Emergence and Enhancement of Ultrasensitivity through Posttranslational Modulation of Protein Stability
Abstract
1. Introduction
1.1. Regulation of Protein Stability through Posttranslational Modifications
1.2. Ultrasensitivity
1.3. Ultrasensitivity through Zero-Order Covalent Modification Cycle
1.4. Caveat of the Goldbeter–Koshland Model Suggests a Mechanism of Signaling Control
2. Methods
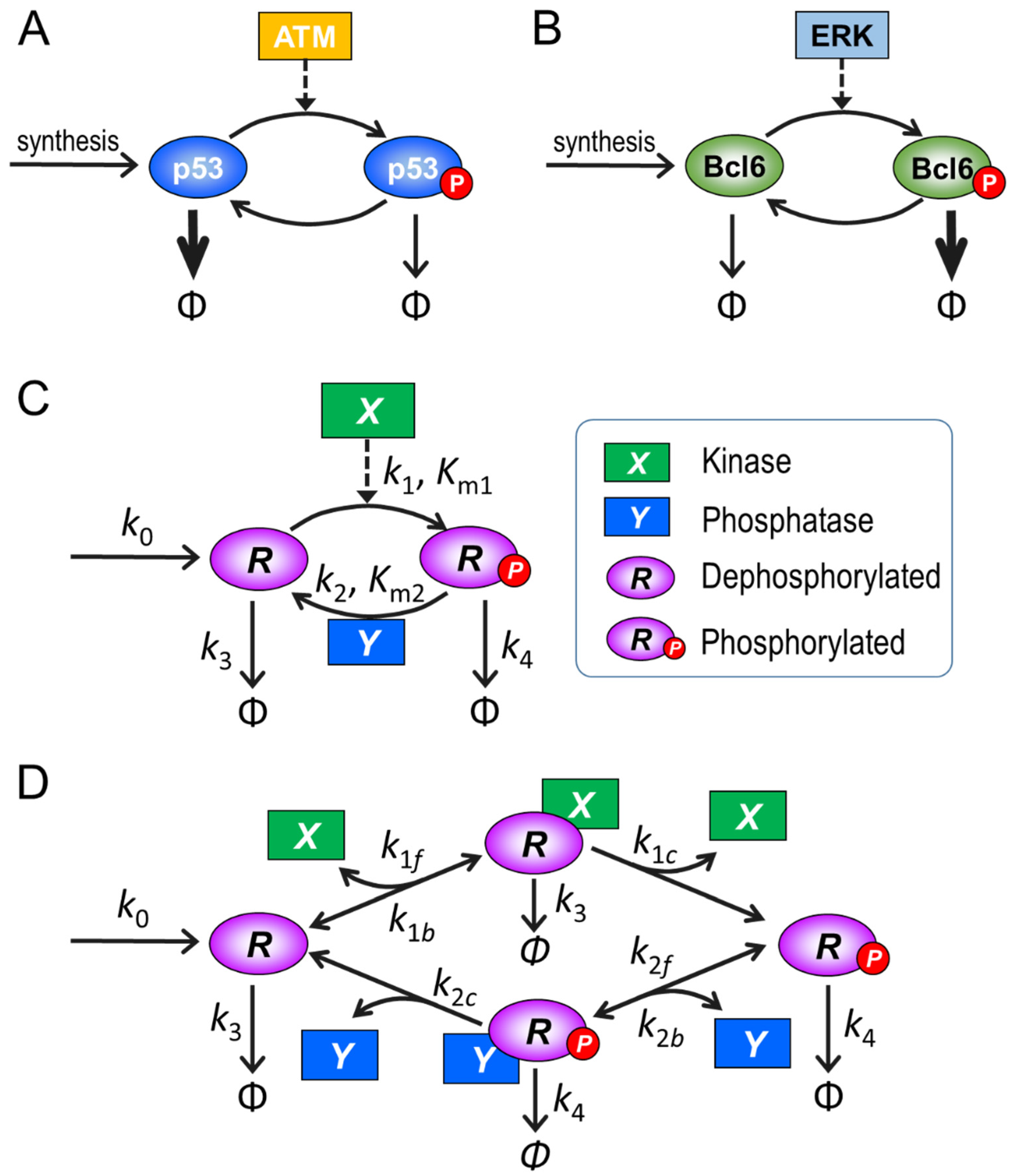
2.1. Model Structure and Parameterization
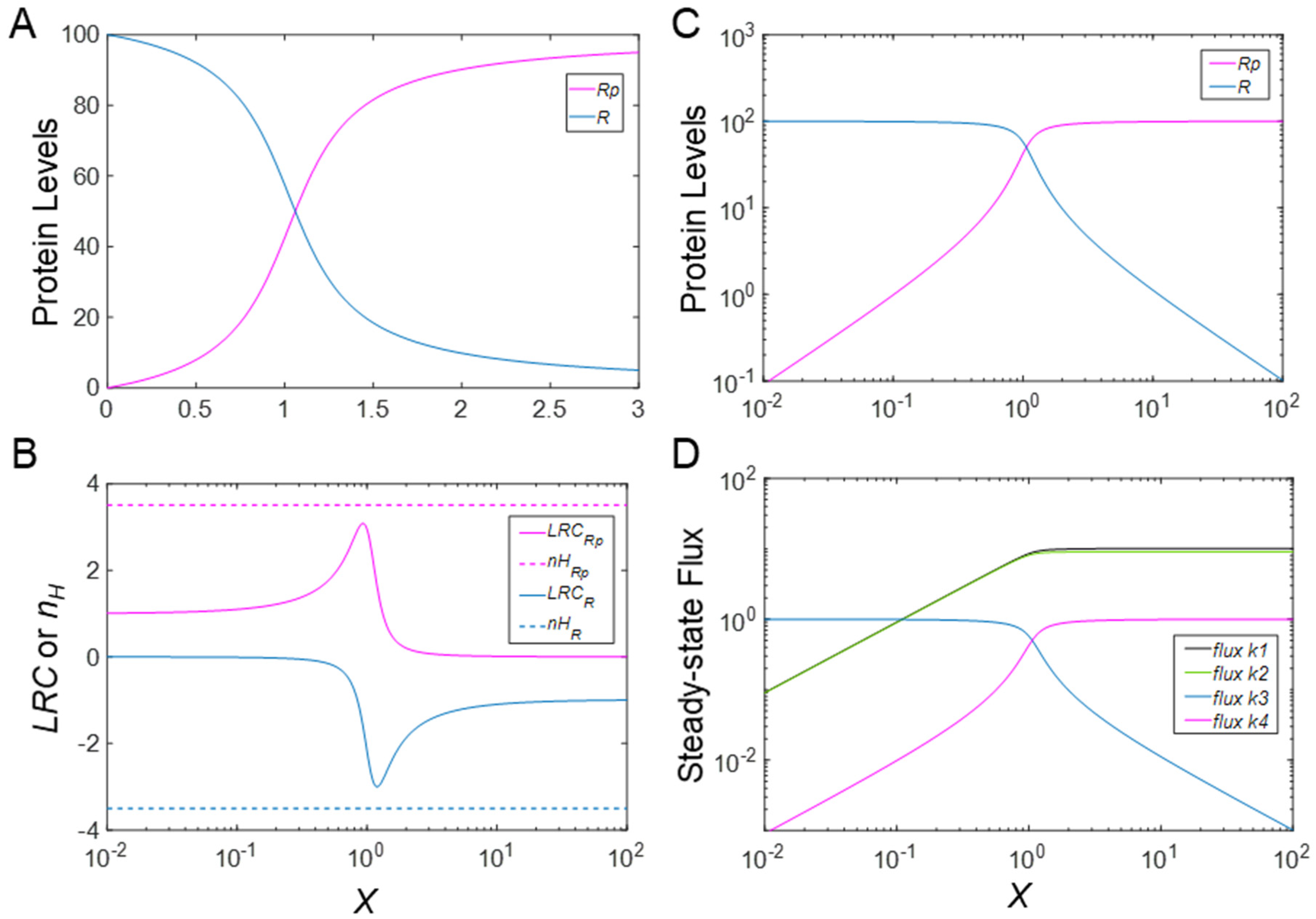
2.2. Metrics of Ultrasensitivity
2.3. Simulation Tools
3. Results
3.1. Ultrasensitivity in the Absence of PTM-Induced Changes in Protein Stability
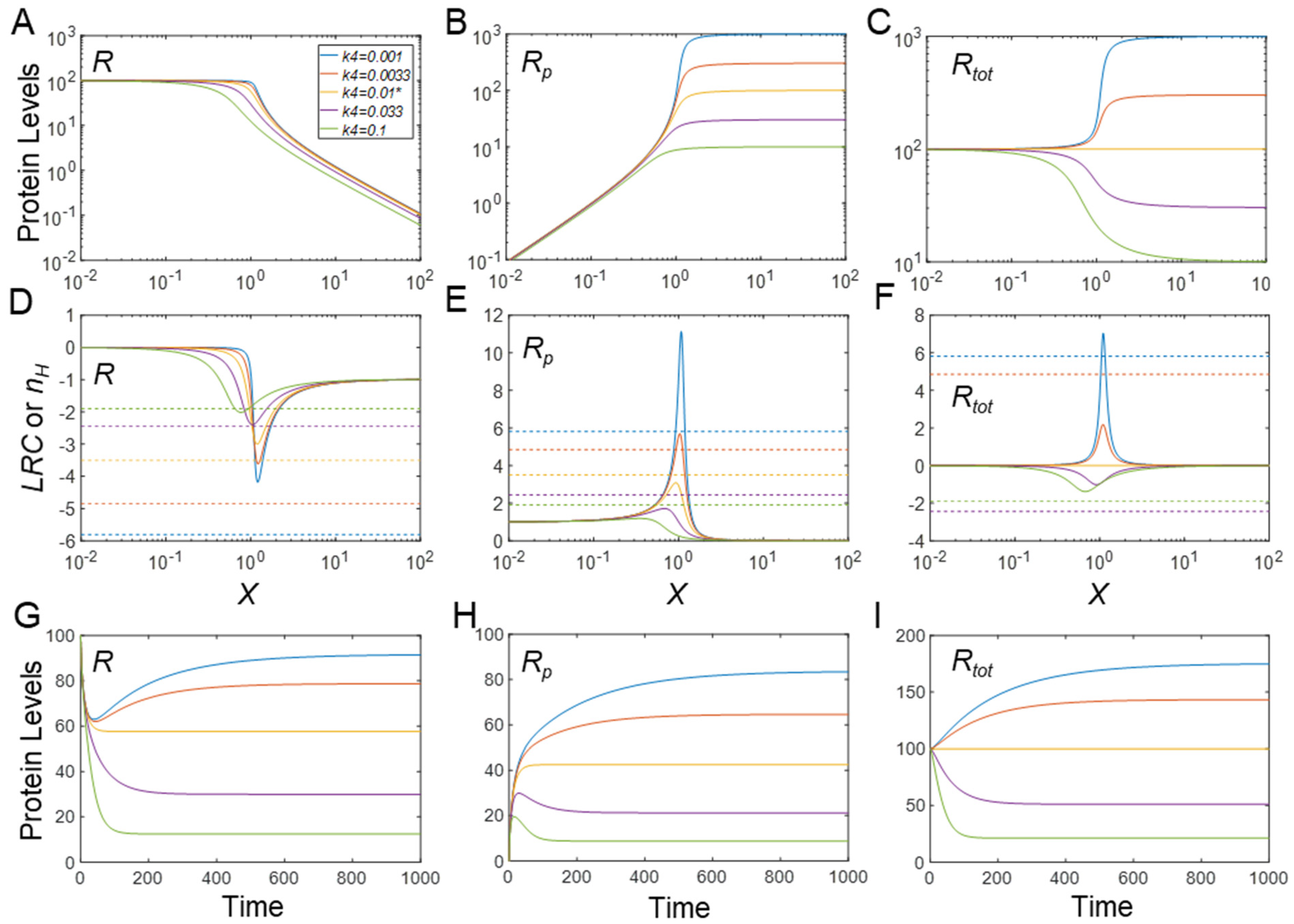
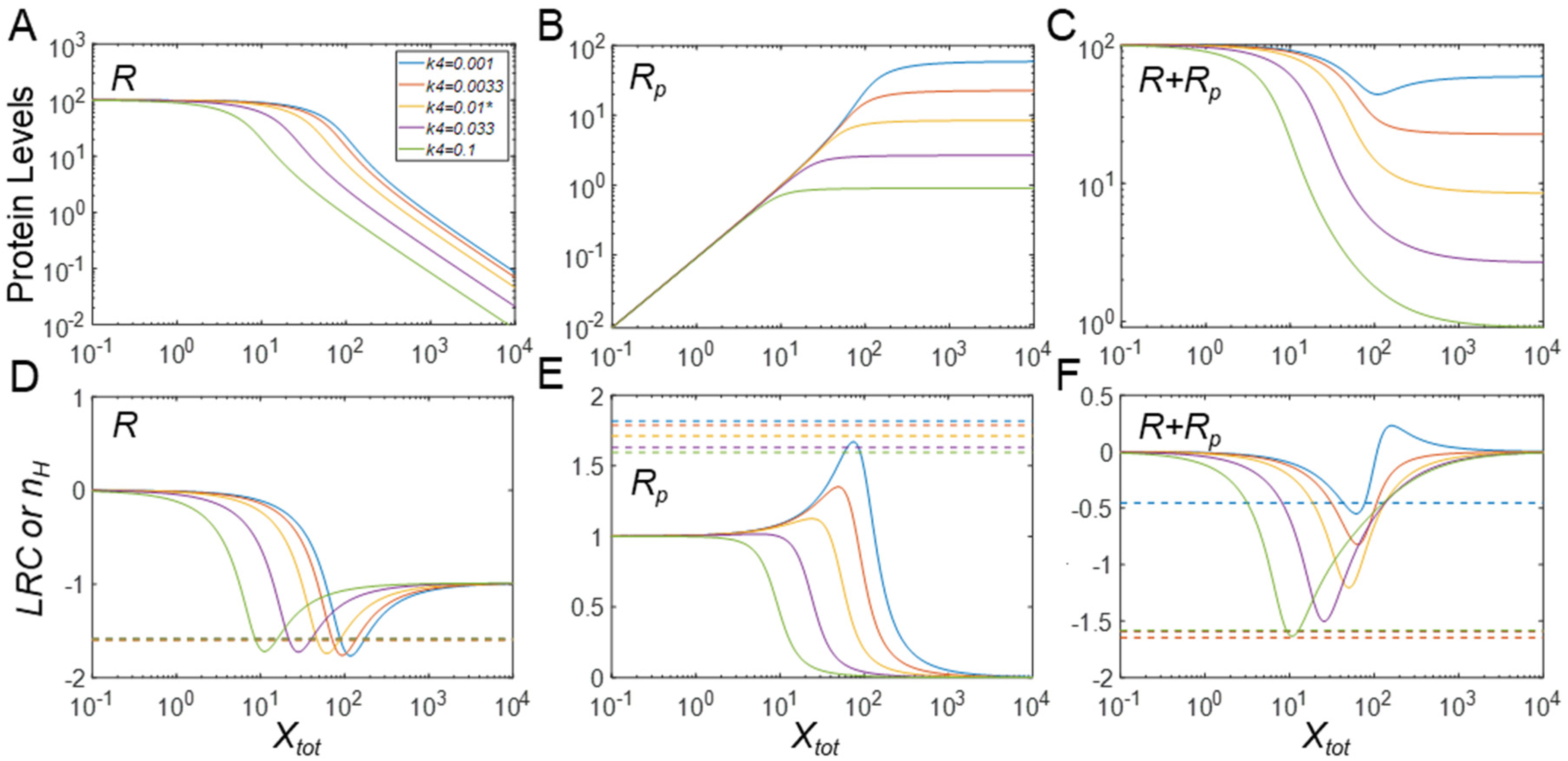
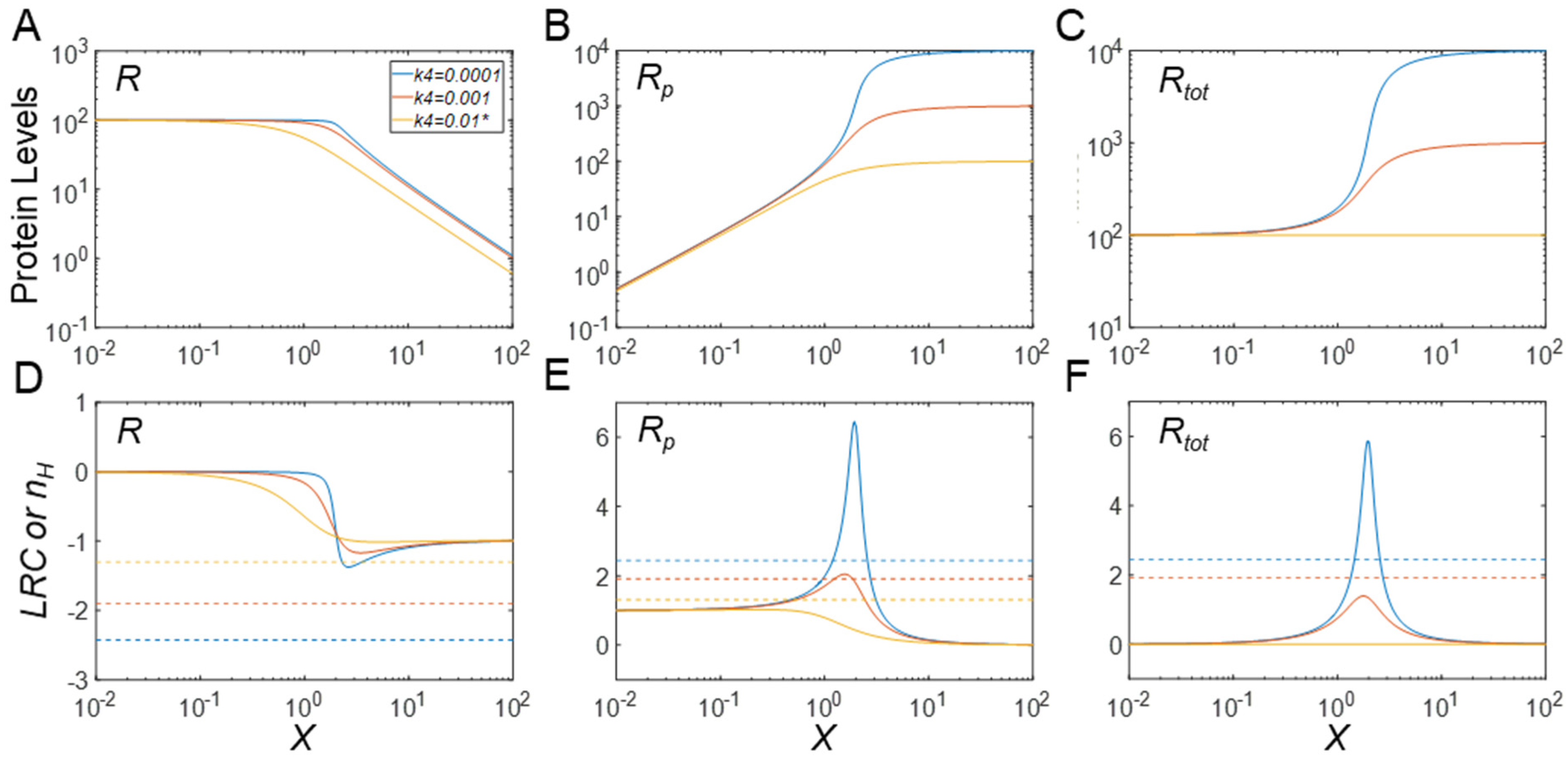
3.2. Effects of Protein Stability on Ultrasensitivity
3.2.1. Effects on Steady-State R
3.2.2. Effects on Steady-State Rp
3.2.3. Effects on Steady-State Rtot
3.2.4. Effects on Timing of Signaling
3.2.5. Behaviors of the Full Model
3.3. Protein Stabilization Can Lead to Emergence of Ultrasensitivity
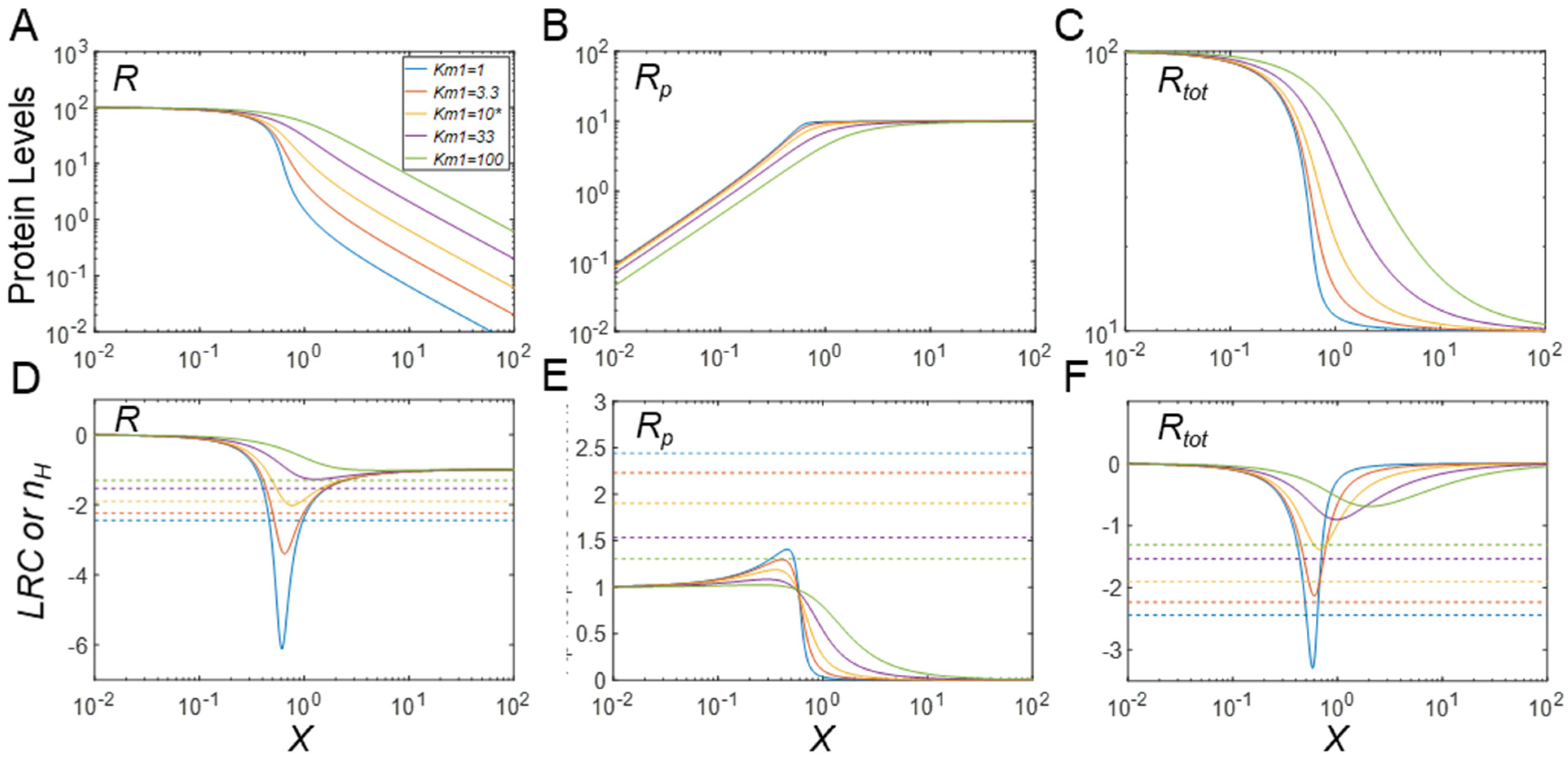
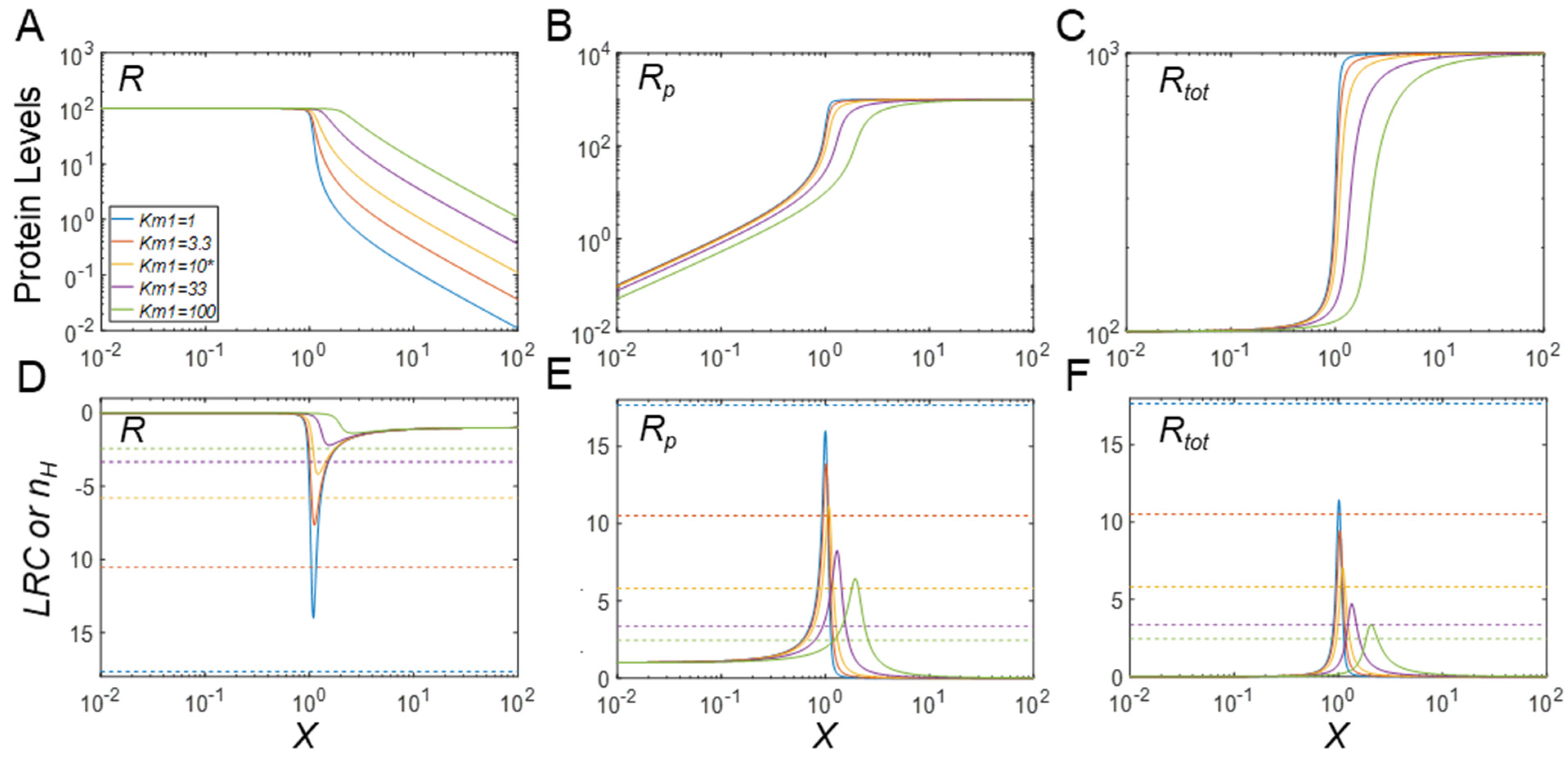
3.4. Regulation of Protein Modification Cycles through Alterations in Enzyme Features
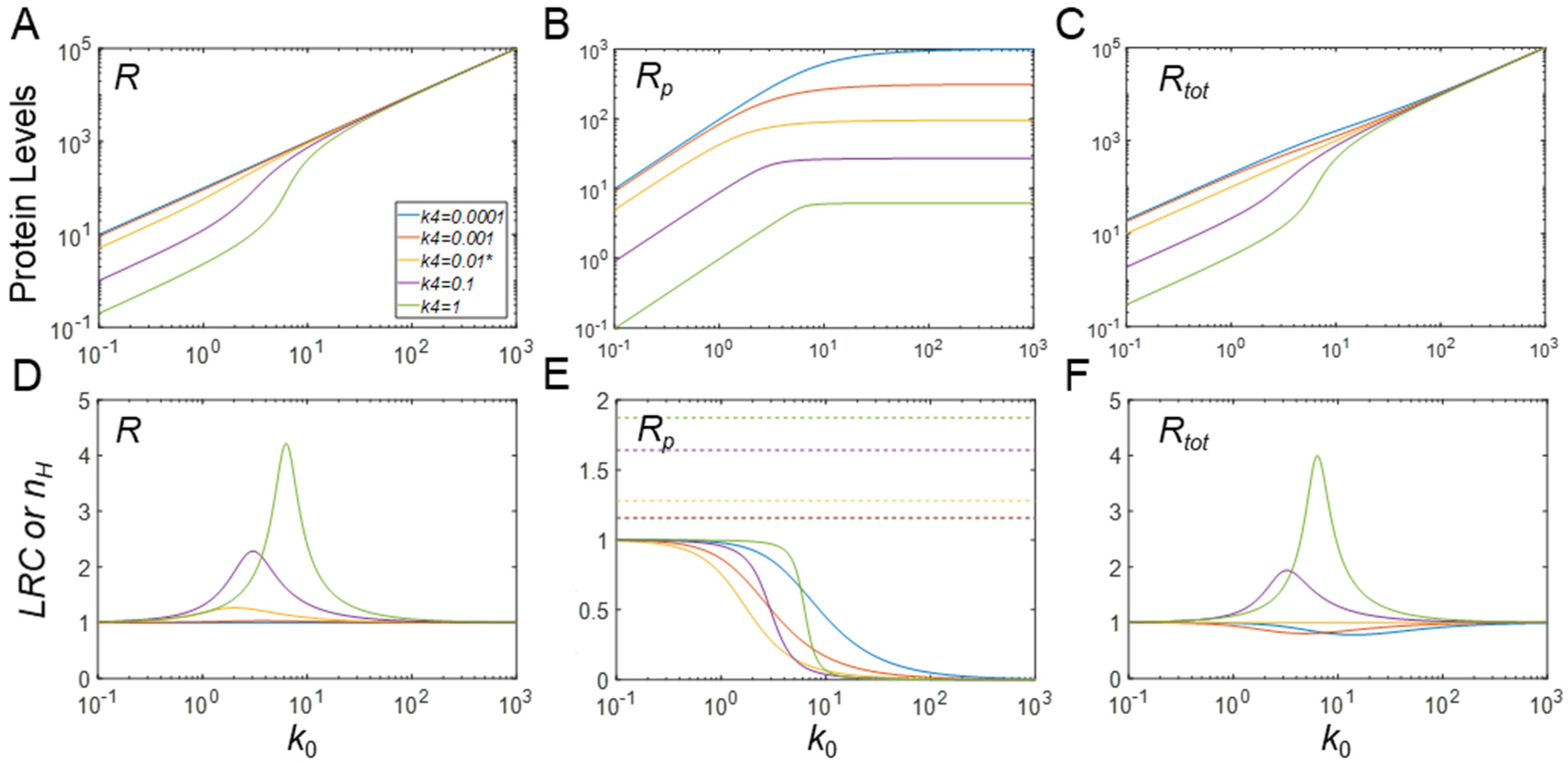
3.5. Ultrasensitivity in Response to Changes in Protein Synthesis Rate
