Abstract
CD44, a non-kinase cell surface transmembrane glycoprotein, has been widely implicated as a cancer stem cell (CSC) marker in several cancers. Cells overexpressing CD44 possess several CSC traits, such as self-renewal and epithelial-mesenchymal transition (EMT) capability, as well as a resistance to chemo- and radiotherapy. The CD44 gene regularly undergoes alternative splicing, resulting in the standard (CD44s) and variant (CD44v) isoforms. The interaction of such isoforms with ligands, particularly hyaluronic acid (HA), osteopontin (OPN) and matrix metalloproteinases (MMPs), drive numerous cancer-associated signalling. However, there are contradictory results regarding whether high or low CD44 expression is associated with worsening clinicopathological features, such as a higher tumour histological grade, advanced tumour stage and poorer survival rates. Nonetheless, high CD44 expression significantly contributes to enhanced tumourigenic mechanisms, such as cell proliferation, metastasis, invasion, migration and stemness; hence, CD44 is an important clinical target. This review summarises current research regarding the different CD44 isoform structures and their roles and functions in supporting tumourigenesis and discusses CD44 expression regulation, CD44-signalling pathways and interactions involved in cancer development. The clinical significance and prognostic value of CD44 and the potential of CD44 as a therapeutic target in cancer are also addressed.
1. Introduction
Cancer manifests as uncontrolled cell proliferation, followed by enhanced migration, invasion, and metastasis to other parts of the body. Numerous investigations support the role of cancer stem cells (CSCs) and their associated markers in tumour malignancies, for example, the cluster of differentiation 44 or CD44 [1]. As a non-kinase cell surface transmembrane glycoprotein that is overexpressed in CSCs and frequently undergoes alternative splicing to support cancer progression, it could severely influence treatment outcomes [2]. In the early 1980s, CD44 was first identified as a glycoprotein expressed on human [3] and murine mesenchymal cells [4]. Subsequently, it was cloned and classified as a cartilage link protein family member [5]. It is a single polypeptide chain encoded by a conserved gene located on either human chromosome 11 [6] or murine chromosome 2 [7] and is also known as In (Lu)-related protein p80, Pgp-1/Ly-24, ECMRIII, HUTCH-1, Hermes antigen, and importantly, hyaluronate receptor. Currently, CD44 is recognised as the main cell surface receptor for hyaluronate, which is the major extracellular matrix (ECM) component [8]. It is a member of the cell adhesion molecules (CAMs) family that plays important roles in cellular communication and adhesion between cells and the ECM [9]. In addition to its role in cellular adhesion and communication, it is essentially involved in several biological and functional processes, such as lymphopoiesis and myelopoiesis, leukocyte activation, angiogenesis and the release of cytokines, as well as many pathological processes including metastasis, epithelial–mesenchymal transition (EMT), cellular growth, proliferation, migration and invasion. Although abundant progress has been made regarding the structure and functional roles of CD44 and its diverse isoforms, the expression level associated with poorer clinicopathological impacts remains unknown. Thus, a significant area of study is aims to further define the functional roles of the different CD44 isoforms in various types of cancers. Increasing evidence suggests that some CD44 isoforms are promising prognostic biomarkers and therapeutic targets for many cancers. This review summarises current insights on CD44 structure and isoforms as well as the CD44-mediated oncogenic signalling pathway in several cancers. We also highlight CD44 interaction with many components in the tumour microenvironment and its functional roles and involvements in tumour progression and aggressiveness, as well as its clinical relevance and the possibility of targeting CD44 for cancer therapy.
2. CD44 Structure and Isoforms
The full-length CD44 gene comprises 20 exons, with the constant exons 1–5 and 16–20 encoding the N-terminal and C-terminal domains respectively, which are homologous domains shared by all CD44 family members [10]. The smallest and the most expressed CD44 isoform is the CD44 standard (CD44s), constructed of ten constant exons with no variant exons [11]. The other isoform, the CD44 variant (CD44v), differs from CD44s by the insertion or excision of alternatively spliced exons between the N-terminal and C-terminal domains [12]. Tolg et al. [13] confirmed that besides ten constant exons, the mouse and rat genome has at least ten variant exons, all of which can be combined alternatively into CD44 mRNA. They suggested that the variant exons be numbered by the exon code v1 to v10. Screaton et al. [14] described the structure of the human CD44 gene, reporting that it contains 19 exons crossing some 50 kilobases of DNA with ten constant exons and nine variant exons coded v2–v10 [15,16]. CD44v isoforms may contain a single variant exon such as CD44v3 or CD44v6, or a combination of variant exons such as CD44v3–v7 and CD44v8-v10. Individual cells can continually alter the splicing of CD44 pre-mRNA, resulting in many possible combinations of these variant exons, giving the potential for great diversity [12].
The CD44 protein has four primary characteristic regions: the extracellular region, the stem region (standard stem region and/or variable stem region), the transmembrane region (TM), and the short C-terminal intracellular/cytoplasmic (CP) region [17]. The extracellular part consists of seven extracellular domains (1–5, 6 and 7 of the constant exons) including N-terminal domains (ligand-binding region). The stem region (alternative splicing area) has an insertion of one or more of the variant exons between exon 5 and exon 6. The transmembrane region is encoded by a single exon (exon 8), whereas the cytoplasmic region is encoded by exon 10 or exon 9. However, exon 9 is spliced out in almost all CD44 cDNA isoforms [12,18]. Several isoforms of the human CD44 molecule are associated with tumour progression and stemness in various cancers, such as breast cancer [19], gliomas [20,21], head and neck squamous cell carcinoma [22], pancreatic cancer [23,24], prostate cancer [25] and colorectal cancer [26,27] (Figure 1 and Table 1). The complexity of the CD44 protein is further augmented by post-translational modifications including variance glycosylation with O-glycans, N-glycans and glycosaminoglycans, such as chondroitin sulphate and heparan sulphate [17]. Due to these side-chain attachments, the conserved format of CD44 (37 kDa) is enlarged to 80–100 kDa with some isoforms surpassing 200 kDa due to a high level of glycosylation [12]. An illustration of CD44 protein structure is shown in Figure 2.
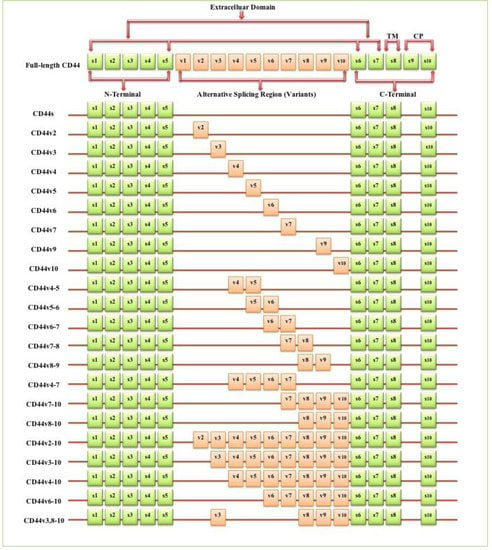
Figure 1.
Schematic diagram of the mouse CD44 gene and most CD44 isoforms involved in cancer progression. The full-length CD44 gene contains 20 exons in mice and 19 exons in humans, with the constant exons 1–5 and 16–20 encoding the N-terminal and the C-terminal domains. CD44 standard (CD44s) is encoded by these ten constant exons and contains no variant exons, whereas the CD44 variant (CD44v) is produced by the alternative splicing of a variable insertion of nine extra exons in humans or ten extra exons in mice. These extra exons are exons 6-15, typically identified as (v1 to v10) in mice and the exons 7-15 identified as (v2 to v10) in humans and are located between the N-terminal and C-terminal domains. CD44v can contain one or multiple variant exons and exon 19 is spliced out in all CD44 isoforms. Abbreviations: CD44s, CD44 standard; CD44v, CD44 variant; s, standard; v, variant; TM, transmembrane; CP, cytoplasmic. Green boxes refer to the constant/standard exons. Orange boxes refer to the variant exons.

Table 1.
CD44 isoforms relevant to cancer progression. Abbreviations: CSCs, cancer stem cells; EMT, epithelial–mesenchymal transition; DFS, disease-free survival; OS, overall survival; TNM stage, tumour (T), node (N), and metastasis (M) stage; FIGO stage, the international federation of gynaecology and obstetrics stage; NHL, Non-Hodgkin’s lymphoma; HPV, human papillomavirus; MAPK, mitogen-activated protein kinase.
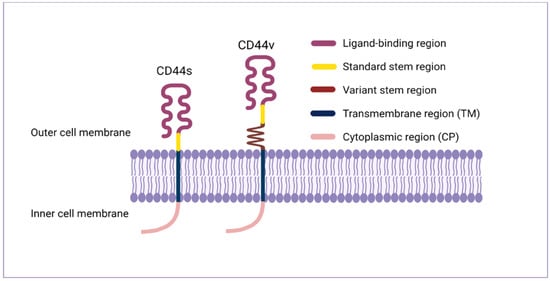
Figure 2.
CD44 protein structure. The CD44 protein has four primary regions: the extracellular region consists of seven extracellular domains including N-terminal domains (ligand-binding region), the stem region (variable stem region and/or standard stem region) which is the alternative splicing area containing an insertion of one or more variant exons, the transmembrane region (TM), and the C-terminal cytoplasmic (CP) region.
3. CD44 Expression in Normal Cells
CD44 is significantly expressed in lymphocytes, smooth muscle, fibroblasts and various types of epithelia and is involved in lymphocyte homing, cell adhesion and aggregation, cell migration, leukocyte activation, lymphopoiesis and myelopoiesis, angiogenesis and cytokine release [12,69]. CD44s was initially isolated from haematopoietic cells even though it is expressed in several other tissues including the liver, lung, pancreas, skin and central nervous system [12]. CD44s is expressed in adult tissues and embryo tissues from day 9.5 post coitum, whereas numerous isoforms of CD44v show a highly specialised expression pattern and are already in the egg cylinder at day 6.5 of development [70]. In contrast to CD44s, CD44v isoforms distribution is more restricted to a selected range of cells during specific stages of activation, maturation or development including macrophages, activated lymphocytes, keratinocytes and some epithelial cells such as in the stomach, bladder and uterine cervix [12] and many carcinomas. In normal tissues, CD44 isoforms play a role in hyaluronic acid (HA) metabolism regulation, whereby loss of CD44 expression disrupts HA metabolism and impairs hair regrowth, wound healing and keratinocyte proliferation [71].
4. CD44 Expression in Tumours
Numerous studies indicated that lymphoma, breast, colon and endometrial cancer have elevated levels of CD44 mRNA [69]. Increasing evidence also suggests that CD44 is extensively overexpressed in other cancer types including gallbladder, prostate, ovarian, oral squamous cell carcinoma and gastric cancer, correlating with aggressive biological behaviour and a poor prognosis [72]. The role of CD44 in tumours is not well defined, however, elevated levels of CD44 are associated with numerous malignant tumours. The physiological functions of CD44 indicate that it is involved in the metastasis of tumours [12]. For instance, lung adenocarcinoma cells show a high expression of CD44v, which correlates with enhanced CSCs characteristics, proliferation and resistance to chemotherapeutics [73], whereas these variants, especially CD44v6, are closely related to metastasis of pancreatic carcinoma cells [69]. Many studies have investigated CD44 expression levels in several cancers in comparison to their adjacent normal tissues and explored the relationship with tumour progression and clinicopathological outcomes by mining various publicly available databases, including The Cancer Genome Atlas (TCGA), Tumour Immune Estimation Resource (TIMER) database, Oncomine database, Gene Expression Profiling Interactive Analysis (GEPIA), In silico Transcriptomics (IST) database, R2 online database, SAGE Genie tools, and Human Gene Expression Map (HGEM) (Table 2 and Figure 3).

Table 2.
Low and high CD44 expression in normal and tumour tissues respectively and association with clinical outcomes.
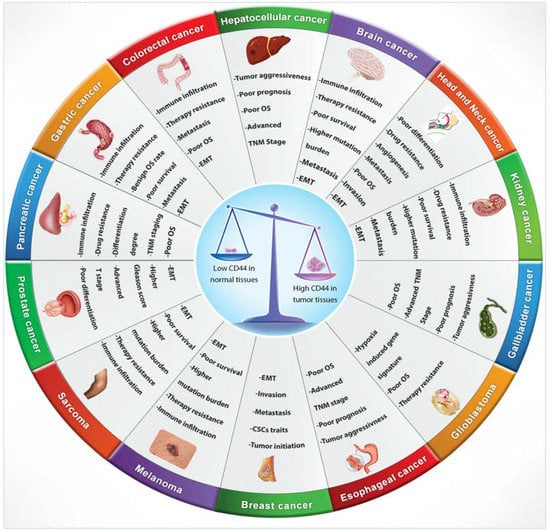
Figure 3.
CD44 distribution in normal versus cancerous tissues and its correlation with clinical outcomes.
5. CD44s VS CD44v: Roles and Functions in Cancer Progression
The expression of CD44 isoforms within the tumour impacts key features of cancer cells such as tumourigenicity, tumour initiating potential, metastasis, chemo and/or radio-resistance, etc. Some researchers concluded that tumours expressing particular isoforms of CD44v are more aggressive compared to tumours expressing CD44s only. Recently, Zhang et al. [28] revealed that CD44s and not CD44v positively promotes tumour initiation and CSCs gene traits by activating the PDGFRβ/STAT3 cascade-signalling pathway. They showed that CD44s is the dominant isoform expressed in breast CSCs and that its elimination impaired CSCs signatures. Conversely, shifting alternative splicing from CD44v to CD44s by manipulating the splicing regulator ESRP1 led to an induction of CSCs traits. Brown et al. [30] also demonstrated that the isoform shift from the variant to the standard isoform was essential for breast cancer cells to undergo EMT, a process often activated during tumour metastasis and recurrence. CD44s accelerates EMT by activating AKT signalling, which results in the formation of EMT-associated recurrent tumours and apoptosis resistance in these tumours. Similarly, CD44s but not CD44v interacts with phosphorylated cortactin activated invadopodia, enabling breast tumour cells to degrade ECM and metastasise to distant organs such as the lungs. Depletion of CD44s eliminates invadopodia activity, prevents ECM degradation and reduces tumour cell invasion and metastasis [29]. CD44s is the predominant isoform highly expressed in hepatocellular carcinoma cells and its expression indicates poor survival in patients with hepatocellular carcinoma. CD44s but not CD44v regulates TGF-β signalling mediated mesenchymal phenotype, which is characterised by stimulated tumour invasiveness and increased expression of the EMT marker, vimentin and low E-cadherin expression [31].
In gallbladder cancer, Miwa et al. [32] found that CD44s+ cells highly expressed ZEB1 and ZEB2 transcription factors that mediate EMT and showed increased invasiveness, chemotaxis and distant metastasis compared to CD44v+ cells. However, CD44s+ cells exhibited decreased tumourigenicity while CD44v+ cells, particularly CD44v9, showed higher tumourigenicity suggesting that both CD44v and CD44s cells play different vital roles in tumour progression and metastasis. Other experimental studies have suggested that CD44v isoforms enhance tumour aggressiveness and bone metastasis in some cancers to a similar extent as CD44s. For instance, in breast and lung cancers, the introduction of ESRP1 prompted the splicing switch from CD44s to CD44v8-10 with no change in the total amount of CD44. This switching did not affect cell proliferation, invasion, migration, sphere formation or bone metastasis, demonstrating that the contribution of CD44v8-10 to tumorigenicity is comparable to that of CD44s [61]. While normal colonic mucosa does not express CD44 isoforms, its tumours present an extensive variation of CD44 isoforms. CD44v isoforms, especially CD44v3 and CD44v6, influence expressing metastasis genes and are involved in metastasis in colorectal cancer [40]. In the same type of cancer, CD44v2 was also highly expressed in both primary and xenografts tumours compared to the normal colonic mucosa, and this overexpression was associated with poor prognosis [36].
Meanwhile, CD44v8-10 was first defined in colon cancer cells, subsequently in gastric CSCs, and was discovered to be upregulated in primary and metastatic tumours [84]. CD44v8-10 expression, but not CD44s, marks gastric CSCs and enhances tumour initiation, probably through augmenting hypoxia or oxidative stress defence [62]. Likewise, CD44v5-6 and CD44v8-9, but not CD44s, are significantly increased in non-small cell lung adenocarcinoma and mediate tumour cell proliferation and poor prognosis by activating the KRAS/MAPK signalling pathway. KRAS-signalling induction additionally stimulates CD44 alternative splicing, resulting in a greater production of CD44v [58]. There are numerous reports that the expression of CD44v6 is an effective progression and prognosis marker in many cancers. For instance, CD44v6 is overexpressed in the mandibular invasive front of oral squamous carcinoma cells in patients with positive cervical lymph node metastasis and additionally, it is linked to the formation of tumour buds [43]. Yanamoto et al. [45] reported that a high intensity of CD44v6 in tongue squamous cell carcinoma was associated with local recurrence. In colorectal cancer, CD44v6 upregulation through nuclear β-catenin signalling activation contributed to tumour budding formation and the identification of those at high risk for locoregional failure in early staged tumours [46]. Ni et al. [44] demonstrated that in prostate cancer, CD44v6 is an important CSCs biomarker and is closely related to tumour cell proliferation, adhesion, invasion, metastasis, chemo- and radio-resistance, EMT induction, and the activation of PI3K/AKT/mTOR and Wnt signalling pathways. In contrast, another study of CD44 isoform expression in prostate cancer concluded that CD44s promoted tumour initiation, cell proliferation, invasion and migration, providing evidence of the correlation between total CD44 expression and prostate cancer progression for CD44s only. Furthermore, alterative splicing from CD44v to CD44s isoform enhanced tumour progression, EMT and stemness [33].
