; Xiao, Y.; Xu, F.; Gao, X.; Cao, S.; Zhang, F.; Wang, G.; Sanders, D.; Chu, C. Cytokinin-Dependent Regulatory Module Underlies the Maintenance of Zinc Nutrition in Rice. New Phytol. 2019, 224, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Vercruyssen, L.; Gonzalez, N.; Werner, T.; Schmülling, T.; Inzé, D. Combining Enhanced Root and Shoot Growth Reveals Cross Talk between Pathways That Control Plant Organ Size in Arabidopsis. Plant Physiol. 2011, 155, 1339–1352. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, J.; Song, J.; Jameson, P.E. Cytokinin Dehydrogenase: A Genetic Target for Yield Improvement in Wheat. Plant Biotechnol. J. 2020, 18, 614–630. [Google Scholar] [CrossRef] [PubMed]
- Heyl, A.; Brault, M.; Frugier, F.; Kuderová, A.; Lindner, A.-C.; Motyka, V.; Rashotte, A.M.; von Schwartzenberg, K.; Vaňková, R.; Schaller, G.E. Nomenclature for Members of the Two-Component Signaling Pathway of Plants. Plant Physiol. 2013, 161, 1063–1065. [Google Scholar] [CrossRef] [PubMed]
- Keshishian, E.A.; Rashotte, A.M. Plant Cytokinin Signalling. Essays Biochem. 2015, 58, 13–27. [Google Scholar] [CrossRef]
- Hothorn, M.; Dabi, T.; Chory, J. Structural Basis for Cytokinin Recognition by Arabidopsis Thaliana Histidine Kinase 4. Nat. Chem. Biol. 2011, 7, 766–768. [Google Scholar] [CrossRef]
- Steklov, M.Y.; Lomin, S.N.; Osolodkin, D.I.; Romanov, G.A. Structural Basis for Cytokinin Receptor Signaling: An Evolutionary Approach. Plant Cell Rep. 2013, 32, 781–793. [Google Scholar] [CrossRef]
- Upadhyay, A.A.; Fleetwood, A.D.; Adebali, O.; Finn, R.D.; Zhulin, I.B. Cache Domains That Are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 2016, 12. [Google Scholar] [CrossRef]
- Wang, F.-F.; Cheng, S.-T.; Wu, Y.; Ren, B.-Z.; Qian, W. A Bacterial Receptor PcrK Senses the Plant Hormone Cytokinin to Promote Adaptation to Oxidative Stress. Cell Rep. 2017, 21, 2940–2951. [Google Scholar] [CrossRef]
- Mähönen, A.P.; Higuchi, M.; Törmäkangas, K.; Miyawaki, K.; Pischke, M.S.; Sussman, M.R.; Helariutta, Y.; Kakimoto, T. Cytokinins Regulate a Bidirectional Phosphorelay Network in Arabidopsis. Curr. Biol. CB 2006, 16, 1116–1122. [Google Scholar] [CrossRef]