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Stommel, J.M.; Wahl, G.M. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004, 23, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Shieh, S.Y.; Ikeda, M.; Taya, Y.; Prives, C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997, 91, 325–334. [Google Scholar] [CrossRef]
- Niu, H.; Ye, B.H.; Dalla-Favera, R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998, 12, 1953–1961. [Google Scholar] [CrossRef]
- Goto, H.; Natsume, T.; Kanemaki, M.T.; Kaito, A.; Wang, S.; Gabazza, E.C.; Inagaki, M.; Mizoguchi, A. Chk1-mediated Cdc25A degradation as a critical mechanism for normal cell cycle progression. J. Cell Sci. 2019, 132, jcs223123. [Google Scholar] [CrossRef] [PubMed]
- Kanarek, N.; Ben-Neriah, Y. Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol. Rev. 2012, 246, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Ulery, P.G.; Rudenko, G.; Nestler, E.J. Regulation of DeltaFosB stability by phosphorylation. J. Neurosci. 2006, 26, 5131–5142. [Google Scholar] [CrossRef]
- Goldbeter, A.; Koshland, D.E., Jr. Sensitivity amplification in biochemical systems. Q. Rev. Biophys. 1982, 15, 555–591. [Google Scholar] [CrossRef]
- Kholodenko, B.N.; Hoek, J.B.; Westerhoff, H.V.; Brown, G.C. Quantification of information transfer via cellular signal transduction pathways. FEBS Lett. 1997, 414, 430–434. [Google Scholar] [CrossRef]