Amongst several molecules that have been extensively described and investigated for their possible roles in pancreatic carcinoma progression and tumourigenesis, CD44 is the most significant [47]. Li et al. [47] confirmed that decreased CD44s and increased CD44v expression in metastatic pancreatic carcinoma in three different cell lines and human tumour tissues. They also showed that CD44s-, CD44v6+ and CD44v9+ were significantly involved in the advanced TNM stage, liver metastasis and lymph node metastasis. In contrast, pancreatic ductal adenocarcinoma cells predominantly expressing high levels of CD44s are associated with an EMT phenotype, extremely invasive, develop gemcitabine resistance tumours and metastasise more rapidly [34]. Similarly, in pancreatic cancer, CD44s is involved in the radio-resistance of cancerous cells and strongly upregulated compared to CD44v after high-dose irradiation resulting in longer-term cell survival via the maintenance of ERK phosphorylation and radiation-stimulated EMT [35]. CD44v isoform expression is commonly believed to be more important for malignancy, especially when EMT is a vital step for metastasis, and is hence associated with a poorer prognosis [85]. However, and as discussed above, clinical data demonstrated that the CD44s isoform also enhanced tumour aggressiveness and metastasis in several cancers to a similar extent as CD44v. It is important to consider that all CD44 isoforms are prognosis markers and potential therapeutic targets for the prevention of metastasis.
6. CD44 Expression Regulation
During tumour progression, CD44 isoform expression is regulated by numerous signalling networks. Since CD44 expression contributes to several signalling pathways involved in modulating multiple cellular processes that support tumourigenesis, it is vital to understand how CD44 expression is regulated and the key signalling networks involved in the regulation of CD44 expression. In this regard, several transcriptional factors, protein kinases, cytokines, epigenetic mechanisms and miRNAs are involved in the regulation of CD44 expression by acting as repressors or activators of CD44. A non-exhaustive list of mechanistic regulators is provided in Figure 4.
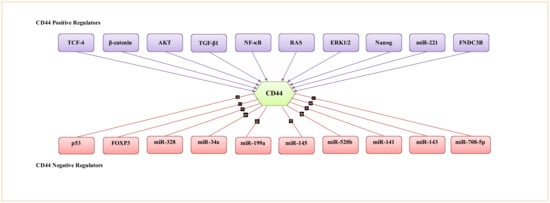
Figure 4.
Representative transcription factors, protein kinases, cytokines and miRNA involved in the regulation of CD44 activity.
6.1. Regulation of CD44 by Transcriptional Factors, Protein Kinases and Cytokines
As CD44 overexpression is an early event in colorectal carcinoma, Wielenga et al. [86] established that CD44 is a target gene of Wnt/β-catenin in mice intestinal tumours, whereby the β-catenin/Tcf-4 signalling pathway mediates transcriptional upregulation of CD44 expression. Thereafter, CD44 expression upregulation by Wnt/β-catenin/Tcf-4 mediated transcription was also confirmed in human colorectal adenocarcinoma cells [87]. Smith et al. [88] demonstrated that the transcription factor (NF-κB) upregulates CD44 expression in triple-negative breast cancer cells, mainly via the interaction with the cis-regulatory element conserved region (CR1) located upstream of the CD44 promoter. The inhibition of NF-κB resulted in reduced CD44 expression. Wang et al. [89] found that CD44 expression was upregulated by activation of the β-catenin signalling pathway in mouse and human pancreatic ductal adenocarcinoma, resulting in EMT phenotype induction characterised by the upregulation of Zeb1 and Snail1 expression. Zhang et al. [28] described CD44s as predominantly expressing CD44 in breast CSCs and found that CD44s promoted CSCs signatures. Meanwhile, manipulating epithelial splicing regulatory protein 1 (ESRP1) suppresses CD44s-mediated induction of CSC traits. Godar et al. [90] showed that p53 inhibited CD44 expression by binding to a noncanonical p53 binding sequence in the CD44 promoter, and the p53 loss resulted in elevated CD44 levels which increased resistance to apoptosis in lung carcinoma cells. Forkhead box protein 3 (Foxp3) bound to the CD44 promoter and significantly inhibited its expression, suppressing the invasion and metastatic capabilities of human breast cancer cells [91]. In cervical cancer and breast cancer cells, the activation of β-catenin along with AKT signalling pathways were correlated with the upregulated expression of CD44. β-catenin knockdown and the inhibition of the AKT pathway significantly suppressed the expression of CD44 [92]. Cheng et al. [93] identified a positive feedback loop between CD44v6 isoforms and the RAS/MAPK signalling pathway. The RAS signalling pathway promoted CD44v6 production, which subsequently acted as co-receptors for several growth factors and tyrosine kinases to further activate RAS signalling. A study by Judd et al. [94] suggested that ERK1/2 signalling regulates CD44 expression in aggressive oral cancer cell lines and that the blockage of ERK1/2 decreased CD44 expression. Furthermore, Huang et al. [95] demonstrated for the first time that the ERK1/2-Nanog signalling pathway played a critical role in the maintenance of cells stemness and tumourigenic abilities by enhancing CD44+ CSCs in head and neck squamous cell carcinomas. Furthermore, Shang et al. [96] revealed that the transforming growth factor-beta 1 (TGF-β1) upregulated CD44 expression in prostate cancer cells. Similarly, crosstalk between the TGF-β pathway and CD44 expression upregulation was observed in oral and oesophageal cancer [97] and hepatocellular carcinoma [98].
6.2. Regulation of CD44 by Epigenetic Mechanisms
To determine whether CD44 expression might be associated with epigenetic regulation in lung cancer, an experiment using TGF-β treatment and DNA methyltransferase inhibitor (AZA) was performed. TGF-β treatment resulted in enhanced CD44 expression levels but AZA treatment did not stimulate CD44 expression, confirming that CD44 activated by TGF-β is not related to the epigenetic mechanism of aberrant promoter demethylation [99]. Other reports have suggested that CD44 expression levels can be epigenetically regulated by DNA methylation at the CD44 promoter region. Eberth et al. [100] showed that CD44 underwent de novo methylation in lymphoma cells, with the hypermethylation of CD44 resulting in transcriptional silencing of this gene, which can be reactivated by AZA treatment. Similarly, in neuroblastoma, Yan et al. [101] demonstrated that CD44 silencing was controlled by aberrant gene hypermethylation. Furthermore, in most prostate cancer cases, the loss of CD44 expression is associated with extensive hypermethylation of the CpG island CD44 promoter region [102,103]. Additionally, the CD44 gene in breast CSCs and CD44 gene hypomethylation was correlated with aggressive features of triple-negative breast cancer [104].
6.3. Regulation of CD44 by miRNAs
Several miRNAs were reported to regulate CD44 expression; for example, miR-328 is a potential regulator of CD44. Through studying the relative luciferase activity with diverse miR-328 mimic concentrations, a negative correlation between miR-328 and CD44 was established; CD44 constructs decreased as the concentration of miR-328 mimics increased [105]. Likewise, CD44 expression was diminished in gastrointestinal cancer cells forced to express miR-328, resulting in cancer cell growth inhibition and impaired resistance to chemotherapy and reactive oxygen species (ROS). In contrast, the stimulation of CD44 expression by a miR-328 inhibitor resulted in gastrointestinal cancer cell growth enhancement [106]. CD44 expression was also found to be suppressed by targeting the 3′-untranslated region (3′-UTR) of the CD44 gene, leading to perturbations of signalling pathways and the suppression of tumourigenesis and metastasis. For instance, miR-34a repressed CD44 expression in prostate CD44+ CSCs, resulting in metastasis and regeneration inhibition [107], and miR-199a-mediated targeting of CD44 in ovarian cancer-initiating cells suppressed tumourigenesis and multidrug resistance [108]. Similarly in gastric cancer, CD44 3′-UTR was found to be directly targeted by miR-145, with miR-145 overexpression repressing CD44 3′-UTR activity, which could be abrogated by blocking the miR-145/CD44 3′-UTR interaction, supporting significant augmentation of chemotherapy resistance and sphere formation [109]. CD44 was also identified as a direct molecular target of miR-520b, whereby miR-520b inhibited tumourigenesis of head and neck cancer through the regulation of cancer stemness conversion [110]. Another study demonstrated that miR-141 suppressed tumour growth and metastasis of prostate CSCs by targeting the CD44 gene [111]. In breast cancer cells, miR-143 inhibited the progression and stemness features by directly targeting CD44 3′-UTR [112]. In contrast, a positive relationship has been found between CD44 and miR-221, both of which are upregulated in hepatocellular carcinoma cells and tumours. Inhibition of miR-221 decreased CD44 protein expression, whereas the miR-221 mimic enhanced protein levels of CD44. Another study described a mechanism of miR-221-CD44 interaction involving the PI3K/AKT/mTOR pathway. Targeting the downstream effector of this pathway by the inhibition of miR-221 reduced CD44 expression [113]. CD44 is also regulated by a different class of non-coding RNA molecules known as the circular RNAs (circRNAs), single-stranded RNAs identified in malignant tumour cells which can join the 3′ end to the 5′ end of the RNA molecule. For example, the circFNDC3B regulates and increases the stability of CD44 expression by forming a ternary complex of circFNDC3B/IGF2BP3/CD44 mRNA, subsequently promoting cell invasion and migration of gastric cancer cells [114].
7. CD44-Downstream Signalling Pathways
The activation of CD44 isoforms modulates the activities of the components of several signalling pathways including enzymes, protein kinase pathways, transcriptional factors and intracellular pathways to contribute to cancer cell proliferation, stemness, invasion, migration and metastasis, as well as drug resistance. Some of these signalling pathways are summarised in Figure 5.
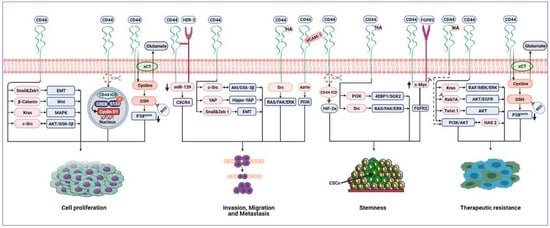
Figure 5.
Cancer-associated signalling pathways modulated by CD44. CD44 can upregulate EMT biomarkers, Snail1 and Zeb1, promoting proliferation and invasion. CD44 activates KRAS through the MAPK pathway, hence promoting tumour cell proliferation and survival. CD44 mediates invasion and metastasis by binding to HER2, leading to the inhibition of miR-139 and upregulation of CXCR4. There is positive crosstalk between CD44 and FGFR2 to maintain cancer stemness. CD44 regulates tumour cell proliferation, invasion and migration by modulating c-Src via AKT/GSK-3β signalling. CD44/VCAM-1 interaction promotes invasion signalling by the ezrin/PI3K pathway. CD44 binds to HA, resulting in stemness development via the PI3K/4EBP1/SOX2 pathway. Also, the HA/CD44 interaction can drive tumour invasion, metastasis and stemness through Src activating RAS/FAK/ERK pathways. Similarly, HA/CD44 stimulates the PI3K/AKT signalling pathway to increase therapeutic resistance. CD44 can sustain EGFR and AKT signalling by inhibition of Rab7A, leading to therapeutic resistance. CD44 promotes the expression of HAS2 by activating the PI3K/AKT signalling pathway. HAS2 further enhances CD44-mediated PI3K/AKT signalling, thus creating a positive feedback loop that drives tumour cell resistance and survival. CD44 can stabilise the cystine/glutamate antiporter (xCT), leading to increased GSH and decreased ROS levels, which, in turn, results in tumour cell proliferation and therapeutic resistance mediated by suppression of the p38 pathway. CD44 regulates tumour cell proliferation by the Wnt/β-catenin signalling pathway. CD44 can promote invasion and migration through the activation of the Hippo-YAP oncogene signalling pathway. CD44 mediates tumour cells resistance by upregulating Twist1 and AKT signalling. CD44-ICD binds to CREB, enhances S133 phosphorylation and enriches CREB recruitment to the cyclin D1 promoter, thus promoting cyclin D1 activity, resulting in cell proliferation. CD44-ICD is released in a hypoxic environment and binds to HIF-2α leading to induced stemness. Abbreviations: EMT, epithelial–mesenchymal transition; HER2, human epidermal growth factor receptor 2; CXCR4, C-X-C chemokine receptor type 4; FGFR2, fibroblast growth factor receptor 2; EGFR, epidermal growth factor receptor; HAS2, hyaluronan synthase 2; GSH, glutathione; ROS, reactive oxygen species; CD44-ICD, CD44 intracellular domain; CREB, cAMP response element-binding; HIF-2α, hypoxia-inducible factors 2 alpha.
In pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma, CD44 is central in promoting the upregulation of EMT biomarkers expression, Snail1 and Zeb1, as well as stimulating migration and invasion. CD44 knockdown using CD44 shRNA in pancreatic ductal adenocarcinoma cells not only efficiently reduced the expression of Zeb1 and Snail1 but also significantly impaired cell proliferation and invasion [89]. CD44 was found to critically contribute to activating the oncogenic KRAS signalling pathway through the MAPK pathway in lung adenocarcinoma cell lines, hence promoting tumour cell proliferation and survival [58]. CD44-mediated gastric cancer cell invasion and metastasis by binding to human epidermal growth factor receptor 2 (HER2) inhibits miR-139 and upregulates the miR-139 target gene, C-X-C chemokine receptor type 4 (CXCR4) [115]. Park et al. [116] demonstrated positive crosstalk between CD44 and fibroblast growth factor receptor 2 (FGFR2). While FGFR2 inhibits transcription of c-Myc, CD44 activates c-Myc expression, in turn enhancing FGFR2 transcription. The cooperative interaction of FGFR2 and CD44 maintains gastric cancer stemness via reciprocally regulating their expression by differentially regulating c-Myc transcription. CD44 in breast CSCs also played a pivotal role in the regulation of tumour cell proliferation, invasion and migration by modulating the levels of c-Src, a master regulator of the MAPK, PI3K, and STAT3 signalling pathways, via the inhibition of c-Jun and degradation by AKT/GSK-3β signalling [117]. In head and neck CSCs, by binding to HA, CD44 mediated stemness and CSCs fraction development via the PI3K/4EBP1/SOX2 pathway, while CD44/VCAM-1 interaction promoted invasion signalling by the ezrin/PI3K pathway [118]. CD44 can directly potentiate receptor tyrosine kinase (RTKs) signalling pathways and act as a coreceptor for several growth factors, such as tyrosine-protein kinase Met (c-Met), vascular endothelial growth factor receptor-2 (VEGFR-2) and epidermal growth factor receptor (EGFR), thus enhancing cancer cell proliferation and correlating with poor prognosis and metastatic potential [119]. Amongst CD44 isoforms, CD44v displays a greater affinity to HA compared to CD44s [120]. By activating RTKs signalling pathways, HA/CD44v6 interaction can drive tumour metastasis [121,122]. In colorectal cancer, this interaction facilitates colorectal CSCs colonisation, invasion and metastasis. Furthermore, the same interaction activates RAS and FAK pathways through Src, resulting in MAPK/ERK signalling pathway activation, and subsequently, enhanced cell proliferation. Similarly, the interaction between HA and CD44v6 stimulates the PI3K/AKT signalling pathway, thus increasing the resistance of colorectal cancer cells to apoptosis [123]. Additionally, in glioblastoma cells, CD44s attenuated EGFR degradation and sustained AKT signalling by the inhibition of Rab7A, which mediated EGFR trafficking for degradation, thus sustaining EGFR levels. CD44 depletion collectively with EGFR inhibition results in synergistic and robust glioblastoma cell death [124]. CD44s additionally promoted the expression of hyaluronan synthase 2 (HAS2) by activating the PI3K/AKT signalling pathway, which further enhanced CD44s-mediated PI3K/AKT signalling, thus creating a positive feedback loop to drive tumour cell survival in breast cancer cells [125]. Thanee et al. [126] revealed that CD44s and CD44v8-10 regulated redox status by stabilising cystine/glutamate antiporter (xCT), leading to increased glutathione (GSH) and subsequently, decreased ROS, which, in turn, suppressed the p38 MAPK pathway that correlates with poor prognosis for cholangiocarcinoma patients.
CD44 signalling plays a pivotal role in regulating the proliferation of chronic myeloid leukaemia cells by modulating the expression and activity of the Wnt/β-catenin signalling pathway. CD44 downregulation reduced β-catenin expression and augmented β-catenin phosphorylation, inhibiting cell proliferation [127]. Through the activation of the Hippo-YAP oncogene signalling pathway, CD44 promoted invasion and migration of docetaxel-resistant prostate cancer cells, which had a larger CD44+ population and positively correlated with poor survival of prostate cancer patients [128]. Zhang et al. [28] revealed that CD44s was preferentially and predominantly expressed in breast CSCs and promotes CSCs activities and tumour initiation through signal transducer and activator of transcription 3 (STAT3) via PDGFRβ/STAT3 signalling pathways. In hepatocellular carcinoma, CD44s also mediated tumour cells anoikis resistance and sphere formation capability by upregulating Twist1 and AKT signalling [129].