- Caesar, K.; Thamm, A.M.K.; Witthöft, J.; Elgass, K.; Huppenberger, P.; Grefen, C.; Horak, J.; Harter, K. Evidence for the Localization of the Arabidopsis Cytokinin Receptors AHK3 and AHK4 in the Endoplasmic Reticulum. J. Exp. Bot. 2011, 62, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Wulfetange, K.; Lomin, S.N.; Romanov, G.A.; Stolz, A.; Heyl, A.; Schmülling, T. The Cytokinin Receptors of Arabidopsis Are Located Mainly to the Endoplasmic Reticulum. Plant Physiol. 2011, 156, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Lomin, S.N.; Schmülling, T. Cytokinin Signaling: From the ER or from the PM? That Is the Question! New Phytol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Savelieva, E.M.; Arkhipov, D.V.; Romanov, G.A. Evidences for Preferential Localization of Cytokinin Receptors of Potato in the Endoplasmic Reticulum. Biochem. Mosc. Suppl. Ser. Membr. Cell Biol. 2020, 14, 146–153. [Google Scholar] [CrossRef]
- Grignon, C.; Sentenac, H. PH and Ionic Conditions in the Apoplast. Ann. Rev. Plant Physiol. Plant. Mol. Biol. 1991, 42, 103–128. [Google Scholar] [CrossRef]
- Geilfus, C.-M. The PH of the Apoplast: Dynamic Factor with Functional Impact under Stress. Mol. Plant. 2017, 10, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Davies, W.J. Modification of Leaf Apoplastic PH in Relation to Stomatal Sensitivity to Root-Sourced Abscisic Acid Signals. Plant Physiol. 2007, 143, 68–77. [Google Scholar] [CrossRef]
- Antoniadi, I.; Novák, O.; Gelová, Z.; Johnson, A.; Plíhal, O.; Simerský, R.; Mik, V.; Vain, T.; Mateo-Bonmatí, E.; Karady, M.; et al. Cell-Surface Receptors Enable Perception of Extracellular Cytokinins. Nat. Commun. 2020, 11, 4284. [Google Scholar] [CrossRef]
- Jaworek, P.; Tarkowski, P.; Hluska, T.; Kouřil, Š.; Vrobel, O.; Nisler, J.; Kopečný, D. Characterization of Five CHASE-Containing Histidine Kinase Receptors from Populus × Canadensis Cv. Robusta Sensing Isoprenoid and Aromatic Cytokinins. Planta 2019, 251, 1. [Google Scholar] [CrossRef]
- Zürcher, E.; Liu, J.; di Donato, M.; Geisler, M.; Müller, B. Plant Development Regulated by Cytokinin Sinks. Science 2016, 353, 1027–1030. [Google Scholar] [CrossRef]
- Zalabák, D.; Johnová, P.; Plíhal, O.; Šenková, K.; Šamajová, O.; Jiskrová, E.; Novák, O.; Jackson, D.; Mohanty, A.; Galuszka, P. Maize Cytokinin Dehydrogenase Isozymes Are Localized Predominantly to the Vacuoles. Plant Physiol. Biochem. 2016, 104, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Gelová, Z.; Ten Hoopen, P.; Novák, O.; Motyka, V.; Pernisová, M.; Dabravolski, S.; Didi, V.; Tillack, I.; Oklešťková, J.; Strnad, M.; et al. Antibody-Mediated Modulation of Cytokinins in Tobacco: Organ-Specific Changes in Cytokinin Homeostasis. J. Exp. Bot. 2018, 69, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Kubiasová, K.; Montesinos, J.C.; Šamajová, O.; Nisler, J.; Mik, V.; Semerádová, H.; Plíhalová, L.; Novák, O.; Marhavý, P.; Cavallari, N.; et al. Cytokinin Fluoroprobe Reveals Multiple Sites of Cytokinin Perception at Plasma Membrane and Endoplasmic Reticulum. Nat. Commun. 2020, 11, 4285. [Google Scholar] [CrossRef]