- Koshland, D.E., Jr.; Goldbeter, A.; Stock, J.B. Amplification and adaptation in regulatory and sensory systems. Science 1982, 217, 220–225. [Google Scholar] [CrossRef]
- Ferrell, J.E., Jr. Tripping the switch fantastic: How a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci. 1996, 21, 460–466. [Google Scholar] [CrossRef]
- Ferrell, J.E., Jr.; Ha, S.H. Ultrasensitivity part III: Cascades, bistable switches, and oscillators. Trends Biochem. Sci. 2014, 39, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bhattacharya, S.; Andersen, M.E. Ultrasensitive response motifs: Basic amplifiers in molecular signalling networks. Open Biol. 2013, 3, 130031. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, J.E., Jr.; Ha, S.H. Ultrasensitivity part I: Michaelian responses and zero-order ultrasensitivity. Trends Biochem. Sci. 2014, 39, 496–503. [Google Scholar] [CrossRef]
- Ferrell, J.E., Jr.; Ha, S.H. Ultrasensitivity part II: Multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends Biochem. Sci. 2014, 39, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Uy, R.; Wold, F. Posttranslational covalent modification of proteins. Science 1977, 198, 890–896. [Google Scholar] [CrossRef]
- Goldbeter, A.; Koshland, D.E., Jr. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. USA 1981, 78, 6840–6844. [Google Scholar] [CrossRef]