CD44 may undergo sequential proteolytic cleavage, releasing the intracellular domain CD44-ICD to translocate into the nucleus and transactivate gene expression. CD44-ICD bound to the cAMP response element-binding protein transcription factor (CREB-TF) and enhanced S133 phosphorylation and CREB-mediated gene transcription. CD44-ICD enriched CREB recruitment to the cyclin D1 promoter, thus promoting cyclin D1 activity, a regulator of protein transcription and cell proliferation, in thyroid carcinoma cells [130]. In glioma cells, CD44-ICD was released in a hypoxic environment and bound to hypoxia-inducible factors 2 alpha (HIF-2α) to induce stemness in glioma [131].
8. CD44 Receptor-Ligand Interactions
CD44 interacts with various ECM components, proteins and cytokines present in the tumour microenvironment. Interaction between ligands such as hyaluronan (HA), osteopontin (OPN) and matrix metalloproteinases (MMPs) with the CD44 receptor can stimulate several cellular signalling pathways [132]. CD44 functions in supporting tumour progression and aggressiveness can be attributed to its diverse binding ligands and interactome (Table 3).
HA is a protuberant molecule present in both the tumour stroma and the pericellular area surrounding tumour cells. It is a linear, negatively charged polysaccharide composed of repeating disaccharides of N-acetylglucosamine and glucuronate [133]. HA is produced by at least three essential intracellular plasma membrane proteins, HA synthase 1-3 (Has1-3) [134]. As the primary ligand of CD44, HA is a major CSCs-associated macromolecule involved in the regulation of cell stemness and drug resistance via the stimulation of EMT, ROS, secretion of extracellular vesicles/exosomes and epigenetic factors regulation (137]. In head and neck CSCs, HA binding to CD44 prompts upregulation of OCT4, Nanog and SOX2 expression, the hallmark of stem cell properties such as spheroid and clone formation, as well as cell growth, self-renewal, poor differentiation and additional chemoresistance in these head and neck CSCs [134].
In addition, HA interacts with CD44 to activate several oncogenic signalling pathways-associated cell surface receptors or domains, such as EGFR, c-Met, HER2, transforming growth factor-beta receptor type 1 (TGFβR1) and non-receptor kinases (Src family). This will consequently promote tumour growth via the activation of MAPK and PI3K/AKT signalling pathways as well as promoting chemoresistance, cell motility and metastasis-related pathways. For instance, HA binding to CD44 recruits RTKs, which in turn promotes cell survival and migration, conferring poor prognostic in pancreatic ductal adenocarcinomas [135]. In malignant pleural mesothelioma, HA and CD44 have been shown to ease cell motility, invasion and consequently, tumour progression [136].
In melanoma cells, the HA–CD44 axis increased cell proliferation [137] and promoted the expression of the inhibitor of differentiation/DNA binding (Id) proteins 1 and 3 via the BMP type II receptor (BMPR) to decrease OS in melanoma patients [138]. Depending on the CD44–HA interaction, liver cancer cells were found to roll on HA enriched endothelial cells, an essential step during metastasis [139]. In acute myeloid leukaemia cells, the binding of CD44 and HA enhanced cell proliferation, and blocking this binding significantly inhibited the growth of CD44+ leukaemia cells [140]. The HA–CD44 axis also activated ERM, ankyrin, Grb2, Gab-1 and Vav2, which drive cell migration via RAS, RhoA and Rac GTPase families [120,133,136,141]. The presence of HA in the tissue can enhance matrix stiffness, and in turn, conjointly modulate cell behaviour. High molecular weight HA concentration significantly impacted some biophysical matrix variations such as viscoelastic properties, specifically reducing the shear storage modulus and increasing compressive resistance [142]. As the inner tumour is disposed to compression, its stiffness is mainly determined by HA due to the fixed negative charges that create hydrated and gel-like regions within the tumour capable of resisting compressive stress [143]. An increase in HA accumulation can be found in advanced tumours and is frequently correlated with aggressive malignancy. Wound healing-associated myofibroblasts and activated cancer-associated fibroblasts (CAF) are presumed to be the main sources of HA in the range of 480 kDa for both cells. Tumours have been proposed to produce very low molecular weight HA, which is specifically regulated by CAF due to the high expression of HAS2 and hyaluronidase (HYAL1) in CAF, which might contribute to greater production of HA in the CAF matrix [144]. In contrast, high molecular mass HA accumulates in naked mole-rat tissues and protects against cancer. Once it is removed by either knocking down HAS2 or overexpressing the HA-degrading enzyme, HYAL2, naked mole-rat cells become susceptible to malignant transformation and easily form tumours [145,146]. The capacity of HA to interact with CD44 also depends on its molecular weight [147]. The amount of HA binding to CD44 increases as a function of HA size, with a half-maximal saturation at ~30 kDa. Furthermore, reversible interaction is confined to the smaller HA (< 10 kDa), whereas binding is irreversible with larger molecular weight HA (≥ 262 kDa) [148]. Recent work demonstrated that the direct correlation of the CD44–HA interaction on proliferation and invasiveness of melanoma cell lines is dependent on the molecular weight and the presentation form (matrix-bound or soluble) of HA. Only soluble low molecular weight HA (30-50 kDa) promoted cell proliferation and invasion in a CD44-dependent manner, while high molecular weight HA (500-750 kDa) and immobilised low molecular weight HA did not affect cell proliferation or invasion [149]. However, another study revealed that immobilised but not soluble HA enhanced co-localisation of CD44 and the receptor for hyaluronic acid-mediated motility (RHAMM), contributing to the aggressive and invasive phenotype in breast cancer cell lines [150].
Another essential ligand of CD44 is osteopontin (OPN), which is also known as early t lymphocyte activation 1 (eta-1), secreted phosphoprotein 1 (SPP1), and bone sialoprotein I (BSPI) [151]. OPN is the ECM component synthesised by osteoblasts, preosteoblasts and osteocytes, and is incorporated into the bone. Other than bone cells, OPN is also produced and secreted by odontoblasts and hypertrophic macrophages, brain, kidney, inner ear, and placenta cells [132,152,153]. The macrophage-secreted OPN in the tumour microenvironment binds to CD44 expressed by the tumour cells, which subsequently promotes clonal growth, invasion and metastasis. These effects require CD44 binding to TIAM1, which activates Rac1. Perturbing the OPN–CD44–TIAM1–Rac1 axis has been proposed as a therapeutic strategy to treat patients with metastatic bladder cancer [153]. Moreover, OPN and CD44 are highly expressed in hepatocellular carcinoma CSCs and are associated with increased incidence of tumour relapse and unfavourable prognosis [154]. Furthermore, the OPN–CD44 axis in hepatocellular carcinoma CD44+ CSCs mediated hepatitis C virus (HCV) replication and interferon (IFN) signalling as well as the expression of CSCs features, suggesting that this signalling pathway is critical for the propagation and pathogenesis of HCV in CSCs, contributing to CSCs phenotype maintenance and promoting aggressive tumour growth [155]. In addition to HA, as mentioned earlier, the dense stroma in pancreatic tumours is also rich in OPN, which sustains oncogenic signalling by interacting with CD44s and CD44v6 to enhance pancreatic cancer cells invasion [136,156]. Amongst the CD44 spliced variants, CD44v6 is highly expressed along with OPN in several cancers. Their interaction drives cancer progression and recurrence in ovarian cancer [157] and tumour cell migration in breast cancer [158]. Additionally, in colorectal cancer cell lines, OPN is overexpressed in response to hypoxic conditions and upregulates the expression of CD44s and its splice variants (specifically CD44v6), resulting in increased colorectal cancer cells radio-resistance [159]. Klement et al. [160] also revealed that both OPN and CD44 were highly expressed in human colon carcinoma compared to the normal colon. They discovered that the OPN/CD44 immune checkpoint controlled cytotoxic T lymphocytes (CD8+ T) cell activation and tumour immune evasion. Various isoforms of OPN have been identified to date, including OPN-a, OPN-b, and OPN-c, with OPN-a predominantly expressed in lung cancer. Through the CD44/NFκB pathway, OPN-a binds to, stabilises and activates CD44 expression, which in turn enhances lung cancer cell growth [161]. OPN also shares a perivascular expression pattern with CD44 in glioma CSCs and activates its signalling, increasing glioma aggressiveness, growth, stemness and radiation resistance [80]. Qiu et al. [162] indicated that the interaction of OPN and CD44 significantly promoted the progression and metastasis of advanced gastric cancer.
Matrix metalloproteinases (MMPs), another important CD44 ligand, are ubiquitously upregulated in many cancers and play important roles in promoting tumour angiogenesis, progression, invasion and metastasis [163]. MMP-14, a member of the MMPs family, has been shown to cleave CD44 to promote migration of osteosarcoma, pancreatic and breast cancer cells [164]. CD44 also induced EMT in colon carcinoma cells by upregulating MPP-14, stimulating cell invasion and migration [165]. In line with this, ovarian cancer patients who co-expressed both CD44 and MMP-14 had a poorer prognosis [166].
The CD44 signalling pathway can upregulate MMP-14 expression in basal-like breast cancers, whereby this upregulation correlates with the induction of basal-like breast cancer cells invasiveness [167]. The upregulation of MMP-2 and MMP-9 is associated with a poor prognosis in glioma and colorectal cancer patients [168,169,170]. Chetty et al. [171] reported that glioblastoma cells overexpressed CD44 and MMP-9 and their interaction controlled cell adhesion, invasion and migration. In non-small cell lung cancer, where CD44 was overexpressed [172], CD44s and MMP-2 co-expression was significantly associated with the lymph node metastasis, higher tumour TNM staging and poor patient prognosis [173]. MMP-2, together with MMP-9, participated in breast cancer cell invasion through their connection with CD44 [174]. The expression of MMP-9 and CD44 is also high in renal cell carcinoma. Through regulation by ribosomal S6 kinase 4 (RSK4), a downstream factor of the RAS/MEK/ERK signalling pathway, CD44 and MMP-9 overexpression is highly associated with the invasion and metastasis grade of metastatic clear cell renal cell carcinoma [175]. Expression of CD44 contributes to the enhanced bone metastasis in the human prostate cancer cell line PC3, grade IV prostate cancer, by promoting tumorigenicity, cell invasion, migration and HA production [176]. The role of CD44 in the invasion and migration of PC3 cells was demonstrated to be through MMP-9 activation. In the same cell line, the highly expressed runt-related transcription factor 2 (RUNX2) forms a complex with the overexpressed CD44, thereby activating many metastasis-related genes, including MMP-9, and consequently contributes to tumour sphere formation and migration [177].

Table 3.
Summary of CD44 interactomes and their effects on cancer progression.
Table 3.
Summary of CD44 interactomes and their effects on cancer progression.
| Effect | Cancer Type | Reference | |
|---|---|---|---|
| Hyaluronan (HA) | Stemness (spheroid and clone formation, self-renewal), cell growth, poor differentiation, chemoresistance | Head and neck CSCs | [134] |
| Hyaluronan (HA) | Cell survival, migration, poor prognosis | Pancreatic cancer | [135] |
| Hyaluronan (HA) | Cell motility, invasion, tumour progression | Pleural mesothelioma | [136] |
| Hyaluronan (HA) | Cell proliferation, poor survival | Melanoma | [137,138] |
| Hyaluronan (HA) | Cancer cells rolling, metastasis | Liver cancer | [139] |
| Hyaluronan (HA) | Cell proliferation | Acute myeloid leukaemia | [140] |
| Osteopontin (OPN) | Metastasis | Bladder cancer | [153] |
| Osteopontin (OPN) | Tumour growth, tumour recurrence, cell survival, metastasis, CSCs phenotype maintenance | Hepatocellular carcinoma | [154] |
| Osteopontin (OPN) | Invasion | Pancreatic cancer | [136,156] |
| Osteopontin (OPN) | Tumour progression and recurrence | Ovarian cancer | [157] |
| Osteopontin (OPN) | Migration | Breast cancer | [158] |
| Osteopontin (OPN) | Radio-resistance | Colorectal carcinoma | [159] |
| Osteopontin (OPN) | Tumour immune evasion | Colon carcinoma | [160] |
| Osteopontin (OPN) | Cell proliferation | Lung cancer | [161] |
| Osteopontin (OPN) | Aggressive growth, stemness, radio-resistance | Glioma CSCs | [80] |
| Osteopontin (OPN) | Tumour progression, metastasis | Gastric cancer | [162] |
| Matrix metalloproteinase14 (MMP-14) | Migration | Pancreatic cancer, breast cancer, osteosarcoma | [164] |
| Matrix metalloproteinase14 (MMP-14) | Invasion, migration | Colon carcinoma | [165] |
| Matrix metalloproteinase 14 (MMP-14) | Poor prognosis | Ovarian cancer | [166] |
| Matrix metalloproteinase 14 (MMP-14) | Poor prognosis, invasion | Breast cancer | [167] |
| Matrix metalloproteinase 9 (MMP-9) | Cell adhesion, invasion, migration | Glioblastoma | [171] |
| Matrix metalloproteinase 9 (MMP-9) | Invasion, metastasis stage | Renal carcinoma | [175] |
| Matrix metalloproteinase 9 (MMP-9) | Invasion, migration, sphere formation | Prostate cancer | [177] |
| Matrix metalloproteinase 2 (MMP-2) | Lymph node metastasis, histopathological grade, TNM stage, poor prognosis | Non-small cell lung cancer | [173] |
| Matrix metalloproteinase 2,9 (MMP-2), (MMP-9) | Invasion | Breast cancer | [174] |
9. The Prognostic and Clinical Value of CD44 Expression in Advanced Cancer
Robust evidence supports that CD44 isoforms are closely related to the clinicopathological features of numerous cancers. CD44 is believed to undergo functional and structural alterations through malignant transformation, which contributes to the detachment of cancer cells from their original site, which then go on to invade the surrounding tissues. Immunohistochemistry analyses of CD44 expression in mucoepidermoid carcinoma revealed that high CD44 expression was significantly correlated with advanced tumours and increased recurrence or metastasis [178]. Similarly, there was a positive correlation between CD44 and tumour grade in salivary gland tumours [179]. Moreover, an analysis of human breast cancer tissues demonstrated that greater CD44 expression is linked to a higher histological tumour grade [180,181,182,183]. However, in patients with invasive breast cancer, CD44 expression was not associated with clinicopathological factors including histological grade, tumour size, tumour stage or metastasis status, except for one positive correlation with HER2 negative status [184]. Recent data by Roosta et al. [185] also demonstrated no relationship between CD44 expression and any clinicopathologic parameters in breast cancer, except with higher tumour stages. Likewise, a meta-analysis to address the prognostic significance of CD44 expression in breast cancer showed no noteworthy association between CD44 and OS or DFS [186]. Since the reported associations are diverse, additional studies with larger prospective cohorts are needed to further evaluate the association between CD44 expression and the prognosis of patients with breast cancer.
Moreover, high CD44 expression was also found in low-grade glioma and glioblastoma tissues compared to normal brain tissues, with the CD44 expression level serving as a predictive marker for OS rate in low-grade glioma patients [187]. Recently, Lee et al. [188] found that the expression of CD44 was positively correlated with a higher histological nuclear and shorter OS in clear renal cell carcinoma and nonclear renal cell carcinoma. Zanjani et al. [189] also reported that CD44 overexpression is statistically associated with more aggressive tumour behaviour, tumour grade and poor survival in clear renal cell carcinoma. However, in patients with chromophobe and papillary renal carcinoma cells, CD44 expression was not significantly correlated with prognosis. He et al. [72] reported that CD44 expression was significantly higher in gallbladder cancer patients with an advanced TNM stage, metastasised and poorly differentiated tumours. Moreover, a Kaplan–Meier analysis confirmed that the OS of patients with high CD44 expression was markedly poorer than those with low CD44 expression. Similarly in cholangiocarcinoma, the positive expression of CD44 was significantly related to large tumour size, high histologic grade, lymph node metastasis and distant metastasis. Taken together, these results demonstrate that the CD44-positive tumours suggest a poorer prognosis [190]. While some studies found no correlation between total CD44 and/or CD44v6 expression and clinical outcomes of patients with gastric cancer [191,192], conversely, the meta-analysis of Fang et al. [193] revealed that overexpression of total CD44 and/or CD44v6 is positively correlated with the TNM stage, T category, N category, invasion and distant metastasis. Moreover, their overexpression predicts a poor OS rate.