- Marhavý, P.; Bielach, A.; Abas, L.; Abuzeineh, A.; Duclercq, J.; Tanaka, H.; Pařezová, M.; Petrášek, J.; Friml, J.; Kleine-Vehn, J.; et al. Cytokinin Modulates Endocytic Trafficking of PIN1 Auxin Efflux Carrier to Control Plant Organogenesis. Dev. Cell 2011, 21, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Pizarro, L.; Leibman-Markus, M.; Marash, I.; Bar, M. Cytokinin Response Induces Immunity and Fungal Pathogen Resistance, and Modulates Trafficking of the PRR LeEIX2 in Tomato. Mol. Plant Pathol. 2020. [Google Scholar] [CrossRef]
- Dortay, H.; Mehnert, N.; Bürkle, L.; Schmülling, T.; Heyl, A. Analysis of Protein Interactions within the Cytokinin-Signaling Pathway of Arabidopsis Thaliana. FEBS J. 2006, 273, 4631–4644. [Google Scholar] [CrossRef]
- Lomin, S.N.; Myakushina, Y.A.; Arkhipov, D.V.; Leonova, O.G.; Popenko, V.I.; Schmülling, T.; Romanov, G.A. Studies of Cytokinin Receptor-Phosphotransmitter Interaction Provide Evidences for the Initiation of Cytokinin Signalling in the Endoplasmic Reticulum. Funct. Plant. Biol. FPB 2018, 45, 192–202. [Google Scholar] [CrossRef]
- Kudla, J.; Bock, R. Lighting the Way to Protein-Protein Interactions: Recommendations on Best Practices for Bimolecular Fluorescence Complementation Analyses. Plant Cell 2016, 28, 1002–1008. [Google Scholar] [CrossRef]
- Capra, E.J.; Laub, M.T. Evolution of Two-Component Signal Transduction Systems. Ann. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef]
- Samanovic, M.I.; Hsu, H.-C.; Jones, M.B.; Jones, V.; McNeil, M.R.; Becker, S.H.; Jordan, A.T.; Strnad, M.; Xu, C.; Jackson, M.; et al. Cytokinin Signaling in Mycobacterium Tuberculosis. mBio 2018, 9. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis Histidine Phosphotransfer Proteins Are Redundant Positive Regulators of Cytokinin Signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef] [PubMed]
- Punwani, J.A.; Kieber, J.J. Localization of the Arabidopsis Histidine Phosphotransfer Proteins Is Independent of Cytokinin. Plant Signal. Behav. 2010, 5, 896–898. [Google Scholar] [CrossRef] [PubMed]
- Mähönen, A.P.; Bishopp, A.; Higuchi, M.; Nieminen, K.M.; Kinoshita, K.; Törmäkangas, K.; Ikeda, Y.; Oka, A.; Kakimoto, T.; Helariutta, Y. Cytokinin Signaling and Its Inhibitor AHP6 Regulate Cell Fate during Vascular Development. Science 2006, 311, 94–98. [Google Scholar] [CrossRef]