- Goldbeter, A.; Koshland, D.E., Jr. Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects. J. Biol. Chem. 1984, 259, 14441–14447. [Google Scholar] [CrossRef]
- Cimino, A.; Hervagault, J.F. Experimental evidence for a zero-order ultrasensitivity in a simple substrate cycle. Biochem. Biophys. Res. Commun. 1987, 149, 615–620. [Google Scholar] [CrossRef]
- Melen, G.J.; Levy, S.; Barkai, N.; Shilo, B.Z. Threshold responses to morphogen gradients by zero-order ultrasensitivity. Mol. Syst. Biol. 2005, 1, 2005.0028. [Google Scholar] [CrossRef]
- Huang, C.Y.; Ferrell, J.E., Jr. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA 1996, 93, 10078–10083. [Google Scholar] [CrossRef]
- Meinke, M.H.; Edstrom, R.D. Muscle glycogenolysis. Regulation of the cyclic interconversion of phosphorylase a and phosphorylase b. J. Biol. Chem. 1991, 266, 2259–2266. [Google Scholar] [CrossRef]
- Meinke, M.H.; Bishop, J.S.; Edstrom, R.D. Zero-order ultrasensitivity in the regulation of glycogen phosphorylase. Proc. Natl. Acad. Sci. USA 1986, 83, 2865–2868. [Google Scholar] [CrossRef] [PubMed]
- LaPorte, D.C.; Koshland, D.E., Jr. Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature 1983, 305, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bhattacharya, S.; Conolly, R.B.; Andersen, M.E. Nonlinearities in cellular dose-response behaviors can be enhanced by protein stabilization. In Proceedings of the Society of Toxicology 54th Annual Meeting, San Diego, CA, USA, 22–26 March 2015. Abstract #719. [Google Scholar]