Many studies have also been conducted to investigate the relationship between CD44 expression and ovarian cancer progression and prognosis with contradictory results. Conic et al. [194] reported that low expression of CD44 was observed more frequently in advanced FIGO stage tumours and higher-grade tumours. Additionally, the mean survival was significantly longer in patients with high CD44 expression compared to those with low or absent CD44 expression. In the same cancer, CD44 overexpression was positively associated with progressive histologic grade and FIGO stage. In addition, multivariate analysis showed that the upregulation of CD44 was an independent predictive and prognostic factor for both OS and DFS of patients with ovarian cancer [195]. A recent meta-analysis also revealed that CD44 expression was significantly associated with a high TNM stage and poor OS in ovarian cancer patients [196]. CD44 is also a strong prognosticator of disease-specific survival (DSS) and nodal invasion in high-grade invasive urothelial carcinoma of the bladder when simultaneously expressed with fibroblast activation protein (FAP) [197]. The expression of CD44 in patients with pancreatic neuroendocrine tumours was positively related to poor tumour differentiation, high histological grade and an advanced stage. Survival analysis showed that CD44 was an important prognostic factor for OS and/or DFS. Moreover, in patients with no or low expression of CD44, a 100% DFS rate was observed, demonstrating a low recurrence risk [198]. In colorectal cancer, there was a strong statistically significant relationship between overexpressed CD44 in the primary colorectal carcinoma cell membrane and tumour grading, the degree of lymphocytic infiltration, lymphovascular invasion, peritumoral budding, lymph node ratio and lymph node metastasis status. CD44 was also correlated with OS reduction, representing an independent prognostic factor [199]. In oral cavity squamous cell carcinoma, some studies indicated no prognostic value of CD44 expression. For instance, Chen et al. [200] found no significant correlation between CD44 and T category, N category, tumour grade or survival. Another study revealed reduced CD44 expression in the advanced grades of oral cavity squamous carcinoma [201], which is in line with the data of Krump and Ehrmann [202]. However, numerous studies on oral carcinoma reported contradictory results, finding that CD44 overexpression was significantly associated with poorer histopathologic differentiation, higher tumour budding, invasion, lymph node status and metastasis. CD44 was also identified as an independent prognostic factor for poor OS, DSS and DFS in patients with advanced oral cancer [203]. High CD44 expression in oral squamous carcinoma cells is associated with increased depth of invasion (DOI), which predicts occult lymph node metastasis [204]. Another recent study revealed that CD44 overexpression was also associated with advanced T classification, lymphovascular invasion, perineural invasion, poor OS, DSS and recurrence-free survival (RFS) in patients with oral cancer [205].
The conflicting data indicate that the role of CD44 is still controversial, with many authors arguing whether CD44 is significantly associated with poorer prognosis in many cancers including breast, gastric, ovarian and oral cancers. However, it seems that larger analyses demonstrate that increased CD44 expression has an unlimited correlation with higher histological tumour grade and an advanced clinical tumour stage. Overexpression of CD44 within numerous tumours, such as mucoepidermoid carcinoma, salivary gland tumours, breast cancer, gliomas, renal carcinoma, gallbladder cancer, cholangiocarcinoma, gastric cancer, ovarian cancer, bladder carcinoma, pancreatic tumours, colorectal cancer and oral cancer has been associated with increased tumorigenicity and decreased overall survival, indicating a poorer prognosis. Figure 6 presents the most relevant pathological features triggered by CD44 in various cancers.
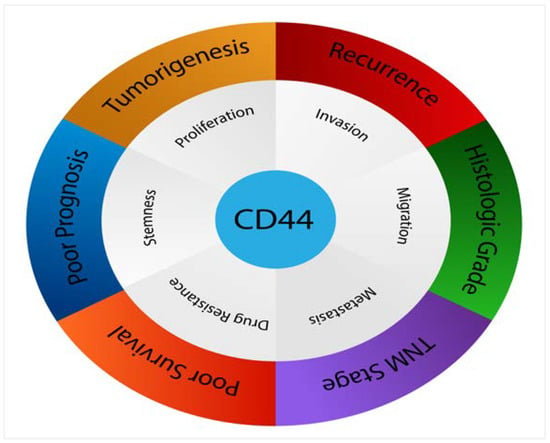
Figure 6.
CD44 affects numerous pathological processes in cancers.
10. Targeting CD44: A Promising Cancer Therapeutic Strategy
Targeted therapies are designed to specifically inhibit or block the aberrantly activated signalling pathways in tumour cells. Due to its widespread roles in promoting tumorigenesis, the inhibition of CD44 could impede tumour growth or sensitise tumour cells to therapy. CD44-targeted therapies include antibodies, peptides, pharmacological and natural inhibitors, HA-modified nanocarriers, siRNAs and CAR T cells therapy.
In the context of CD44-targeted antibody treatment, Arabi et al. [206] compared the antitumour activity of Doxil and monoclonal antibody (mAb)-modified Doxil against CD44, showing a significant improvement in cellular uptake of CD44-targeted, (mAb)-modified Doxil in CD44+ murine colon carcinoma cells compared to Doxil. Additionally, CD44-targeted, mAb-modified Doxil mice demonstrated a higher doxorubicin concentration inside the tumour cells compared to Doxil-treated mice. However, CD44- mouse embryonic fibroblast cells showed similar uptake and cytotoxicity between the CD44-targeted, mAb-modified Doxil and Doxil treatments. Another antibody, the humanised mAb (RG7356), was shown to be cytotoxic in chronic lymphocytic leukaemia cells, especially the leukaemia B cells that overexpress CD44, and had little effect on normal B cells. Moreover, RG7356 induced rapid internalisation of CD44 in chronic lymphocytic leukaemia cells expressing the zeta-associated protein of 70 kDa (ZAP-70), resulting in ZAP-70 inhibition and subsequently promoting caspase-dependent apoptosis [207]. The transcriptomic profiling of human breast tumours and mouse stroma cells revealed that RG7356 induced a significant immune-stimulatory effect by binding to CD44+ tumour cells, resulting in the secretion of chemoattractants that are essential for immune cell recruitment (i.e., macrophages) to the tumour site, finally leading to antibody-dependent cellular phagocytosis (ADCP) of the cancerous cells by macrophages [208]. In the PC3 prostate cancer cell line, CD44 was discovered to carry the mAb F77 (a developed prostate cancer-specific mAb) epitope at the exon 14 region, in which F77 induced apoptosis in this cell line in a CD44-dependent manner. Meanwhile, CD44 knockdown almost completely inhibited F77-induced apoptosis [209]. In addition, the anti-CD44 mAb A3D8 enhanced apoptosis in acute myeloid leukaemia cells through caspase-8 activation by binding to CD44s protein [210]. In human ovarian cancer cell lines overexpressing CD44, the encapsulated glycosylated paclitaxel liposomes (gPTX-L) conjugated with anti-CD44 antibody efficiently enhanced cytotoxicity in vitro and in vivo, suppressing tumour growth in vivo [211].
In addition to antibody-mediated treatment strategy, potent synthetic peptides can also selectively bind to CD44 and serve as blocking agents. Peptides could be superior to antibodies for diagnostic and therapeutic purposes because of their robust physicochemical properties. Park et al. [212] developed a novel detection peptide as an alternative to antibodies for detecting CD44 in breast CSCs. They discovered seven different peptides (P1–P7) that bind comparably to CD44, in which P7 (FNLPLPSRPLLR) exhibited the highest specificity and affinity for CD44. Similarly, a polyvalent-directed peptide polymer (PDPP) was fabricated by conjugating the combinational peptides P6 and P7 on the poly-D-lysine (PDL) polymer to replace antibodies to recognise breast CSCs. PDPP had elevated affinity and thus inhibition potential against the CD44 biomarker in breast CSCs [213]. By targeting CD44, a short cationic antimicrobial peptide (CM11) loaded in HA/chitosan nanoparticles showed significantly higher cytotoxicity and apoptosis against several cancer cells including lung adenocarcinoma, neuroblastoma and pancreatic carcinoma cell lines compared to nanoparticles without HA coating [214].
Other than directly targeting CD44, several natural compounds and chemotherapeutic agents can indirectly inhibit the overexpressed CD44 isoforms in cancer cells and CSCs. Salinomycin (SLM) is a monocarboxylic polyether antibiotic isolated from Streptomyces albus, which, when incorporated with HA, targets and reduces the CD44+ CSCs population. Furthermore, a combination of HA-coated SLM nanoparticles and paclitaxel (PTX) nanoparticles showed higher cytotoxicity against CD44+ CSCs [215]. Also, sulfasalazine (SSZ), an inhibitor of the cystine-glutamate transporter subunit (xCT) interacts with CD44v and reduces the survival of human gastric CD44v+ CSCs both in vitro and in vivo [216]. Zerumbone (ZER), a monocyclic terpene derived from Southeast Asian ginger, suppressed CD44 expression in breast cancer cells through the inhibition of the STAT3 pathway [217]. Likewise, combined treatment of epigallocatechin gallate (EGCG) and curcumin-suppressed breast CSCs by reducing the CD44+ CSCs population and inhibiting STAT3 and NFκB signalling pathways [218]. Silibinin, a natural standardised extract of the milk thistle seeds, together with 5-fluorouracil (5-FU), inhibited CD44v6+ subpopulation proliferation in human colon carcinoma cells, and when CD44v6 was knocked down, cell sphere formation and migration were suppressed whereas apoptotic and autophagic cell death pathways were induced [219]. In glioblastoma cells, Galangin (3,5,7-trihydroxyflavone), a natural flavonoid in plants, was observed to inhibit CD44 and EMT through vascular endothelial growth factor (VEGF) downregulation, suppressing the proliferation, invasion migration and angiogenesis of tumour cells [220]. Apigenin (4′,5,7-trihydroxyflavone), another flavonoid compound present in many plants, induced apoptosis and inhibited prostate CD44+ CSCs and PC3 cell survival and migration mainly through the PI3K/AKT/NF-κB signalling pathway [221].
Researchers have taken advantage of the ability of HA to target and bind to CD44 in targeting CD44-overexpressing cancer cells. Eliaz and Szoka [222] provided evidence of the influential delivery of chemotherapeutic agents to cancer cells highly expressing CD44 by HA-modified liposomes. In this study, HA-targeted liposomes bound to the CD44+o-overexpressing B16F10 murine melanoma cell line but not to the CV-1 African green monkey kidney cell line, which expressed low levels of CD44. Moreover, doxorubicin (DOX), when encapsulated in HA-targeted liposomes, was significantly more potent than the nonencapsulated form in killing the cells, expressing high levels of CD44. Spadea et al. [223] evaluated the expression of CD44 isoforms and HA-internalisation efficacy in human dermal fibroblasts (HDFs) and cancer cell lines including prostate, thyroid, head and neck, breast, ovarian, pancreatic, colorectal and endometrial cancers. They found a positive correlation between the expression of CD44s and HA uptake level. Additionally, CD44s+ HDFs were less effective in the uptake of HA compared to CD44s+ cancer cells, indicating that HA targets CD44s expressed on cancer cells better than CD44s expressed on non-cancer cells. In gastric cancer, CD44 and HER2 are considered key molecules that participate in many crucial cellular processes. SN38 (7-ethyl-10-hydroxy-camptothecin) was successfully delivered to human gastric solid tumours through encapsulation in hybrid NPs comprised of a nanoparticle core made of PLGA and a lipoid shell synthesised by conjugating the AHNP peptides and n-hexadecylamine (HDA) to the carboxyl groups of HA. HA and AHNP on the nanoparticle surface allowed superior delivery of SN38 to gastric cancer cells by targeting CD44 and HER2, leading to repressed relative signalling cascades and inhibition of cell growth and invasion [224]. In addition, HA modification efficiently facilitated the delivery of curcumin (CUR)/DOX nanoparticles to hepatocellular carcinoma and human non-small cell lung cancer (NSCLC) for the treatment of multidrug resistance (MDR) cells through CD44 receptor-mediated targeted delivery [225].
Small interfering RNAs (siRNAs) are silencing RNAs that cause gene silencing through the repression of translation. A significant knockdown of CD44 expression was achieved by transfecting a designed siRNA into NSCLC cells. The inhibition of CD44 expression in these cells suppressed cell proliferation and colony formation ability [172]. Similarly, siRNA was used to inhibit CD44 expression in EGFR wild-type NSCLC cells and the downregulation of CD44 attenuated cell growth, promoted cell cycle arrest at the G0/G1 stage and stimulated cell apoptosis. Furthermore, CD44 inhibition significantly augmented the degradation of EGFR and enhanced the sensitivity of cells to cisplatin [226]. HA/PEI and HA/PEG nanoparticles were used to deliver multidrug resistance 1 (MDR1) siRNA in CD44+ ovarian cancer cells in combination with PTX, resulting in MDR1 downregulation, increasing apoptosis and the suppression of ovarian cancer growth [227]. In colon cancer, selective targeting of CD44+ cells was achieved via delivery of anti-KRAS siRNA loaded in poly hexamethylene biguanide (PHMB) and a chitosan complex coated with HA [228]. In the same cancer, CD44 was targeted directly by ON-TARGET plus human CD44 siRNA or indirectly by silencing mucin (MUC5AC) gene expression using a small hairpin RNA construct (pSUPER-Retro-shMUC5AC), resulting in decreased expression of CD44 cell migratory and invasion downstream signalling molecules, such as phosphorylated Src, AKT and integrin-β4 [229].
Recently, CD44 has been considered an attractive target for chimeric antigen receptor T cell (CAR T cells) therapy. Porcellini et al. [230] investigated the antitumour activity of the CD44v6-CAR T cells in some solid tumours broadly expressing CD44v6 in most cell lines including lung and ovarian carcinomas. They found that the generated CD44v6-CAR T cells controlled tumour growth and extend OS in the abovementioned two solid tumours in vivo. The antitumour activity of CD44-CAR T cells was also investigated for hepatocellular carcinoma in vitro and in vivo. CD44-CAR T cells had stronger tumour growth suppression capacity and prolonged survival in CD44+ hepatocellular carcinoma xenograft mice compared to normal and mock T cells [231]. The constructed bispecific tumour-targeted T cell engager (BiTE) molecule specific for CD44v6 was incorporated into an oncolytic helper binary adenovirus (CAdDuo) encoding an immune checkpoint blocker (PD-L1Ab) and immunostimulatory cytokine (interleukin [IL]-12) to form CAdTrio. This CD44-CAdTrio allowed HER2-CAR T cells to kill numerous CD44v6+ head and neck carcinoma cell lines and improve tumour control and survival [232]. A recent review by Alhabbab [233] concluded that the CAR T cells presently in use have a great success rate in leukaemia and, to some extent, in patients with solid tumours. However, according to the author, to date, no clinical trial has reported CD44-CAR T cells for the treatment of solid tumours. Based on these reports, CD44-CAR T cells induce remarkable tumour growth inhibition in several CD44-positive carcinoma cells and xenograft mice with no reported signs of CD44-CAR T cells mediating toxicity towards healthy tissues. Such findings sound promising; however, CD44-CAR T cell therapy has yet to be applied clinically. Since xenograft mice models do not fully reflect the nature of the human immune system, one major reason could be that CD44 is indeed expressed by many healthy cells, including T cells, fibroblasts and macrophages, and its targeting using CAR T cells might mediate toxicity in cancer patients.
11. Conclusions
Expansive evidence indicates that both CD44s and CD44v isoforms are overexpressed in a variety of cancers and regulated by several signalling networks. These isoforms play crucial roles in the development of various cancers, for instance, through their interaction with ligands such as hyaluronan, osteopontin and matrix metalloproteinases. Extensive studies revealed that both CD44s and CD44v play different vital roles in enhancing several carcinogenic processes including EMT, cell growth, proliferation, invasion, migration, metastasis, tumour initiation, stemness and therapeutic resistance. However, the conflicting data reported show that further investigation is required to explore what isoform has more impact on the key features of tumorigenicity and to define the fundamental mechanisms by which these isoforms promote tumorigenicity and tumour aggressiveness.
Regarding the conflicting reports, it is also debatable whether CD44 is significantly associated with a poorer prognosis in many cancers; however, more robust analyses demonstrate that increased CD44 expression has an unlimited correlation with higher histological tumour grade, advanced clinical tumour stage and shorter survival and indicates a poor prognosis. For these reasons, CD44 is a promising target for cancer therapy, particularly for tumours overexpressing CD44. Targeting CD44 isoforms may reverse some malignant behaviours and sensitise tumour cells to therapy. Current CD44-targeted therapies include antibodies, peptides, pharmacological and natural inhibitors, HA-modified nanocarriers, siRNAs and CAR T cells therapy. Despite the success of these approaches, along with mediated tumour growth inhibition in several CD44+ carcinoma cells and xenograft mice, they have yet to be translated into clinical application in human trials. Since xenograft mice models do not fully reflect the nature of the human body, one major reason could be that CD44 is indeed expressed by many healthy cells and its targeting might mediate toxicity in cancer patients. Accordingly, further analyses are still required before the translation to clinic trial. A summary of carcinogenic mechanisms and signalling pathways induced by CD44, as well as CD44 regulators, ligands, prognostic value and possible targeting strategies is shown in Figure 7.
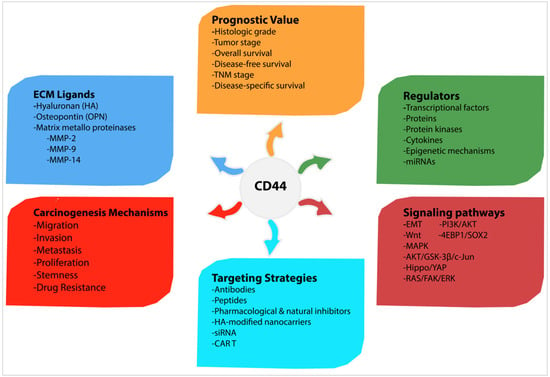
Figure 7.