- Müller, B. Generic Signal-Specific Responses: Cytokinin and Context-Dependent Cellular Responses. J. Exp. Bot. 2011, 62, 3273–3288. [Google Scholar] [CrossRef] [PubMed]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.-H.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B Response Regulators of Arabidopsis Play Key Roles in Cytokinin Signaling and Plant Development. Plant Cell 2008, 20, 2102–2116. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Carson, S.D.B.; To, J.P.C.; Kieber, J.J. Expression Profiling of Cytokinin Action in Arabidopsis. Plant Physiol. 2003, 132, 1998–2011. [Google Scholar] [CrossRef]
- Kiba, T.; Aoki, K.; Sakakibara, H.; Mizuno, T. Arabidopsis Response Regulator, ARR22, Ectopic Expression of Which Results in Phenotypes Similar to the Wol Cytokinin-Receptor Mutant. Plant Cell Physiol. 2004, 45, 1063–1077. [Google Scholar] [CrossRef]
- Gattolin, S.; Alandete-Saez, M.; Elliott, K.; Gonzalez-Carranza, Z.; Naomab, E.; Powell, C.; Roberts, J.A. Spatial and Temporal Expression of the Response Regulators ARR22 and ARR24 in Arabidopsis Thaliana. J. Exp. Bot. 2006, 57, 4225–4233. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A Subset of Arabidopsis AP2 Transcription Factors Mediates Cytokinin Responses in Concert with a Two-Component Pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef]
- Rashotte, A.M.; Goertzen, L.R. The CRF Domain Defines Cytokinin Response Factor Proteins in Plants. BMC Plant Biol. 2010, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Cutcliffe, J.W.; Hellmann, E.; Heyl, A.; Rashotte, A.M. CRFs Form Protein–Protein Interactions with Each Other and with Members of the Cytokinin Signalling Pathway in Arabidopsis via the CRF Domain. J. Exp. Bot. 2011, 62, 4995–5002. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; Shi, X.; Robinson, B.R.; Gupta, S.; Compton, M.A.; Gerken, D.M.; Goertzen, L.R.; Rashotte, A.M. Vascular Expression and C-Terminal Sequence Divergence of Cytokinin Response Factors in Flowering Plants. Plant Cell Physiol. 2012, 53, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Gupta, S.; Rashotte, A.M. Solanum Lycopersicum Cytokinin Response Factor (SlCRF) Genes: Characterization of CRF Domain-Containing ERF Genes in Tomato. J. Exp. Bot. 2012, 63, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Zwack, P.J.; Robinson, B.R.; Risley, M.G.; Rashotte, A.M. Cytokinin Response Factor 6 Negatively Regulates Leaf Senescence and Is Induced in Response to Cytokinin and Numerous Abiotic Stresses. Plant Cell Physiol. 2013, 54, 971–981. [Google Scholar] [CrossRef]