- Mallela, A.; Nariya, M.K.; Deeds, E.J. Crosstalk and ultrasensitivity in protein degradation pathways. PLoS Comput. Biol. 2020, 16, e1008492. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Tyson, J.J. Misuse of the Michaelis-Menten rate law for protein interaction networks and its remedy. PLoS Comput. Biol. 2020, 16, e1008258. [Google Scholar] [CrossRef]
- Blüthgen, N.; Bruggeman, F.J.; Legewie, S.; Herzel, H.; Westerhoff, H.V.; Kholodenko, B.N. Effects of sequestration on signal transduction cascades. FEBS J. 2006, 273, 895–906. [Google Scholar] [CrossRef]
- Fujioka, A.; Terai, K.; Itoh, R.E.; Aoki, K.; Nakamura, T.; Kuroda, S.; Nishida, E.; Matsuda, M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2006, 281, 8917–8926. [Google Scholar] [CrossRef]
- Jeynes-Smith, C.; Araujo, R.B. Ultrasensitivity and bistability in covalent-modification cycles with positive autoregulation. Proc. R. Soc. A 2021, 477, 20210069. [Google Scholar] [CrossRef]
- Verma, R.; McDonald, H.; Yates, J.R., 3rd; Deshaies, R.J. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Mol. Cell 2001, 8, 439–448. [Google Scholar] [CrossRef]
- Johnson, E.S.; Gonda, D.K.; Varshavsky, A. Cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature 1990, 346, 287–291. [Google Scholar] [CrossRef]