Summary of carcinogenic mechanisms and signalling pathways induced by CD44, as well as CD44 regulators, ligands, prognostic value and possible targeting strategies.
Author Contributions
Conceptualization, A.S. and M.H.M.; methodology, S.E.S. and M.A.M.; validation, A.S.; investigation, M.H.M.; resources, A.S.; writing—original draft preparation, M.H.M.; writing—review and editing, S.E.S. and M.A.M.; visualization, M.H.M.; supervision, A.S.; project administration, M.H.M.; funding acquisition, A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universiti Putra Malaysia under Geran Putra Berimpak grant number UPM/700-1/2/GPPI/2017/9541700 and the APC was funded by Research Management Center Universiti Putra Malaysia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Thapa, R.; Wilson, G.D. The Importance of CD44 as a Stem Cell Biomarker and Therapeutic Target in Cancer. Stem Cells Int. 2016, 2016, 2087204. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Dalchau, R.; Kirkley, J.; Fabre, J.W. Monoclonal antibody to a human brain-granulocyte-T lymphocyte antigen probably homologous to the W 3/13 antigen of the rat. Eur. J. Immunol. 1980, 10, 745–749. [Google Scholar] [CrossRef]
- Hughes, E.N.; Mengod, G.; August, J.T. Murine cell surface glycoproteins. Characterization of a major component of 80,000 daltons as a polymorphic differentiation antigen of mesenchymal cells. J. Biol. Chem. 1981, 256, 7023–7027. [Google Scholar] [CrossRef]
- Stamenkovic, I.; Amiot, M.; Pesando, J.M.; Seed, B. A lymphocyte molecule implicated in lymph node homing is a member of the cartilage link protein family. Cell 1989, 56, 1057–1062. [Google Scholar] [CrossRef]
- Goodfellow, P.N.; Banting, G.; Wiles, M.V.; Tunnacliffe, A.; Parkar, M.; Solomon, E.; Dalchau, R.; Fabre, J.W. The gene, MIC4, which controls expression of the antigen defined by monoclonal antibody F10.44.2, is on human chromosome 11. Eur. J. Immunol. 1982, 12, 659–663. [Google Scholar] [CrossRef]
- Colombatti, A.; Hughes, E.N.; Taylor, B.A.; August, J.T. Gene for a major cell surface glycoprotein of mouse macrophages and other phagocytic cells is on chromosome 2. Proc. Natl. Acad. Sci. USA 1982, 79, 1926–1929. [Google Scholar] [CrossRef] [PubMed]
- Zeilstra, J. CD44 Isoforms in Intestinal Cancer: Identity and Functions. Ph.D. Thesis, Faculty of Medicine (AMC), University of Amsterdam, Amsterdam, The Netherlands, 2020. [Google Scholar]
- Ouhtit, A.; Rizeq, B.; Saleh, H.A.; Rahman, M.M.; Zayed, H. Novel CD44-downstream signaling pathways mediating breast tumor invasion. Int. J. Biol. Sci. 2018, 14, 1782–1790. [Google Scholar] [CrossRef]
- Loh, T.J.; Moon, H.; Cho, S.; Jang, H.; Liu, Y.C.; Tai, H.; Jung, D.W.; Williams, D.R.; Kim, H.R.; Shin, M.G.; et al. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol. Rep. 2015, 34, 1231–1238. [Google Scholar] [CrossRef]
- Rall, C.J.; Rustgi, A.K. CD44 isoform expression in primary and metastatic pancreatic adenocarcinoma. Cancer Res. 1995, 55, 1831–1835. [Google Scholar]
- Mishra, M.N.; Chandavarkar, V.; Sharma, R.; Bhargava, D. Structure, function and role of CD44 in neoplasia. J. Oral Maxillofac. Pathol. 2019, 23, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Tölg, C.; Hofmann, M.; Herrlich, P.; Ponta, H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993, 21, 1225–1229. [Google Scholar] [CrossRef]
- Screaton, G.R.; Bell, M.V.; Jackson, D.G.; Cornelis, F.B.; Gerth, U.; Bell, J.I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 1992, 89, 12160–12164. [Google Scholar] [CrossRef] [PubMed]
- Zeilstra, J.; Joosten, S.P.; van Andel, H.; Tolg, C.; Berns, A.; Snoek, M.; Van De Wetering, M.; Spaargaren, M.; Clevers, H.; Pals, S.T. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene 2014, 33, 665–670. [Google Scholar] [CrossRef]
- Goodison, S.; Urquidi, V.; Tarin, D. CD44 cell adhesion molecules. Mol. Pathol. 1999, 52, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.L.; Li, D.; Lu, T.X.; Chang, S.W. Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms. Int. J. Mol. Sci. 2020, 21, 336. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef]
- Wolf, K.J.; Shukla, P.; Springer, K.; Lee, S.; Coombes, J.D.; Choy, C.J.; Kenny, S.J.; Xu, K.; Kumar, S. A mode of cell adhesion and migration facilitated by CD44-dependent microtentacles. Proc. Natl. Acad. Sci. USA 2020, 117, 11432–11443. [Google Scholar] [CrossRef]
- Hassn Mesrati, M.; Behrooz, A.B.; Abuhamad, A.Y.; Syahir, A. Understanding Glioblastoma Biomarkers: Knocking a Mountain with a Hammer. Cells 2020, 9, 1236. [Google Scholar] [CrossRef]
- Ludwig, N.; Szczepanski, M.J.; Gluszko, A.; Szafarowski, T.; Azambuja, J.H.; Dolg, L.; Gellrich, N.-C.; Kampmann, A.; Whiteside, T.L.; Zimmerer, R.M. CD44(+) tumor cells promote early angiogenesis in head and neck squamous cell carcinoma. Cancer Lett. 2019, 467, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Gzil, A.; Zarębska, I.; Bursiewicz, W.; Antosik, P.; Grzanka, D.; Szylberg, Ł. Markers of pancreatic cancer stem cells and their clinical and therapeutic implications. Mol. Biol. Rep. 2019, 46, 6629–6645. [Google Scholar] [CrossRef] [PubMed]
- Durko, L.; Wlodarski, W.; Stasikowska-Kanicka, O.; Wagrowska-Danilewicz, M.; Danilewicz, M.; Hogendorf, P.; Strzelczyk, J.; Malecka-Panas, E. Expression and Clinical Significance of Cancer Stem Cell Markers CD24, CD44, and CD133 in Pancreatic Ductal Adenocarcinoma and Chronic Pancreatitis. Dis. Markers 2017, 2017, 3276806. [Google Scholar] [PubMed]
- Li, W.; Qian, L.; Lin, J.; Huang, G.; Hao, N.; Wei, X.; Wang, W.; Liang, J. CD44 regulates prostate cancer proliferation, invasion and migration via PDK1 and PFKFB4. Oncotarget 2017, 8, 65143–65151. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, Y.; Xie, L.; Huang, A.; Xue, C.; Gu, Z.; Wang, K.; Zong, S. The Prognostic and Clinical Value of CD44 in Colorectal Cancer: A Meta-Analysis. Front. Oncol. 2019, 9, 309. [Google Scholar] [CrossRef]
- Holah, N.S.; Aiad, H.A.; Asaad, N.Y.; Elkhouly, E.A.; Lasheen, A.G. Evaluation of the role of CD44 as a cancer stem cell marker in colorectal carcinoma: Immunohistochemical study. Menoufia Med. J. 2017, 30, 174–183. [Google Scholar]
- Zhang, H.; Brown, R.L.; Wei, Y.; Zhao, P.; Liu, S.; Liu, X.; Deng, Y.; Hu, X.; Zhang, J.; Gao, X.D.; et al. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019, 33, 166–179. [Google Scholar] [CrossRef]
- Zhao, P.; Xu, Y.; Wei, Y.; Qiu, Q.; Chew, T.L.; Kang, Y.; Cheng, C. The CD44s splice isoform is a central mediator for invadopodia activity. J. Cell Sci. 2016, 129, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.L.; Reinke, L.M.; Damerow, M.S.; Perez, D.; Chodosh, L.A.; Yang, J.; Cheng, C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Investig. 2011, 121, 1064–1074. [Google Scholar] [CrossRef]
- Mima, K.; Okabe, H.; Ishimoto, T.; Hayashi, H.; Nakagawa, S.; Kuroki, H.; Watanabe, M.; Beppu, T.; Tamada, M.; Nagano, O.; et al. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 2012, 72, 3414–3423. [Google Scholar] [CrossRef]
- Miwa, T.; Nagata, T.; Kojima, H.; Sekine, S.; Okumura, T. Isoform switch of CD44 induces different chemotactic and tumorigenic ability in gallbladder cancer. Int. J. Oncol. 2017, 51, 771–780. [Google Scholar] [CrossRef]
- Chen, Q.; Gu, M.; Cai, Z.k.; Zhao, H.; Sun, S.C.; Liu, C.; Zhan, M.; Chen, Y.B.; Wang, Z. TGF-β1 promotes epithelial-to-mesenchymal transition and stemness of prostate cancer cells by inducing PCBP1 degradation and alternative splicing of CD44. Cell. Mol. Life Sci. 2021, 78, 949–962. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, C.; Chang, K.; Karnad, A.; Jagirdar, J.; Kumar, A.P.; Freeman, J.W. CD44 Expression Level and Isoform Contributes to Pancreatic Cancer Cell Plasticity, Invasiveness, and Response to Therapy. Clin. Cancer Res. 2016, 22, 5592–5604. [Google Scholar] [CrossRef]
- Tsubouchi, K.; Minami, K.; Hayashi, N.; Yokoyama, Y.; Mori, S.; Yamamoto, H.; Koizumi, M. The CD44 standard isoform contributes to radioresistance of pancreatic cancer cells. J. Radiat. Res. 2017, 58, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Ichikawa, Y.; Zheng, Y.W.; Oshima, T.; Miyata, H.; Nakazawa, K.; Guan, H.B.; Shiozawa, M.; Akaike, M.; Watanabe, K.; et al. Prognostic significance of CD44 variant 2 upregulation in colorectal cancer. Br. J. Cancer 2014, 111, 365–374. [Google Scholar] [CrossRef]
- Gotoda, T.; Matsumura, Y.; Kondo, H.; Saitoh, D.; Shimada, Y.; Kosuge, T.; Kanai, Y.; Kakizoe, T. Expression of CD44 variants and its association with survival in pancreatic cancer. Jpn. J. Cancer Res. 1998, 89, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Todoroki, K.; Ogasawara, S.; Akiba, J.; Nakayama, M.; Naito, Y.; Seki, N.; Kusukawa, J.; Yano, H. CD44v3+/CD24- cells possess cancer stem cell-like properties in human oral squamous cell carcinoma. Int. J. Oncol. 2016, 48, 99–109. [Google Scholar] [CrossRef]
- Holm, F.; Hellqvist, E.; Mason, C.N.; Ali, S.A.; Delos-Santos, N.; Barrett, C.L.; Chun, H.J.; Minden, M.D.; Moore, R.A.; Marra, M.A.; et al. Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. Proc. Natl. Acad. Sci. USA 2015, 112, 15444–15449. [Google Scholar] [CrossRef]
- Bánky, B.; Rásó-Barnett, L.; Barbai, T.; Tímár, J.; Becságh, P.; Rásó, E. Characteristics of CD44 alternative splice pattern in the course of human colorectal adenocarcinoma progression. Mol. Cancer 2012, 11, 83. [Google Scholar] [CrossRef] [PubMed]
- Spiegelberg, D.; Kuku, G.; Selvaraju, R.; Nestor, M. Characterization of CD44 variant expression in head and neck squamous cell carcinomas. Tumour Biol. 2014, 35, 2053–2062. [Google Scholar] [CrossRef]
- Endo, K.; Terada, T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: Relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J. Hepatol. 2000, 32, 78–84. [Google Scholar] [CrossRef]
- Okuyama, K.; Fukushima, H.; Naruse, T.; Yanamoto, S.; Tsuchihashi, H.; Umeda, M. CD44 Variant 6 Expression and Tumor Budding in the Medullary Invasion Front of Mandibular Gingival Squamous Cell Carcinoma Are Predictive Factors for Cervical Lymph Node Metastasis. Pathol. Oncol. Res. 2019, 25, 603–609. [Google Scholar] [CrossRef]
- Ni, J.; Cozzi, P.J.; Hao, J.L.; Beretov, J.; Chang, L.; Duan, W.; Shigdar, S.; Delprado, W.J.; Graham, P.H.; Bucci, J.; et al. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate 2014, 74, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Yanamoto, S.; Yamada, S.; Takahashi, H.; Naruse, T.; Matsushita, Y.; Ikeda, H.; Shiraishi, T.; Seki, S.; Fujita, S.; Ikeda, T.; et al. Expression of the cancer stem cell markers CD44v6 and ABCG2 in tongue cancer: Effect of neoadjuvant chemotherapy on local recurrence. Int. J. Oncol. 2014, 44, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Goto, A.; Sugiyama, M.; Matsuoka, H.; Abe, N.; Sakamoto, A.; Atomi, Y. Possible contribution of CD44 variant 6 and nuclear beta-catenin expression to the formation of budding tumor cells in patients with T1 colorectal carcinoma. Cancer 2001, 92, 2539–2546. [Google Scholar] [CrossRef]
- Li, Z.; Chen, K.; Jiang, P.; Zhang, X.; Li, X.; Li, Z. CD44v/CD44s expression patterns are associated with the survival of pancreatic carcinoma patients. Diagn. Pathol. 2014, 9, 79. [Google Scholar] [CrossRef]
- Kainz, C.; Kohlberger, P.; Tempfer, C.; Sliutz, G.; Gitsch, G.; Reinthaller, A.; Breitenecker, G. Prognostic value of CD44 splice variants in human stage III cervical cancer. Eur. J. Cancer 1995, 31, 1706–1709. [Google Scholar] [CrossRef]
- Yakushijin, Y.; Steckel, J.; Kharbanda, S.; Hasserjian, R.; Neuberg, D.; Jiang, W.; Anderson, I.; Shipp, M.A. A directly spliced exon 10-containing CD44 variant promotes the metastasis and homotypic aggregation of aggressive non-Hodgkin’s lymphoma. Blood 1998, 91, 4282–4291. [Google Scholar] [CrossRef]
- Li, D.M.; Li, S.S.; Zhang, Y.H.; Zhang, H.J.; Gao, D.L.; Wang, Y.X. Expression of human chorionic gonadotropin, CD44v6 and CD44v4/5 in esophageal squamous cell carcinoma. World J. Gastroenterol. 2005, 11, 7401–7404. [Google Scholar] [CrossRef]
- Lourenço, B.N.; Springer, N.L.; Ferreira, D.; Oliveira, C.; Granja, P.L.; Fischbach, C. CD44v6 increases gastric cancer malignant phenotype by modulating adipose stromal cell-mediated ECM remodeling. Integr. Biol. 2018, 10, 145–158. [Google Scholar] [CrossRef]
- Suwannakul, N.; Ma, N.; Midorikawa, K.; Oikawa, S.; Kobayashi, H.; He, F.; Kawanishi, S.; Murata, M. CD44v9 Induces Stem Cell-Like Phenotypes in Human Cholangiocarcinoma. Front. Cell Dev. Biol. 2020, 8, 417. [Google Scholar] [CrossRef]
- Kobayashi, K.; Matsumoto, H.; Matsuyama, H.; Fujii, N.; Inoue, R.; Yamamoto, Y.; Nagao, K. Clinical significance of CD44 variant 9 expression as a prognostic indicator in bladder cancer. Oncol. Rep. 2016, 36, 2852–2860. [Google Scholar] [CrossRef]
- Li, N.; Tsuji, M.; Kanda, K.; Murakami, Y.; Kanayama, H.; Kagawa, S. Analysis of CD44 isoform v10 expression and its prognostic value in renal cell carcinoma. BJU Int. 2000, 85, 514–518. [Google Scholar] [CrossRef]
- Iida, N.; Bourguignon, L.Y. Coexpression of CD44 variant (v10/ex14) and CD44S in human mammary epithelial cells promotes tumorigenesis. J. Cell. Physiol. 1997, 171, 152–160. [Google Scholar] [CrossRef]
- Iida, J.; Clancy, R.; Dorchak, J.; Somiari, R.I.; Somiari, S.; Cutler, M.L.; Mural, R.J.; Shriver, C.D. DNA aptamers against exon v10 of CD44 inhibit breast cancer cell migration. PLoS ONE 2014, 9, e88712. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.A.; Kallakury, B.V.; Sheehan, C.E.; Ross, J.S. Expression of CD44 standard form and variant isoforms in non-small cell lung carcinomas. Hum. Pathol. 1997, 28, 809–814. [Google Scholar] [CrossRef]
- Zhao, P.; Damerow, M.S.; Stern, P.; Liu, A.H.; Sweet-Cordero, A.; Siziopikou, K.; Neilson, J.R.; Sharp, P.A.; Cheng, C. CD44 promotes Kras-dependent lung adenocarcinoma. Oncogene 2013, 32, 5186–5190. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dall, P.; Hekele, A.; Ikenberg, H.; Göppinger, A.; Bauknecht, T.; Pfleiderer, A.; Moll, J.; Hofmann, M.; Ponta, H.; Herrlich, P. Increasing incidence of CD44v7/8 epitope expression during uterine cervical carcinogenesis. Int. J. Cancer 1996, 69, 79–85. [Google Scholar] [CrossRef]