- Zwack, P.J.; De Clercq, I.; Howton, T.C.; Hallmark, H.T.; Hurny, A.; Keshishian, E.A.; Parish, A.M.; Benkova, E.; Mukhtar, M.S.; Van Breusegem, F.; et al. Cytokinin Response Factor 6 Represses Cytokinin-Associated Genes during Oxidative Stress. Plant Physiol. 2016, 172, 1249–1258. [Google Scholar] [CrossRef]
- Zwack, P.J.; Compton, M.A.; Adams, C.I.; Rashotte, A.M. Cytokinin Response Factor 4 (CRF4) Is Induced by Cold and Involved in Freezing Tolerance. Plant Cell Rep. 2016, 35, 573–584. [Google Scholar] [CrossRef]
- Jeon, J.; Cho, C.; Lee, M.R.; Van Binh, N.; Kim, J. CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 Regulate Lateral Root Development in Response to Cold Stress in Arabidopsis. Plant Cell 2016, 28, 1828–1843. [Google Scholar] [CrossRef]
- Qin, L.; Wang, L.; Guo, Y.; Li, Y.; Ümüt, H.; Wang, Y. An ERF Transcription Factor from Tamarix Hispida, ThCRF1, Can Adjust Osmotic Potential and Reactive Oxygen Species Scavenging Capability to Improve Salt Tolerance. Plant Sci. Int. J. Exp. Plant. Biol. 2017, 265, 154–166. [Google Scholar] [CrossRef]
- Melton, A.E.; Zwack, P.J.; Rashotte, A.M.; Goertzen, L.R. Identification and Functional Characterization of the Marshallia (Asteraceae) Clade III Cytokinin Response Factor (CRF). Plant Signal. Behav. 2019, 1–6. [Google Scholar] [CrossRef]
- Chevalier, F.; Perazza, D.; Laporte, F.; Le Hénanff, G.; Hornitschek, P.; Bonneville, J.-M.; Herzog, M.; Vachon, G. GeBP and GeBP-like Proteins Are Noncanonical Leucine-Zipper Transcription Factors That Regulate Cytokinin Response in Arabidopsis. Plant Physiol. 2008, 146, 1142–1154. [Google Scholar] [CrossRef][Green Version]
- Takei, K.; Sakakibara, H.; Taniguchi, M.; Sugiyama, T. Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species That Induces Gene Expression of Maize Response Regulator. Plant Cell Physiol. 2001, 42, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Weiler, E.W.; Ziegler, H. Determination of Phytohormones in Phloem Exudate from Tree Species by Radioimmunoassay. Planta 1981, 152, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Marsh, E.L.; Schroeder, S.G.; Schachtman, D.P. Metabolomic and Proteomic Changes in the Xylem Sap of Maize under Drought. Plant Cell Environ. 2008, 31, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Field, S.K.; Smith, J.P.; Morrison, E.N.; Emery, R.J.N.; Holzapfel, B.P. Soil Temperature Prior to Veraison Alters Grapevine Carbon Partitioning, Xylem Sap Hormones, and Fruit Set. Am. J. Enol. Vitic. 2019. [Google Scholar] [CrossRef]