- Prakash, S.; Inobe, T.; Hatch, A.J.; Matouschek, A. Substrate selection by the proteasome during degradation of protein complexes. Nat. Chem. Biol. 2009, 5, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Legewie, S.; Bluthgen, N.; Herzel, H. Quantitative analysis of ultrasensitive responses. FEBS J. 2005, 272, 4071–4079. [Google Scholar] [CrossRef]
- Altszyler, E.; Ventura, A.C.; Colman-Lerner, A.; Chernomoretz, A. Ultrasensitivity in signaling cascades revisited: Linking local and global ultrasensitivity estimations. PLoS ONE 2017, 12, e0180083. [Google Scholar] [CrossRef]
- Breitschopf, K.; Haendeler, J.; Malchow, P.; Zeiher, A.M.; Dimmeler, S. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: Molecular characterization of the involved signaling pathway. Mol. Cell Biol. 2000, 20, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.E.; Gu, J.; Schau, M.; Bunn, H.F. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 7987–7992. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Zeisel, A.; Kostler, W.J.; Molotski, N.; Tsai, J.M.; Krauthgamer, R.; Jacob-Hirsch, J.; Rechavi, G.; Soen, Y.; Jung, S.; Yarden, Y.; et al. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Mol. Syst. Biol. 2011, 7, 529. [Google Scholar] [CrossRef]
- Buchler, N.E.; Louis, M. Molecular titration and ultrasensitivity in regulatory networks. J. Mol. Biol. 2008, 384, 1106–1119. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M. Tunable ultrasensitivity: Functional decoupling and biological insights. Sci. Rep. 2016, 6, 20345. [Google Scholar] [CrossRef]