- Omara-Opyene, A.L.; Qiu, J.; Shah, G.V.; Iczkowski, K.A. Prostate cancer invasion is influenced more by expression of a CD44 isoform including variant 9 than by Muc18. Lab. Investig. 2004, 84, 894–907. [Google Scholar] [CrossRef][Green Version]
- Hiraga, T.; Nakamura, H. Comparable roles of CD44v8-10 and CD44s in the development of bone metastases in a mouse model. Oncol. Lett. 2016, 12, 2962–2969. [Google Scholar] [CrossRef]
- Lau, W.M.; Teng, E.; Chong, H.S.; Lopez, K.A.; Tay, A.Y.; Salto-Tellez, M.; Shabbir, A.; So, J.B.; Chan, S.L. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014, 74, 2630–2641. [Google Scholar] [CrossRef]
- Yae, T.; Tsuchihashi, K.; Ishimoto, T.; Motohara, T.; Yoshikawa, M.; Yoshida, G.J.; Wada, T.; Masuko, T.; Mogushi, K.; Tanaka, H.; et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 2012, 3, 883. [Google Scholar] [CrossRef]
- Choi, E.S.; Kim, H.; Kim, H.P.; Choi, Y.; Goh, S.H. CD44v8-10 as a potential theranostic biomarker for targeting disseminated cancer cells in advanced gastric cancer. Sci. Rep. 2017, 7, 4930. [Google Scholar] [CrossRef] [PubMed]
- Kagami, T.; Yamade, M.; Suzuki, T.; Uotani, T.; Tani, S.; Hamaya, Y.; Iwaizumi, M.; Osawa, S.; Sugimoto, K.; Baba, S.; et al. High expression level of CD44v8-10 in cancer stem-like cells is associated with poor prognosis in esophageal squamous cell carcinoma patients treated with chemoradiotherapy. Oncotarget 2018, 9, 34876–34888. [Google Scholar] [CrossRef]
- Hagiwara, M.; Kikuchi, E.; Tanaka, N.; Kosaka, T.; Mikami, S.; Saya, H.; Oya, M. Variant isoforms of CD44 involves acquisition of chemoresistance to cisplatin and has potential as a novel indicator for identifying a cisplatin-resistant population in urothelial cancer. BMC Cancer 2018, 18, 113. [Google Scholar] [CrossRef]
- Olsson, E.; Honeth, G.; Bendahl, P.O.; Saal, L.H.; Gruvberger-Saal, S.; Ringnér, M.; Vallon-Christersson, J.; Jönsson, G.; Holm, K.; Lövgren, K.; et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer 2011, 11, 418. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Zhu, H.; Shao, L.; Zhu, D.; Chen, Y.W. Rho-kinase (ROK) promotes CD44v (3,8-10)-ankyrin interaction and tumor cell migration in metastatic breast cancer cells. Cell Motil. Cytoskelet. 1999, 43, 269–287. [Google Scholar] [CrossRef]
- Fujita, N.; Yaegashi, N.; Ide, Y.; Sato, S.; Nakamura, M.; Ishiwata, I.; Yajima, A. Expression of CD44 in normal human versus tumor endometrial tissues: Possible implication of reduced expression of CD44 in lymph-vascular space involvement of cancer cells. Cancer Res. 1994, 54, 3922–3928. [Google Scholar]
- Ruiz, P.; Schwärzler, C.; Günthert, U. CD44 isoforms during differentiation and development. BioEssays 1995, 17, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Stamenkovic, I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev. 1999, 13, 35–48. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xue, C.; Yu, Y.; Chen, J.; Chen, X.; Ren, F.; Ren, Z.; Cui, G.; Sun, R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag. Res. 2018, 10, 3857–3865. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Ozaki, M.; Hegab, A.E.; Hamamoto, J.; Kagawa, S.; Arai, D.; Yasuda, H.; Naoki, K.; Soejima, K.; Saya, H.; et al. Variant CD44 expression is enriching for a cell population with cancer stem cell-like characteristics in human lung adenocarcinoma. J. Cancer 2017, 8, 1774–1785. [Google Scholar] [CrossRef]
- Xia, P.; Xu, X.Y. Prognostic significance of CD44 in human colon cancer and gastric cancer: Evidence from bioinformatic analyses. Oncotarget 2016, 7, 45538–45546. [Google Scholar] [CrossRef] [PubMed]
- Li, X.P.; Zhang, X.W.; Zheng, L.Z.; Guo, W.J. Expression of CD44 in pancreatic cancer and its significance. Int. J. Clin. Exp. Pathol. 2015, 8, 6724–6731. [Google Scholar]
- Mishra, A.; Sriram, H.; Chandarana, P.; Tanavde, V.; Kumar, R.V.; Gopinath, A.; Govindarajan, R.; Ramaswamy, S.; Sadasivam, S. Decreased expression of cell adhesion genes in cancer stem-like cells isolated from primary oral squamous cell carcinomas. Tumour Biol. 2018, 40, 1010428318780859. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, I.; Quintela-Vieira, F.; Lobo, J.; Moreira-Barbosa, C.; Menezes, F.D.; Martins, A.T.; Oliveira, J.; Silva, R.; Jerónimo, C.; Henrique, R. Expression of EMT-Related Genes CAMK2N1 and WNT5A is increased in Locally Invasive and Metastatic Prostate Cancer. J. Cancer 2019, 10, 5915–5925. [Google Scholar] [CrossRef]
- Dzobo, K.; Senthebane, D.A.; Ganz, C.; Thomford, N.E. The significance of Cancer stem cell markers’ gene expression and Relevance for Survival Outcomes. OMICS J. Integr. Biol. 2020. Preprint. [Google Scholar] [CrossRef]
- Rajarajan, A.; Stokes, A.; Bloor, B.K.; Ceder, R.; Desai, H.; Grafström, R.C.; Odell, E.W. CD44 expression in oro-pharyngeal carcinoma tissues and cell lines. PLoS ONE 2012, 7, e28776. [Google Scholar] [CrossRef] [PubMed]
- Pietras, A.; Katz, A.M.; Ekström, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef]
- Wang, F.; Zheng, Z.; Guan, J.; Qi, D.; Zhou, S.; Shen, X.; Wang, F.; Wenkert, D.; Kirmani, B.; Solouki, T.; et al. Identification of a panel of genes as a prognostic biomarker for glioblastoma. EBioMedicine 2018, 37, 68–77. [Google Scholar] [CrossRef]
- Liu, M.Y.; Su, H.; Huang, H.L.; Chen, J.Q. Cancer stem-like cells with increased expression of NY-ESO-1 initiate breast cancer metastasis. Oncol. Lett. 2019, 18, 3664–3672. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K.; Sinkala, M. The Cancer Stem Cell Marker CD44 Play Key Roles in Immune Suppression/Evasion, Drug Resistance, Epithelial-Mesenchymal Transition, and Metastasis in Several Human Cancers. OMICS J. Integr. Biol. 2021, 2021, 2021010490. [Google Scholar]
- Tanabe, K.K.; Ellis, L.M.; Saya, H. Expression of CD44R1 adhesion molecule in colon carcinomas and metastases. Lancet 1993, 341, 725–726. [Google Scholar] [CrossRef]
- Prochazka, L.; Tesarik, R.; Turanek, J. Regulation of alternative splicing of CD44 in cancer. Cell. Signal. 2014, 26, 2234–2239. [Google Scholar] [CrossRef]
- Wielenga, V.J.; Smits, R.; Korinek, V.; Smit, L.; Kielman, M.; Fodde, R.; Clevers, H.; Pals, S.T. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am. J. Pathol. 1999, 154, 515–523. [Google Scholar] [CrossRef]
- Van de Wetering, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef]
- Smith, S.M.; Lyu, Y.L.; Cai, L. NF-κB affects proliferation and invasiveness of breast cancer cells by regulating CD44 expression. PLoS ONE 2014, 9, e106966. [Google Scholar] [CrossRef]
- Wang, L.; Yang, H.; Abel, E.V.; Ney, G.M.; Palmbos, P.L.; Bednar, F.; Zhang, Y.; Leflein, J.; Waghray, M.; Owens, S.; et al. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev. 2015, 29, 171–183. [Google Scholar] [CrossRef]
- Godar, S.; Ince, T.A.; Bell, G.W.; Feldser, D.; Donaher, J.L.; Bergh, J.; Liu, A.; Miu, K.; Watnick, R.S.; Reinhardt, F.; et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell 2008, 134, 62–73. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, Y.; Hao, Q.; Wang, S.; Li, H.; Li, J.; Gao, Y.; Li, M.; Li, W.; Xue, X.; et al. FOXP3 suppresses breast cancer metastasis through downregulation of CD44. Int. J. Cancer 2015, 137, 1279–1290. [Google Scholar] [CrossRef]
- Li, J.; Zhou, B.P. Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011, 11, 49. [Google Scholar] [CrossRef]
- Cheng, C.; Yaffe, M.B.; Sharp, P.A. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006, 20, 1715–1720. [Google Scholar] [CrossRef]
- Judd, N.P.; Winkler, A.E.; Murillo-Sauca, O.; Brotman, J.J.; Law, J.H.; Lewis, J.S.; Jr Dunn, G.P.; Bui, J.D.; Sunwoo, J.B.; Uppaluri, R. ERK1/2 regulation of CD44 modulates oral cancer aggressiveness. Cancer Res. 2012, 72, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yoon, C.; Zhou, X.H.; Zhou, Y.C.; Zhou, W.W.; Liu, H.; Yang, X.; Lu, J.; Lee, S.Y.; Huang, K. ERK1/2-Nanog signaling pathway enhances CD44(+) cancer stem-like cell phenotypes and epithelial-to-mesenchymal transition in head and neck squamous cell carcinomas. Cell Death Dis. 2020, 11, 266. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Cai, Q.; Zhang, M.; Zhu, S.; Ma, Y.; Sun, L.; Jiang, N.; Tian, J.; Niu, X.; Chen, J.; et al. A switch from CD44⁺ cell to EMT cell drives the metastasis of prostate cancer. Oncotarget 2015, 6, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, H.; Whelan, K.A.; Tanaka, K.; Natsuizaka, M.; Long, A.; Guo, A.; Chang, S.; Kagawa, S.; Srinivasan, S.; Guha, M.; et al. Mitochondrial SOD2 regulates epithelial–mesenchymal transition and cell populations defined by differential CD44 expression. Oncogene 2015, 34, 5229–5239. [Google Scholar] [CrossRef] [PubMed]
- Fernando, J.; Malfettone, A.; Cepeda, E.B.; Vilarrasa-Blasi, R.; Bertran, E.; Raimondi, G.; Fabra, À.; Alvarez-Barrientos, A.; Fernández-Salguero, P.; Fernández-Rodríguez, C.M.; et al. A mesenchymal-like phenotype and expression of CD44 predict lack of apoptotic response to sorafenib in liver tumor cells. Int. J. Cancer 2015, 136, E161–E172. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.N.; Ahn, D.H.; Kang, N.; Yeo, C.D.; Kim, Y.K.; Lee, K.Y.; Kim, T.J.; Lee, S.H.; Park, M.S.; Yim, H.W.; et al. TGF-β induced EMT and stemness characteristics are associated with epigenetic regulation in lung cancer. Sci. Rep. 2020, 10, 10597. [Google Scholar] [CrossRef]
- Eberth, S.; Schneider, B.; Rosenwald, A.; Hartmann, E.M.; Romani, J.; Zaborski, M.; Siebert, R.; Drexler, H.G.; Quentmeier, H. Epigenetic regulation of CD44 in Hodgkin and non-Hodgkin lymphoma. BMC Cancer 2010, 10, 517. [Google Scholar] [CrossRef]
- Yan, P.; Mühlethaler, A.; Bourloud, K.B.; Beck, M.N.; Gross, N. Hypermethylation-mediated regulation of CD44 gene expression in human neuroblastoma. Genes Chromosomes Cancer 2003, 36, 129–138. [Google Scholar] [CrossRef]
- Verkaik, N.S.; van Steenbrugge, G.J.; van Weerden, W.M.; Bussemakers, M.J.; van der Kwast, T.H. Silencing of CD44 expression in prostate cancer by hypermethylation of the CD44 promoter region. Lab. Investig. 2000, 80, 1291–1298. [Google Scholar] [CrossRef]
- Lou, W.; Krill, D.; Dhir, R.; Becich, M.J.; Dong, J.T.; Frierson, H.F.; Jr Isaacs, W.B.; Isaacs, J.T.; Gao, A.C. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res. 1999, 59, 2329–2331. [Google Scholar] [PubMed]
- Kagara, N.; Huynh, K.T.; Kuo, C.; Okano, H.; Sim, M.S.; Elashoff, D.; Chong, K.; Giuliano, A.E.; Hoon, D.S. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am. J. Pathol. 2012, 181, 257–267. [Google Scholar] [CrossRef]
- Xue, H.G.; Yang, A.H.; Sun, X.G.; Lu, Y.Y.; Tian, Z.B. Expression of microRNA-328 Functions as a Biomarker for Recurrence of Early Gastric Cancer (EGC) After Endoscopic Submucosal Dissection (ESD) by Modulating CD44. Med Sci. Monit. 2016, 22, 4779–4785. [Google Scholar] [CrossRef][Green Version]
- Ishimoto, T.; Sugihara, H.; Watanabe, M.; Sawayama, H.; Iwatsuki, M.; Baba, Y.; Okabe, H.; Hidaka, K.; Yokoyama, N.; Miyake, K.; et al. Macrophage-derived reactive oxygen species suppress miR-328 targeting CD44 in cancer cells and promote redox adaptation. Carcinogenesis 2014, 35, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef]
- Cheng, W.; Liu, T.; Wan, X.; Gao, Y.; Wang, H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012, 279, 2047–2059. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.F.; Ma, X.Q.; Wang, L.P.; Wang, W. MicroRNA-145 exerts tumor-suppressive and chemo-resistance lowering effects by targeting CD44 in gastric cancer. World J. Gastroenterol. 2017, 23, 2337–2345. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Cheng, A.J.; Lee, L.Y.; You, G.R.; Li, Y.L.; Chen, H.Y.; Chang, J.T. MiR-520b as a novel molecular target for suppressing stemness phenotype of head-neck cancer by inhibiting CD44. Sci. Rep. 2017, 7, 2042. [Google Scholar] [CrossRef]
- Liu, C.; Liu, R.; Zhang, D.; Deng, Q.; Liu, B.; Chao, H.P.; Rycaj, K.; Takata, Y.; Lin, K.; Lu, Y.; et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017, 8, 14270. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, D.; Nie, J.; Zhou, S.; Wang, J.; Tang, Q.; Yang, X. MicroRNA-143 targets CD44 to inhibit breast cancer progression and stem cell-like properties. Mol. Med. Rep. 2016, 13, 5193–5199. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jiang, J.; Badawi, M.; Schmittgen, T.D. miR-221 regulates CD44 in hepatocellular carcinoma through the PI3K-AKT-mTOR pathway. Biochem. Biophys. Res. Commun. 2017, 487, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Qin, H.; Li, Y.; Zhang, Y.; Zhuang, X.; Liu, L.; Lu, K.; Li, L.; Deng, X.; Liu, F.; et al. FNDC3B circular RNA promotes the migration and invasion of gastric cancer cells via the regulation of E-cadherin and CD44 expression. J. Cell. Physiol. 2019, 234, 19895–19910. [Google Scholar] [CrossRef]
- Bao, W.; Fu, H.J.; Xie, Q.S.; Wang, L.; Zhang, R.; Guo, Z.Y.; Zhao, J.; Meng, Y.L.; Ren, X.L.; Wang, T.; et al. HER2 interacts with CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139 in gastric cancer cells. Gastroenterology 2011, 141, 2076–2087.e6. [Google Scholar] [CrossRef]
- Park, J.; Kim, S.Y.; Kim, H.J.; Kim, K.M.; Choi, E.Y.; Kang, M.S. A reciprocal regulatory circuit between CD44 and FGFR2 via c-myc controls gastric cancer cell growth. Oncotarget 2016, 7, 28670–28683. [Google Scholar] [CrossRef]
- Nam, K.; Oh, S.; Lee, K.; Yoo, S.; Shin, I. CD44 regulates cell proliferation, migration, and invasion via modulation of c-Src transcription in human breast cancer cells. Cell. Signal. 2015, 27, 1882–1894. [Google Scholar] [CrossRef]