- Foo, E.; Morris, S.E.; Parmenter, K.; Young, N.; Wang, H.; Jones, A.; Rameau, C.; Turnbull, C.G.N.; Beveridge, C.A. Feedback Regulation of Xylem Cytokinin Content Is Conserved in Pea and Arabidopsis. Plant Physiol. 2007, 143, 1418–1428. [Google Scholar] [CrossRef]
- Matsumoto-Kitano, M.; Kusumoto, T.; Tarkowski, P.; Kinoshita-Tsujimura, K.; Václavíková, K.; Miyawaki, K.; Kakimoto, T. Cytokinins Are Central Regulators of Cambial Activity. Proc. Natl. Acad. Sci. USA 2008, 105, 20027–20031. [Google Scholar] [CrossRef]
- Zhang, R.; Letham, D.S.; Willcocks, D.A. Movement to Bark and Metabolism of Xylem Cytokinins in Stems of Lupinus Angustifolius. Phytochemistry 2002, 60, 483–488. [Google Scholar] [CrossRef]
- Gillissen, B.; Bürkle, L.; André, B.; Kühn, C.; Rentsch, D.; Brandl, B.; Frommer, W.B. A New Family of High-Affinity Transporters for Adenine, Cytosine, and Purine Derivatives in Arabidopsis. Plant Cell 2000, 12, 291–300. [Google Scholar] [CrossRef]
- Bürkle, L.; Cedzich, A.; Döpke, C.; Stransky, H.; Okumoto, S.; Gillissen, B.; Kühn, C.; Frommer, W.B. Transport of Cytokinins Mediated by Purine Transporters of the PUP Family Expressed in Phloem, Hydathodes, and Pollen of Arabidopsis. Plant J. Cell Mol. Biol. 2003, 34, 13–26. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, D.; Zhang, G.; Gao, S.; Liu, L.; Xu, F.; Che, R.; Wang, Y.; Tong, H.; Chu, C. Big Grain3, Encoding a Purine Permease, Regulates Grain Size via Modulating Cytokinin Transport in Rice. J. Integr. Plant Biol. 2019, 61, 581–597. [Google Scholar] [CrossRef]
- Pastor-Anglada, M.; Pérez-Torras, S. Who Is Who in Adenosine Transport. Front. Pharmacol. 2018, 9, 627. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Makita, N.; Yamaya, T.; Sakakibara, H. Functional Characterization and Expression Analysis of a Gene, OsENT2, Encoding an Equilibrative Nucleoside Transporter in Rice Suggest a Function in Cytokinin Transport. Plant Physiol. 2005, 138, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Hirose, N.; Wang, X.; Wen, P.; Xue, L.; Sakakibara, H.; Zuo, J. Arabidopsis SOI33/AtENT8 Gene Encodes a Putative Equilibrative Nucleoside Transporter That Is Involved in Cytokinin Transport In Planta. J. Integr. Plant Biol. 2005, 47, 588–603. [Google Scholar] [CrossRef]
- Wormit, A.; Traub, M.; Flörchinger, M.; Neuhaus, H.E.; Möhlmann, T. Characterization of Three Novel Members of the Arabidopsis Thaliana Equilibrative Nucleoside Transporter (ENT) Family. Biochem. J. 2004, 383, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Kiba, T.; Sakakibara, H. Metabolism and Long-Distance Translocation of Cytokinins. J. Integr. Plant Biol. 2010, 52, 53–60. [Google Scholar] [CrossRef]