- Xu, Y.; Gunawardena, J. Realistic enzymology for post-translational modification: Zero-order ultrasensitivity revisited. J. Theor. Biol. 2012, 311, 139–152. [Google Scholar] [CrossRef][Green Version]
- Jithinraj, P.K.; Roy, U.; Gopalakrishnan, M. Zero-order ultrasensitivity: A study of criticality and fluctuations under the total quasi-steady state approximation in the linear noise regime. J. Theor. Biol. 2014, 344, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, T.; Croll, D.H.; Owen, J.A.; Vander Heiden, M.G.; Locasale, J.W.; Alon, U.; Cantley, L.C.; Gunawardena, J. A fundamental trade-off in covalent switching and its circumvention by enzyme bifunctionality in glucose homeostasis. J. Biol. Chem. 2014, 289, 13010–13025. [Google Scholar] [CrossRef] [PubMed]
- Soyer, O.S.; Kuwahara, H.; Csikasz-Nagy, A. Regulating the total level of a signaling protein can vary its dynamics in a range from switch like ultrasensitivity to adaptive responses. FEBS J. 2009, 276, 3290–3298. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Description | Default Value |
|---|---|---|
| k0 | Rate constant of synthesis of R | 1 (concentration/time) |
| k1 | Catalytic rate constant for phosphorylation | 10 (1/time) |