- Le, P.N.; Nieto, C.; Chowdhury, F.N.; Tyagi, A.; Lyons, T.R.; Young, C.D.; Zhou, H.; Somerset, H.L.; Wang, X.J.; Jimeno, A. Cancer cell CD44 mediates macrophage/monocyte-driven regulation of head and neck cancer stem cells. Cancer Res. 2020, 80, 1079. [Google Scholar]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Götte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Heldin, P.; Hascall, V.C.; Karamanos, N.K.; Skandalis, S.S.; Markwald, R.R.; Ghatak, S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011, 278, 1429–1443. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Sleeman, J. CD44 is a multidomain signaling platform that integrates extracellular matrix cues with growth factor and cytokine signals. Adv. Cancer Res. 2014, 123, 231–254. [Google Scholar]
- Orian-Rousseau, V. CD44, a therapeutic target for metastasising tumours. Eur. J. Cancer 2010, 46, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 30. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, H.; Liu, S.; Kim, C.K.; Xu, Y.; Hurley, L.A.; Nishikawa, R.; Nagane, M.; Hu, B.; Stegh, A.H.; et al. Internalized CD44s splice isoform attenuates EGFR degradation by targeting Rab7A. Proc. Natl. Acad. Sci. USA 2017, 114, 8366–8371. [Google Scholar] [CrossRef]
- Liu, S.; Cheng, C. Akt Signaling Is Sustained by a CD44 Splice Isoform-Mediated Positive Feedback Loop. Cancer Res. 2017, 77, 3791–3801. [Google Scholar] [CrossRef] [PubMed]
- Thanee, M.; Loilome, W.; Techasen, A.; Sugihara, E.; Okazaki, S.; Abe, S.; Ueda, S.; Masuko, T.; Namwat, N.; Khuntikeo, N.; et al. CD44 variant-dependent redox status regulation in liver fluke-associated cholangiocarcinoma: A target for cholangiocarcinoma treatment. Cancer Sci. 2016, 107, 991–1000. [Google Scholar] [CrossRef]
- Chang, G.; Zhang, H.; Wang, J.; Zhang, Y.; Xu, H.; Wang, C.; Zhang, H.; Ma, L.; Li, Q.; Pang, T. CD44 targets Wnt/β-catenin pathway to mediate the proliferation of K562 cells. Cancer Cell Int. 2013, 13, 117. [Google Scholar] [CrossRef]
- Lai, C.J.; Lin, C.Y.; Liao, W.Y.; Hour, T.C.; Wang, H.D.; Chuu, C.P. CD44 Promotes Migration and Invasion of Docetaxel-Resistant Prostate Cancer Cells Likely via Induction of Hippo-Yap Signaling. Cells 2019, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Okabe, H.; Ishimoto, T.; Mima, K.; Nakagawa, S.; Hayashi, H.; Kuroki, H.; Imai, K.; Nitta, H.; Saito, S.; Hashimoto, D.; et al. CD44s signals the acquisition of the mesenchymal phenotype required for anchorage-independent cell survival in hepatocellular carcinoma. Br. J. Cancer 2014, 110, 958–966. [Google Scholar] [CrossRef]
- De Falco, V.; Tamburrino, A.; Ventre, S.; Castellone, M.D.; Malek, M.; Manié, S.N.; Santoro, M. CD44 proteolysis increases CREB phosphorylation and sustains proliferation of thyroid cancer cells. Cancer Res. 2012, 72, 1449–1458. [Google Scholar] [CrossRef]
- Johansson, E.; Grassi, E.S.; Pantazopoulou, V.; Tong, B.; Lindgren, D.; Berg, T.J.; Pietras, E.J.; Axelson, H.; Pietras, A. CD44 Interacts with HIF-2α to Modulate the Hypoxic Phenotype of Perinecrotic and Perivascular Glioma Cells. Cell Rep. 2017, 20, 1641–1653. [Google Scholar] [CrossRef]
- Morath, I.; Hartmann, T.N.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81 Pt A, 166–173. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan-CD44 Interactions in Cancer: Paradoxes and Possibilities. Clin. Cancer Res. 2009, 15, 7462–7468. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.; Earle, C.; Shiina, M. Activation of Matrix Hyaluronan-Mediated CD44 Signaling, Epigenetic Regulation and Chemoresistance in Head and Neck Cancer Stem Cells. Int. J. Mol. Sci. 2017, 18, 1849. [Google Scholar] [CrossRef]
- Franklin, O.; Billing, O.; Öhlund, D.; Berglund, A.; Herdenberg, C.; Wang, W.; Hellman, U.; Sund, M. Novel prognostic markers within the CD44-stromal ligand network in pancreatic cancer. J. Pathology. Clin. Res. 2019, 5, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Dericks, L.; Schmid, R.A. CD44 and its ligand hyaluronan as potential biomarkers in malignant pleural mesothelioma: Evidence and perspectives. Respir. Res. 2017, 18, 58. [Google Scholar] [CrossRef]
- Ahrens, T.; Assmann, V.; Fieber, C.; Termeer, C.; Herrlich, P.; Hofmann, M.; Simon, J.C. CD44 is the principal mediator of hyaluronic-acid-induced melanoma cell proliferation. J. Investig. Dermatol. 2001, 116, 93–101. [Google Scholar]
- Wu, R.L.; Sedlmeier, G.; Kyjacova, L.; Schmaus, A.; Philipp, J.; Thiele, W.; Garvalov, B.K.; Sleeman, J.P. Hyaluronic acid-CD44 interactions promote BMP4/7-dependent Id1/3 expression in melanoma cells. Sci. Rep. 2018, 8, 14913. [Google Scholar] [CrossRef]
- Hanke-Roos, M.; Fuchs, K.; Maleschlijski, S.; Sleeman, J.; Orian-Rousseau, V.; Rosenhahn, A. CD44 mediates the catch-bond activated rolling of HEPG2Iso epithelial cancer cells on hyaluronan. Cell Adhes. Migr. 2017, 11, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Pęcak, A.; Skalniak, Ł.; Pels, K.; Książek, M.; Madej, M.; Krzemień, D.; Malicki, S.; Władyka, B.; Dubin, A.; Holak, T.A.; et al. Anti-CD44 DNA Aptamers Selectively Target Cancer Cells. Nucleic Acid Ther. 2020, 30, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell. Signal. 2019, 63, 109377. [Google Scholar] [CrossRef]
- Kreger, S.T.; Voytik-Harbin, S.L. Hyaluronan concentration within a 3D collagen matrix modulates matrix viscoelasticity, but not fibroblast response. Matrix Biol. 2009, 28, 336–346. [Google Scholar] [CrossRef]
- Gkretsi, V.; Stylianopoulos, T. Cell Adhesion and Matrix Stiffness: Coordinating Cancer Cell Invasion and Metastasis. Front. Oncol. 2018, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Sapudom, J.; Müller, C.D.; Nguyen, K.T.; Martin, S.; Anderegg, U.; Pompe, T. Matrix Remodeling and Hyaluronan Production by Myofibroblasts and Cancer-Associated Fibroblasts in 3D Collagen Matrices. Gels 2020, 6, 33. [Google Scholar] [CrossRef]
- Takasugi, M.; Firsanov, D.; Tombline, G.; Ning, H.; Ablaeva, J.; Seluanov, A.; Gorbunova, V. Naked mole-rat very-high-molecular-mass hyaluronan exhibits superior cytoprotective properties. Nat. Commun. 2020, 11, 2376. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Reviews. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Amorim, S.; Pashkuleva, I.; Reis, C.A.; Reis, R.L.; Pires, R.A. Tunable layer-by-layer films containing hyaluronic acid and their interactions with CD44. J. Mater. Chem. B 2020, 8, 3880–3885. [Google Scholar] [CrossRef] [PubMed]
- Wolny, P.M.; Banerji, S.; Gounou, C.; Brisson, A.R.; Day, A.J.; Jackson, D.G.; Richter, R.P. Analysis of CD44-hyaluronan interactions in an artificial membrane system: Insights into the distinct binding properties of high and low molecular weight hyaluronan. J. Biol. Chem. 2010, 285, 30170–30180. [Google Scholar] [CrossRef]
- Sapudom, J.; Ullm, F.; Martin, S.; Kalbitzer, L.; Naab, J.; Möller, S.; Schnabelrauch, M.; Anderegg, U.; Schmidt, S.; Pompe, T. Molecular weight specific impact of soluble and immobilized hyaluronan on CD44 expressing melanoma cells in 3D collagen matrices. Acta Biomater. 2017, 50, 259–270. [Google Scholar] [CrossRef]
- Carvalho, A.M.; Soares da Costa, D.; Paulo, P.M.R.; Reis, R.L.; Pashkuleva, I. Co-localization and crosstalk between CD44 and RHAMM depend on hyaluronan presentation. Acta Biomater. 2020, 8, 19. [Google Scholar]
- Mirzaei, A.; Mohammadi, S.; Ghaffari, S.H.; Yaghmaie, M.; Vaezi, M.; Alimoghaddam, K.; Ghavamzadeh, A. Osteopontin b and c Splice isoforms in Leukemias and Solid Tumors: Angiogenesis Alongside Chemoresistance. Asian Pac. J. Cancer Prev. 2018, 19, 615–623. [Google Scholar]
- Butler, W.T. The nature and significance of osteopontin. Connect. Tissue Res. 1989, 23, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Sottnik, J.L.; Dancik, G.M.; Sahu, D.; Hansel, D.E.; Theodorescu, D.; Schwartz, M.A. An Osteopontin/CD44 axis in RhoGDI2-mediated metastasis suppression. Cancer Cell 2016, 30, 432–443. [Google Scholar] [CrossRef]
- Yang, G.H.; Fan, J.; Xu, Y.; Qiu, S.J.; Yang, X.R.; Shi, G.M.; Wu, B.; Dai, Z.; Liu, Y.K.; Tang, Z.Y.; et al. Osteopontin combined with CD44, a novel prognostic biomarker for patients with hepatocellular carcinoma undergoing curative resection. Oncologist 2008, 13, 1155–1165. [Google Scholar] [CrossRef]
- Shirasaki, T.; Honda, M.; Yamashita, T.; Nio, K.; Shimakami, T.; Shimizu, R.; Nakasyo, S.; Murai, K.; Shirasaki, N.; Okada, H.; et al. The osteopontin-CD44 axis in hepatic cancer stem cells regulates IFN signaling and HCV replication. Sci. Rep. 2018, 8, 13143. [Google Scholar] [CrossRef]
- Kolb, A.; Kleeff, J.; Guweidhi, A.; Esposito, I.; Giese, N.A.; Adwan, H.; Giese, T.; Büchler, M.W.; Berger, M.R.; Friess, H. Osteopontin influences the invasiveness of pancreatic cancer cells and is increased in neoplastic and inflammatory conditions. Cancer Biol. Ther. 2005, 4, 740–746. [Google Scholar] [CrossRef]
- Shi, J.; Zhou, Z.; Di, W.; Li, N. Correlation of CD44v6 expression with ovarian cancer progression and recurrence. BMC Cancer 2013, 13, 182. [Google Scholar] [CrossRef]
- Khan, S.A.; Cook, A.C.; Kappil, M.; Günthert, U.; Chambers, A.F.; Tuck, A.B.; Denhardt, D.T. Enhanced cell surface CD44 variant (v6, v9) expression by osteopontin in breast cancer epithelial cells facilitates tumor cell migration: Novel post-transcriptional, post-translational regulation. Clin. Exp. Metastasis 2005, 22, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Wohlleben, G.; Hauff, K.; Gasser, M.; Waaga-Gasser, A.M.; Grimmig, T.; Flentje, M.; Polat, B. Hypoxia induces differential expression patterns of osteopontin and CD44 in colorectal carcinoma. Oncol. Rep. 2018, 39, 442–448. [Google Scholar] [CrossRef]
- Klement, J.D.; Paschall, A.V.; Redd, P.S.; Ibrahim, M.L.; Lu, C.; Yang, D.; Celis, E.; Abrams, S.I.; Ozato, K.; Liu, K. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Investig. 2018, 128, 5549–5560. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.J.; Wu, C.C.; Sheu, G.T.; Chang, H.Y.; Chen, M.Y.; Lin, Y.Y.; Chuang, C.Y.; Hsu, S.L.; Chang, J.T. Integrin β3 and CD44 levels determine the effects of the OPN-a splicing variant on lung cancer cell growth. Oncotarget 2016, 7, 55572–55584. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Hu, Y.; Zhang, Z.Y.; Ye, L.; Xu, F.H.; Schneider, M.E.; Ma, X.L.; Du, Y.X.; Zuo, X.B.; Zhou, F.S.; et al. Genetic association of osteopontin (OPN) and its receptor CD44 genes with susceptibility to Chinese gastric cancer patients. J. Cancer Res. Clin. Oncol. 2014, 140, 2143–2156. [Google Scholar] [CrossRef]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Park, Y.S.; Kim, H.J.; Kim, C.H.; Lim, S.W.; Huh, J.W.; Lee, J.H.; Kim, H.R. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int. J. Oncol. 2012, 41, 211–218. [Google Scholar] [PubMed]
- Vos, M.C.; Hollemans, E.; Ezendam, N.; Feijen, H.; Boll, D.; Pijlman, B.; van der Putten, H.; Klinkhamer, P.; van Kuppevelt, T.H.; van der Wurff, A.A.; et al. MMP-14 and CD44 in Epithelial-to-Mesenchymal Transition (EMT) in ovarian cancer. J. Ovarian Res. 2016, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef]
- Montgomery, N.; Hill, A.; McFarlane, S.; Neisen, J.; O’Grady, A.; Conlon, S.; Jirstrom, K.; Kay, E.W.; Waugh, D.J. CD44 enhances invasion of basal-like breast cancer cells by upregulating serine protease and collagen-degrading enzymatic expression and activity. Breast Cancer Res. 2012, 14, R84. [Google Scholar] [CrossRef]
- Langers, A.M.; Verspaget, H.W.; Hawinkels, L.J.; Kubben, F.J.; van Duijn, W.; van der Reijden, J.J.; Hardwick, J.C.; Hommes, D.W.; Sier, C.F. MMP-2 and MMP-9 in normal mucosa are independently associated with outcome of colorectal cancer patients. Br. J. Cancer 2012, 106, 1495–1498. [Google Scholar] [CrossRef]
- Zhou, W.; Yu, X.; Sun, S.; Zhang, X.; Yang, W.; Zhang, J.; Zhang, X.; Jiang, Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed. Pharmacother. 2019, 118, 109369. [Google Scholar] [CrossRef]
- Levicar, N.; Nuttall, R.K.; Lah, T.T. Acta Neurochir. 2003, 145, 825. [CrossRef]
- Chetty, C.; Vanamala, S.K.; Gondi, C.S.; Dinh, D.H.; Gujrati, M.; Rao, J.S. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell. Signal. 2012, 24, 549–559. [Google Scholar] [CrossRef]
- Hu, B.; Ma, Y.; Yang, Y.; Zhang, L.; Han, H.; Chen, J. CD44 promotes cell proliferation in non-small cell lung cancer. Oncol. Lett. 2018, 15, 5627–5633. [Google Scholar] [CrossRef]
- Zhang, L.; Li, N.; Yan, H.C.; Jiang, H.; Fang, X.J. Expression of Novel CD44st and MMP2 in NSCLC Tissues and Their Clinical Significance. Oncol. Res. Treat. 2017, 40, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, F.; Oliveira, V.M.; Silva, M.A.; Taromaru, G.C.; Rinaldi, J.F.; Aoki, T. The interaction between aromatase, metalloproteinase 2,9 and CD44 in breast cancer. Rev. Da Assoc. Med. Bras. 2010, 56, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, P.; Liu, Y.; Zhao, D.; Xu, J.; Yu, K.; Yan, Q.; Guo, S.; Wang, Z.; Fan, L. Expression of RSK4, CD44 and MMP-9 is upregulated and positively correlated in metastatic ccRCC. Diagn. Pathol. 2020, 15, 28. [Google Scholar]
- Hiraga, T.; Ito, S.; Nakamura, H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013, 73, 4112–4122. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; AlJohani, H.; Majumdar, S.; Chellaiah, M.A. Characterization of CD44 intracellular domain interaction with RUNX2 in PC3 human prostate cancer cells. Cell Commun. Signal. 2019, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Binmadi, N.; Elsissi, A.; Elsissi, N. Expression of cell adhesion molecule CD44 in mucoepidermoid carcinoma and its association with the tumor behavior. Head Face Med. 2016, 12, 8. [Google Scholar] [CrossRef]
- Fok, T.C.; Lapointe, H.; Tuck, A.B.; Chambers, A.F.; Jackson-Boeters, L.; Daley, T.D.; Darling, M.R. Expression and localization of osteopontin, homing cell adhesion molecule/CD44, and integrin αvβ3 in pleomorphic adenoma, polymorphous low-grade adenocarcinoma, and adenoid cystic carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2013, 116, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wu, K.; Tian, Y.; Liu, Q.; Han, N.; Yuan, X.; Zhang, L.; Wu, G.S.; Wu, K. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int. J. Oncol. 2016, 49, 1343–1350. [Google Scholar] [CrossRef]