- Zhang, K.; Novák, O.; Wei, Z.; Gou, M.; Zhang, X.; Yu, Y.; Yang, H.; Cai, Y.; Strnad, M.; Liu, C.-J. Arabidopsis ABCG14 Protein Controls the Acropetal Translocation of Root-Synthesized Cytokinins. Nat. Commun. 2014, 5, 3274. [Google Scholar] [CrossRef]
- Ko, D.; Kang, J.; Kiba, T.; Park, J.; Kojima, M.; Do, J.; Kim, K.Y.; Kwon, M.; Endler, A.; Song, W.-Y.; et al. Arabidopsis ABCG14 Is Essential for the Root-to-Shoot Translocation of Cytokinin. Proc. Natl. Acad. Sci. USA 2014, 111, 7150–7155. [Google Scholar] [CrossRef]
- Zhao, J.; Yu, N.; Ju, M.; Fan, B.; Zhang, Y.; Zhu, E.; Zhang, M.; Zhang, K. ABC Transporter OsABCG18 Controls the Shootward Transport of Cytokinins and Grain Yield in Rice. J. Exp. Bot. 2019, 70, 6277–6291. [Google Scholar] [CrossRef]
- Feng, Y.; Sun, Q.; Zhang, G.; Wu, T.; Zhang, X.; Xu, X.; Han, Z.; Wang, Y. Genome-Wide Identification and Characterization of ABC Transporters in Nine Rosaceae Species Identifying MdABCG28 as a Possible Cytokinin Transporter Linked to Dwarfing. Int. J. Mol. Sci. 2019, 20, 5783. [Google Scholar] [CrossRef]
- Poitout, A.; Crabos, A.; Petřík, I.; Novák, O.; Krouk, G.; Lacombe, B.; Ruffel, S. Responses to Systemic Nitrogen Signaling in Arabidopsis Roots Involve Trans-Zeatin in Shoots. Plant Cell 2018, 30, 1243–1257. [Google Scholar] [CrossRef]
- Kim, A.; Chen, J.; Khare, D.; Jin, J.-Y.; Yamaoka, Y.; Maeshima, M.; Zhao, Y.; Martinoia, E.; Hwang, J.-U.; Lee, Y. Non-Intrinsic ATP-Binding Cassette Proteins ABCI19, ABCI20 and ABCI21 Modulate Cytokinin Response at the Endoplasmic Reticulum in Arabidopsis Thaliana. Plant Cell Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Benková, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins Act Directly on Lateral Root Founder Cells to Inhibit Root Initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef]
- Tessi, T.M.; Brumm, S.; Winklbauer, E.; Schumacher, B.; Pettinari, G.; Lescano, I.; González, C.A.; Wanke, D.; Maurino, V.G.; Harter, K.; et al. Arabidopsis AZG2 Transports Cytokinins in Vivo and Regulates Lateral Root Emergence. New Phytol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tessi, T.M.; Shahriari, M.; Maurino, V.G.; Meissner, E.; Novák, O.; Pasternak, T.; Schumacher, B.S.; Flubacher, N.S.; Nautscher, M.; Williams, A.; et al. The Auxin Transporter PIN1 and the Cytokinin Transporter AZG1 Interact to Regulate the Root Stress Response. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mansfield, T.A.; Schultes, N.P.; Mourad, G.S. AtAzg1 and AtAzg2 Comprise a Novel Family of Purine Transporters in Arabidopsis. FEBS Lett. 2009, 583, 481–486. [Google Scholar] [CrossRef] [PubMed]