- Xu, H.; Tian, Y.; Yuan, X.; Liu, Y.; Wu, H.; Liu, Q.; Wu, G.S.; Wu, K. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. OncoTargets Ther. 2016, 9, 431–444. [Google Scholar]
- McFarlane, S.; Coulter, J.A.; Tibbits, P.; O’Grady, A.; McFarlane, C.; Montgomery, N.; Hill, A.; McCarthy, H.O.; Young, L.S.; Kay, E.W.; et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget 2015, 6, 11465–11476. [Google Scholar] [CrossRef] [PubMed]
- Louhichi, T.; Ziadi, S.; Saad, H.; Dhiab, M.B.; Mestiri, S.; Trimeche, M. Clinicopathological significance of cancer stem cell markers CD44 and ALDH1 expression in breast cancer. Breast Cancer 2018, 25, 698–705. [Google Scholar] [CrossRef]
- Jang, M.H.; Kang, H.J.; Jang, K.S.; Paik, S.S.; Kim, W.S. Clinicopathological analysis of CD44 and CD24 expression in invasive breast cancer. Oncol. Lett. 2016, 12, 2728–2733. [Google Scholar] [CrossRef] [PubMed]
- Roosta, Y.; Sanaat, Z.; Nikanfar, A.R.; Dolatkhah, R.; Fakhrjou, A. Predictive Value of CD44 for Prognosis in Patients with Breast Cancer. Asian Pac. J. Cancer Prev. 2020, 21, 2561–2567. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Q.; Wang, Q.; Wang, Y.; Chen, J. Prognostic Significance of CD24 and CD44 in Breast Cancer: A Meta-Analysis. Int. J. Biol. Markers 2017, 32, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Li, Q.; Wang, M.; Hu, J.; Dai, J.; Niu, L.; Yuan, G.; Pan, Y. Elevated CD44 expression predicts poor prognosis in patients with low-grade glioma. Oncol. Lett. 2019, 18, 3698–3704. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Kim, J.M.; Lee, H.J.; Seong, I.O.; Kim, K.H. Immunohistochemical expression of CD44, matrix metalloproteinase2 and matrix metalloproteinase9 in renal cell carcinomas. Urol. Oncol. 2019, 37, 742–748. [Google Scholar] [CrossRef]
- Zanjani, L.S.; Madjd, Z.; Abolhasani, M.; Rasti, A.; Fodstad, O.; Andersson, Y.; Asgari, M. Increased expression of CD44 is associated with more aggressive behavior in clear cell renal cell carcinoma. Biomark. Med. 2018, 12, 45–61. [Google Scholar] [CrossRef]
- Wattanawongdon, W.; Simawaranon Bartpho, T.; Tongtawee, T. Expression of CD44 and MDM2 in cholangiocarcinoma is correlated with poor clinicopathologic characteristics. Int. J. Clin. Exp. Pathol. 2019, 12, 3961–3967. [Google Scholar]
- Kim, J.Y.; Bae, B.N.; Kim, K.S.; Shin, E.; Park, K. Osteopontin, CD44, and NFkappaB expression in gastric adenocarcinoma. Cancer Res. Treat. 2009, 41, 29–35. [Google Scholar] [CrossRef][Green Version]
- Fanelli, M.F.; Chinen, L.T.; Begnami, M.D.; Costa, W.L.; Jr Fregnami, J.H.; Soares, F.A.; Montagnini, A.L. The influence of transforming growth factor-α, cyclooxygenase-2, matrix metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in epithelial-mesenchymal transition on overall survival of patients with gastric cancer. Histopathology 2012, 61, 153–161. [Google Scholar] [CrossRef]
- Fang, M.; Wu, J.; Lai, X.; Ai, H.; Tao, Y.; Zhu, B.; Huang, L. CD44 and CD44v6 are Correlated with Gastric Cancer Progression and Poor Patient Prognosis: Evidence from 42 Studies. Cell. Physiol. Biochem. 2016, 40, 567–578. [Google Scholar] [CrossRef]
- Conić, I.; Stojnev, S.; Dimitrijević, A.; Jankovič-Veličković, L.; Djordjević, B.; Djordjević, I.; Pejčić, I.; Vrbić, S. Clinical significant of CD44 expression in serous ovarian cancer. Acta Med. Median. 2020, 59, 26–33. [Google Scholar] [CrossRef]
- Zhou, J.; Du, Y.; Lu, Y.; Luan, B.; Xu, C.; Yu, Y.; Zhao, H. CD44 Expression Predicts Prognosis of Ovarian Cancer Patients Through Promoting Epithelial-Mesenchymal Transition (EMT) by Regulating Snail, ZEB1, and Caveolin-1. Front. Oncol. 2019, 9, 802. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Ding, D. The prognostic role of the cancer stem cell marker CD44 in ovarian cancer: A meta-analysis. Cancer Cell Int. 2017, 17, 8. [Google Scholar] [CrossRef]
- Calvete, J.; Larrinaga, G.; Errarte, P.; Martín, A.M.; Dotor, A.; Esquinas, C.; Nunes-Xavier, C.E.; Pulido, R.; López, J.I.; Angulo, J.C. The coexpression of fibroblast activation protein (FAP) and basal-type markers (CK 5/6 and CD44) predicts prognosis in high-grade invasive urothelial carcinoma of the bladder. Hum. Pathol. 2019, 91, 61–68. [Google Scholar] [CrossRef]
- Sun, Z.; Li, D.; Wu, H.; Hou, B. Tumour stem cell markers CD133 and CD44 are useful prognostic factors after surgical resection of pancreatic neuroendocrine tumours. Oncol. Lett. 2020, 20, 341. [Google Scholar] [CrossRef]
- Mohamed, S.Y.; Kaf, R.M.; Ahmed, M.M.; Elwan, A.; Ashour, H.R.; Ibrahim, A. The Prognostic Value of Cancer Stem Cell Markers (Notch1, ALDH1, and CD44) in Primary Colorectal Carcinoma. J. Gastrointest. Cancer 2019, 50, 824–837. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, J.; Lu, J.; Xiong, H.; Shi, X.; Gong, L. Significance of CD44 expression in head and neck cancer: A systemic review and meta-analysis. BMC Cancer 2014, 14, 15. [Google Scholar] [CrossRef]
- Kaza, S.; Kantheti, L.P.; Poosarla, C.; Gontu, S.R.; Kattappagari, K.K.; Baddam, V.R. A study on the expression of CD44 adhesion molecule in oral squamous cell carcinoma and its correlation with tumor histological grading. J. Orofac. Sci. 2018, 10, 42–49. [Google Scholar]
- Krump, M.; Ehrmann, J. Differences in CD44s expression in HNSCC tumours of different areas within the oral cavity. Biomed. Pap. 2013, 157, 280–283. [Google Scholar] [CrossRef]
- Boxberg, M.; Götz, C.; Haidari, S.; Dorfner, C.; Jesinghaus, M.; Drecoll, E.; Boskov, M.; Wolff, K.D.; Weichert, W.; Haller, B.; et al. Immunohistochemical expression of CD44 in oral squamous cell carcinoma in relation to histomorphological parameters and clinicopathological factors. Histopathology 2018, 73, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Morand, G.B.; Ikenberg, K.; Vital, D.G.; Cardona, I.; Moch, H.; Stoeckli, S.J.; Huber, G.F. Preoperative assessment of CD44-mediated depth of invasion as predictor of occult metastases in early oral squamous cell carcinoma. Head Neck 2019, 41, 950–958. [Google Scholar] [CrossRef]
- Lee, J.R.; Roh, J.L.; Lee, S.M.; Park, Y.; Cho, K.J.; Choi, S.H.; Nam, S.Y.; Kim, S.Y. Overexpression of cysteine-glutamate transporter and CD44 for prediction of recurrence and survival in patients with oral cavity squamous cell carcinoma. Head Neck 2018, 40, 2340–2346. [Google Scholar] [CrossRef] [PubMed]
- Arabi, L.; Badiee, A.; Mosaffa, F.; Jaafari, M.R. Targeting CD44 expressing cancer cells with anti-CD44 monoclonal antibody improves cellular uptake and antitumor efficacy of liposomal doxorubicin. J. Control. Release 2015, 220 Pt A, 275–286. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, C.C.; Fecteau, J.F.; Cui, B.; Chen, L.; Zhang, L.; Wu, R.; Rassenti, L.; Lao, F.; Weigand, S.; et al. Targeting chronic lymphocytic leukemia cells with a humanized monoclonal antibody specific for CD44. Proc. Natl. Acad. Sci. USA 2013, 110, 6127–6132. [Google Scholar] [CrossRef]
- Maisel, D.; Birzele, F.; Voss, E.; Nopora, A.; Bader, S.; Friess, T.; Goller, B.; Laifenfeld, D.; Weigand, S.; Runza, V. Targeting Tumor Cells with Anti-CD44 Antibody Triggers Macrophage-Mediated Immune Modulatory Effects in a Cancer Xenograft Model. PLoS ONE 2016, 11, e0159716. [Google Scholar] [CrossRef]
- Chen, X.; Nagai, Y.; Zhu, Z.; Ruan, H.; Peehl, D.M.; Greene, M.I.; Zhang, H. A spliced form of CD44 expresses the unique glycan that is recognized by the prostate cancer specific antibody F77. Oncotarget 2017, 9, 3631–3640. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Xia, L.; Ling, P.; Waxman, S.; Jing, Y. CD44 ligation with A3D8 antibody induces apoptosis in acute myeloid leukemia cells through binding to CD44s and clustering lipid rafts. Cancer Biol. Ther. 2012, 13, 1276–1283. [Google Scholar] [CrossRef]
- Khayrani, A.C.; Mahmud, H.; Oo, A.; Zahra, M.H.; Oze, M.; Du, J.; Alam, M.J.; Afify, S.M.; Quora, H.; Shigehiro, T.; et al. Targeting Ovarian Cancer Cells Overexpressing CD44 with Immunoliposomes Encapsulating Glycosylated Paclitaxel. Int. J. Mol. Sci. 2019, 20, 1042. [Google Scholar] [CrossRef]
- Park, H.Y.; Lee, K.J.; Lee, S.J.; Yoon, M.Y. Screening of peptides bound to breast cancer stem cell specific surface marker CD44 by phage display. Mol. Biotechnol. 2012, 51, 212–220. [Google Scholar] [CrossRef]
- Cho, J.H.; Lee, S.C.; Ha, N.R.; Lee, S.J.; Yoon, M.Y. A novel peptide-based recognition probe for the sensitive detection of CD44 on breast cancer stem cells. Mol. Cell. Probes 2015, 29, 492–499. [Google Scholar] [CrossRef]
- Taghipour-Sabzevar, V.; Sharifi, T.; Bagheri-Khoulenjani, S.; Goodarzi, V.; Kooshki, H.; Halabian, R.; Moghaddam, M.M. Targeted delivery of a short antimicrobial peptide against CD44-overexpressing tumor cells using hyaluronic acid-coated chitosan nanoparticles: An in vitro study. J. Nanoparticle Res. 2020, 22, 99. [Google Scholar] [CrossRef]
- Muntimadugu, E.; Kumar, R.; Saladi, S.; Rafeeqi, T.A.; Khan, W. CD44 targeted chemotherapy for co-eradication of breast cancer stem cells and cancer cells using polymeric nanoparticles of salinomycin and paclitaxel. Colloids Surfaces. B Biointerfaces 2016, 143, 532–546. [Google Scholar] [CrossRef]
- Shitara, K.; Doi, T.; Nagano, O.; Imamura, C.K.; Ozeki, T.; Ishii, Y.; Tsuchihashi, K.; Takahashi, S.; Nakajima, T.E.; Hironaka, S.; et al. Dose-escalation study for the targeting of CD44v+ cancer stem cells by sulfasalazine in patients with advanced gastric cancer (EPOC1205). Gastric Cancer 2017, 20, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kil, W.H.; Lee, J.; Oh, S.J.; Han, J.; Jeon, M.; Jung, T.; Lee, S.K.; Bae, S.Y.; Lee, H.C.; et al. Zerumbone suppresses EGF-induced CD44 expression through the inhibition of STAT3 in breast cancer cells. Oncol. Rep. 2014, 32, 2666–2672. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.S.; Vadgama, J.V. Curcumin and epigallocatechin gallate inhibit the cancer stem cell phenotype via down-regulation of STAT3-NFκB signaling. Anticancer. Res. 2015, 35, 39–46. [Google Scholar] [PubMed]
- Patel, S.; Waghela, B.; Shah, K.; Vaidya, F.; Mirza, S.; Patel, S.; Pathak, C.; Rawal, R. Publisher Correction: Silibinin, A Natural Blend In Polytherapy Formulation For Targeting Cd44v6 Expressing Colon Cancer Stem Cells. Sci. Rep. 2018, 8, 17992. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, D.; Xu, X.B.; Qiu, S.; Luo, S.; Qiu, E.; Rong, Z.; Zhang, J.; Zheng, D. Galangin inhibits epithelial-mesenchymal transition and angiogenesis by downregulating CD44 in glioma. J. Cancer 2019, 10, 4499–4508. [Google Scholar] [CrossRef]
- Erdogan, S.; Doganlar, O.; Doganlar, Z.B.; Serttas, R.; Turkekul, K.; Dibirdik, I.; Bilir, A. The flavonoid apigenin reduces prostate cancer CD44(+) stem cell survival and migration through PI3K/Akt/NF-κB signaling. Life Sci. 2016, 162, 77–86. [Google Scholar] [CrossRef]
- Eliaz, R.E.; Szoka, F.C., Jr. Liposome-encapsulated doxorubicin targeted to CD44: A strategy to kill CD44-overexpressing tumor cells. Cancer Res. 2001, 61, 2592–2601. [Google Scholar]
- Spadea, A.; Rios de la Rosa, J.M.; Tirella, A.; Ashford, M.B.; Williams, K.J.; Stratford, I.J.; Tirelli, N.; Mehibel, M. Evaluating the Efficiency of Hyaluronic Acid for Tumor Targeting via CD44. Mol. Pharm. 2019, 16, 2481–2493. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Luo, H.; Cao, Z.; Chen, Y.; Gao, J.; Li, Y.; Jiang, Q.; Xu, R.; Liu, J. Dual-targeting hybrid nanoparticles for the delivery of SN38 to Her2 and CD44 overexpressed human gastric cancer. Nanoscale 2016, 8, 11543–11558. [Google Scholar] [CrossRef] [PubMed]
- Diao, L.; Shen, A.; Yang, Y.; Tao, J.; Hu, Y. CD44-targeted hyaluronic acid–curcumin reverses chemotherapeutics resistance by inhibiting P-gp and anti-apoptotic pathways. RSC Adv. 2019, 9, 40873–40882. [Google Scholar] [CrossRef]
- Yin, J.; Zhang, H.; Wu, X.; Zhang, Y.; Li, J.; Shen, J.; Zhao, Y.; Xiao, Z.; Lu, L.; Huang, C.; et al. CD44 inhibition attenuates EGFR signaling and enhances cisplatin sensitivity in human EGFR wild-type non-small-cell lung cancer cells. Int. J. Mol. Med. 2020, 45, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Iyer, A.K.; Singh, A.; Choy, E.; Hornicek, F.J.; Amiji, M.M.; Duan, Z. MDR1 siRNA loaded hyaluronic acid-based CD44 targeted nanoparticle systems circumvent paclitaxel resistance in ovarian cancer. Sci. Rep. 2015, 5, 8509. [Google Scholar] [CrossRef]
- Tirella, A.; Kloc-Muniak, K.; Good, L.; Ridden, J.; Ashford, M.; Puri, S.; Tirelli, N. CD44 targeted delivery of siRNA by using HA-decorated nanotechnologies for KRAS silencing in cancer treatment. Int. J. Pharm. 2019, 561, 114–123. [Google Scholar] [CrossRef]
- Pothuraju, R.; Rachagani, S.; Krishn, S.R.; Chaudhary, S.; Nimmakayala, R.K.; Ganguly, K.; Lakshmanan, I.; Cox, J.L.; Mallya, K.; Kaur, S.; et al. Molecular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol. Cancer 2020, 19, 37. [Google Scholar] [CrossRef]
- Porcellini, S.; Asperti, C.; Corna, S.; Cicoria, E.; Valtolina, V.; Stornaiuolo, A.; Valentinis, B.; Bordignon, C.; Traversari, C. CAR T Cells Redirected to CD44v6 Control Tumor Growth in Lung and Ovary Adenocarcinoma Bearing Mice. Front. Immunol. 2020, 11, 99. [Google Scholar] [CrossRef]
- Wang, H.; Ye, X.; Ju, Y.; Cai, Z.; Wang, X.; Du, P.; Zhang, M.; Li, Y.; Cai, J. Minicircle DNA-Mediated CAR T Cells Targeting CD44 Suppressed Hepatocellular Carcinoma Both in vitro and in vivo. OncoTargets Ther. 2020, 13, 3703–3716. [Google Scholar] [CrossRef]
- Porter, C.E.; Rosewell Shaw, A.; Jung, Y.; Yip, T.; Castro, P.D.; Sandulache, V.C.; Sikora, A.; Gottschalk, S.; Ittman, M.M.; Brenner, M.K.; et al. Oncolytic Adenovirus Armed with BiTE, Cytokine, and Checkpoint Inhibitor Enables CAR T Cells to Control the Growth of Heterogeneous Tumors. Mol. Ther. 2020, 28, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Alhabbab, R.Y. Targeting Cancer Stem Cells by Genetically Engineered Chimeric Antigen Receptor T Cells. Front. Genet. 2020, 11, 312. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).