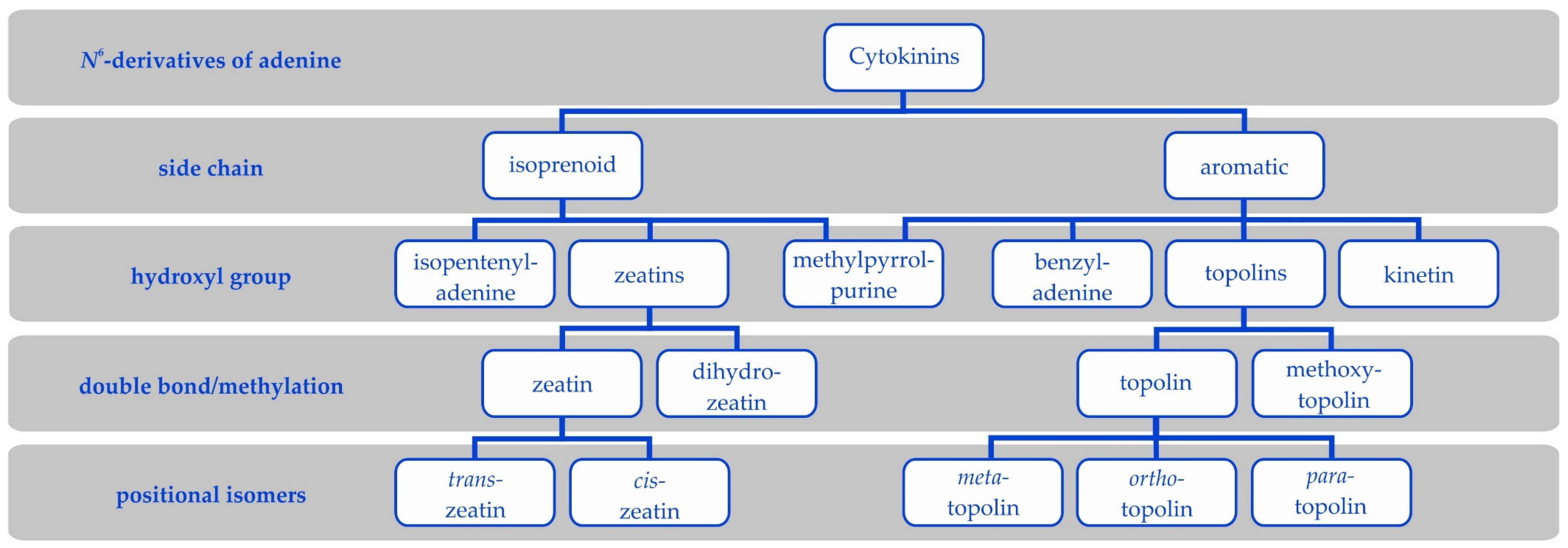
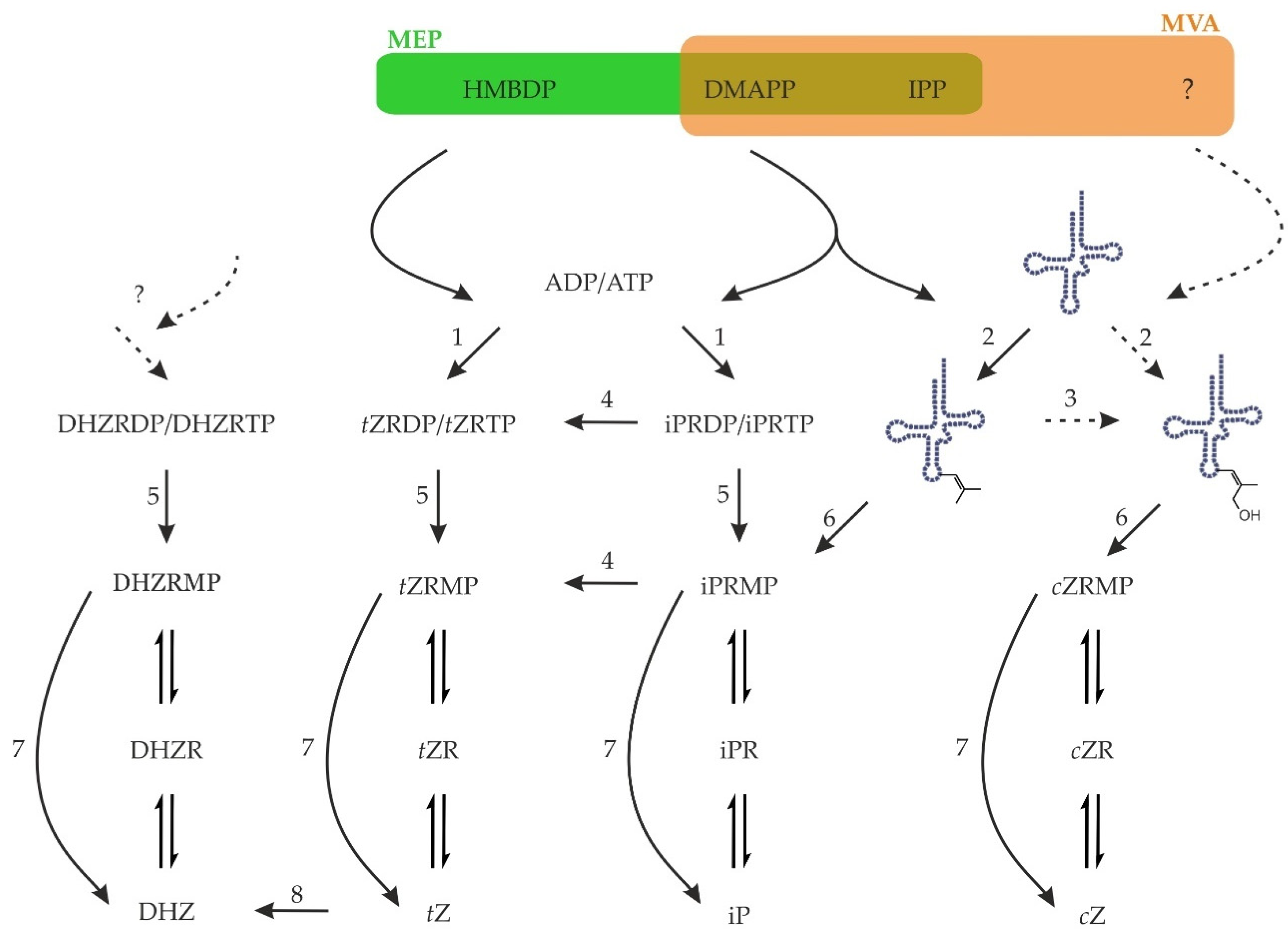
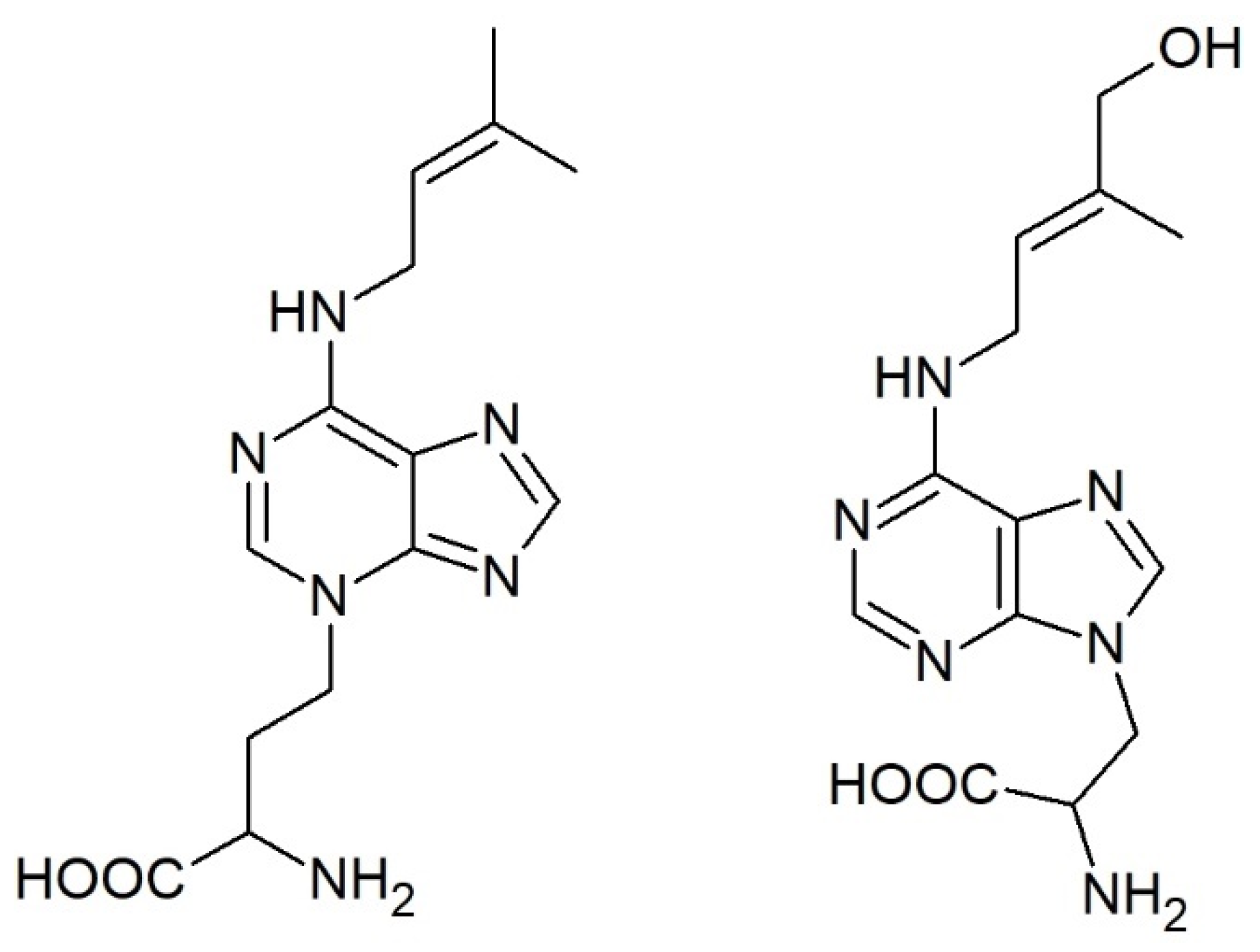
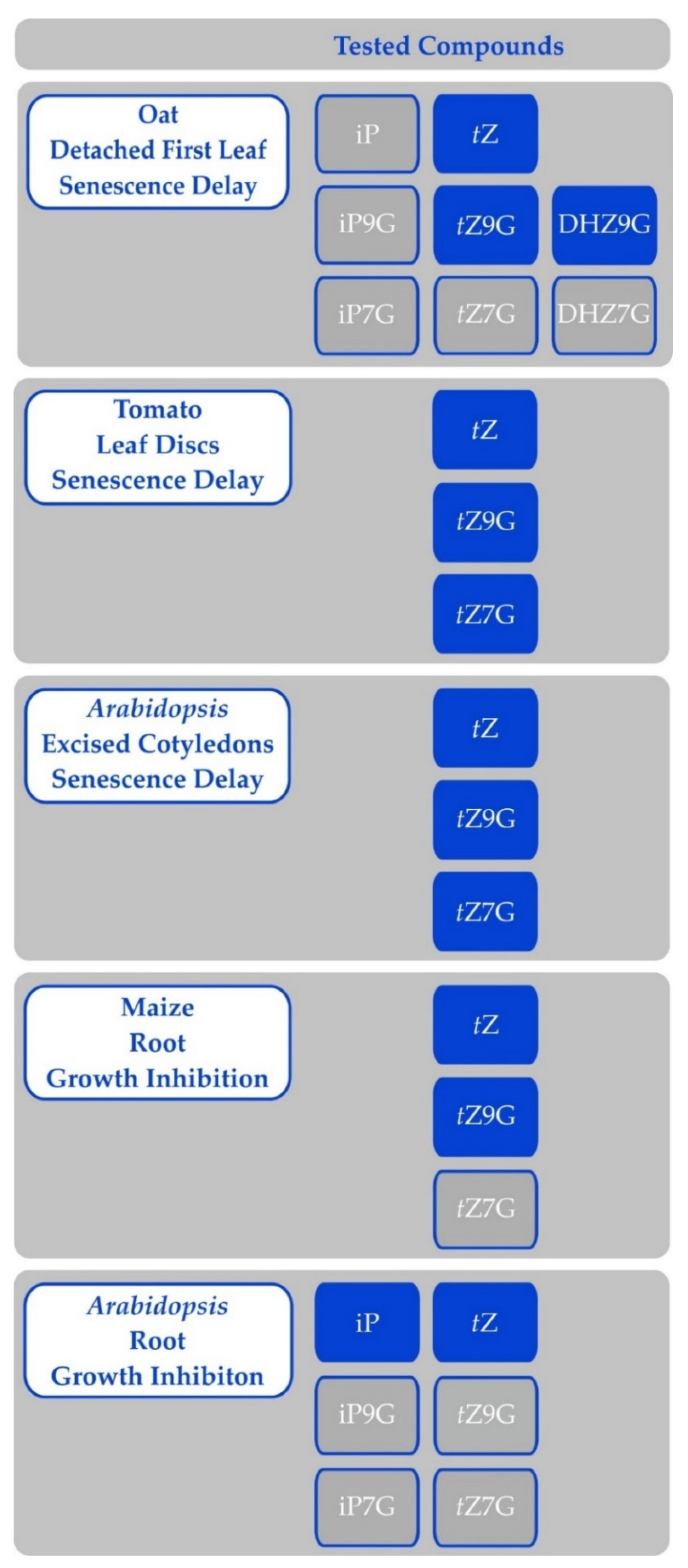
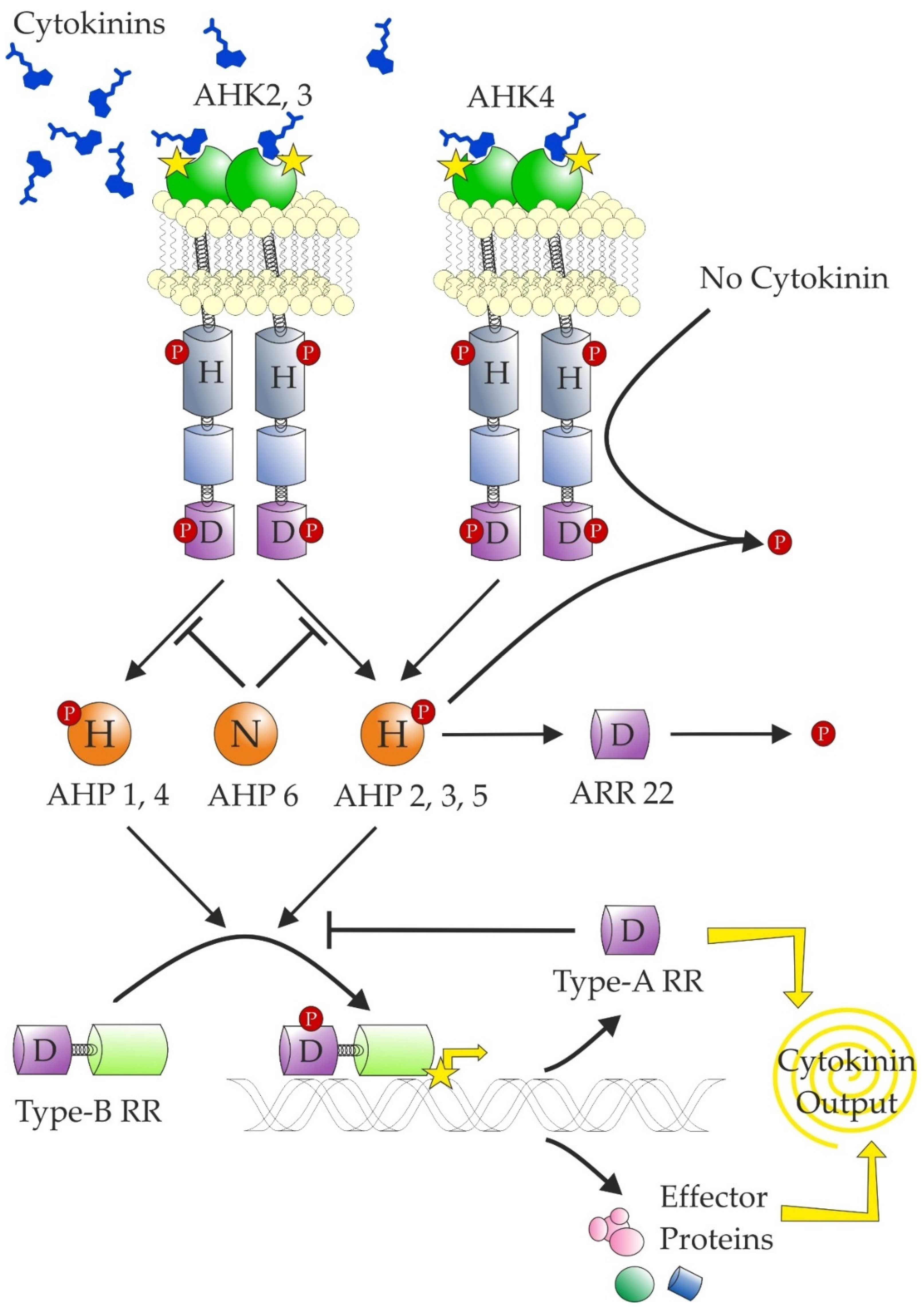
| Hulks | Deadpools | Commentary |
|---|---|---|
| tZ, iP | cZ aromatic cytokinins | While tZ and iP induce large changes in plant physiology, cZ and aromatic cytokinins are “slow” with weak CK activity, but they are resistant and thus persistent. |
| AtIPT3 | AtIPT5, SlIPT3 and PpIPT5a tRNA IPTs | AtIPT3 is the main enzyme synthesising CKs with spatio-temporally focused production; on the other hand, IPT5 orthologues are expressed widely but they have only low activity, thus producing small less focused amounts of CKs. tRNA IPTs usually exert wide expression and they produce the Deadpool cZ. |
| receptors on PM | receptors on ER | PM-localised receptors may perceive the Hulk CKs tZ and iP with very high activity. To diminish Hulk signalling when Hulk CKs are not present, AHK4 also possesses a phosphatase activity. On the other hand, ER-localised receptors perceive autocrine Deadpool signals. |
| LOGs AtLOG7 | two-step activation AtLOG8 | LOGs activate CKs (almost) in a single step from biosynthesis products specifically in regions of high cell division activities, while the two-step activation functions throughout the plant body at low speed. Among LOGs, one can distinguish AtLOG7 with high activity and expression focused to high activity hotspots and AtLOG8 with minimal activity but expression throughout the plant. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hluska, T.; Hlusková, L.; Emery, R.J.N. The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction. Biomolecules 2021, 11, 209. https://doi.org/10.3390/biom11020209
Hluska T, Hlusková L, Emery RJN. The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction. Biomolecules. 2021; 11(2):209. https://doi.org/10.3390/biom11020209
Chicago/Turabian StyleHluska, Tomáš, Lucia Hlusková, and R. J. Neil Emery. 2021. "The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction" Biomolecules 11, no. 2: 209. https://doi.org/10.3390/biom11020209
APA StyleHluska, T., Hlusková, L., & Emery, R. J. N. (2021). The Hulks and the Deadpools of the Cytokinin Universe: A Dual Strategy for Cytokinin Production, Translocation, and Signal Transduction. Biomolecules, 11(2), 209. https://doi.org/10.3390/biom11020209