| Km1 | Michaelis constant for phosphorylation | 10 (concentration) |
| X | Kinase | 0 (concentration) |
| k2 | Catalytic rate constant for dephosphorylation | 10 (1/time) |
| Km2 | Michaelis constant for dephosphorylation | 10 (concentration) |
| k3 | Degradation rate constant of R | 0.01 (1/time) |
| k4 | Degradation rate constant of Rp | 0.01 (1/time) |
| Y | Phosphatase | 1 (concentration) |
| Parameter | Description | Default Value |
|---|---|---|
| k0 | Rate constant of synthesis of R | 1 (concentration/time) |
| k1f | Association rate constant for R and X binding | 10 (1/concentration/time) |
| k1b | Dissociation rate constant for RX complex | 90 (1/time) |
| k1c | Catalytic rate constant for phosphorylation | 10 (1/time) |
| Xtot | Total kinase | 0 (concentration) |
| k2f | Association rate constant for Rp and Y binding | 10 (1/concentration/time) |
| k2b | Dissociation rate constant for RpY complex | 90 (1/time) |
| k2c | Catalytic rate constant for dephosphorylation | 10 (1/time) |
| k3 | Degradation rate constant of R and RX | 0.01 (1/time) |
| k4 | Degradation rate constant of Rp and RpY | 0.01 (1/time) |
| Ytot | Total phosphatase | 1 (concentration) |
| Parameter Varied | Stability Condition | Effects on |LRC|max | ||
|---|---|---|---|---|
| R vs. Xtot | Rp vs. Xtot | R + Rp vs. Xtot | ||
| ↓k4 | k3 > k4 | ↑#, – * | ↑ | ↑#, ↓ * |
| ↑k4 | k3 < k4 | ↓#, – * | ↓ | ↑ |
| ↓Km1, ↓Km2 | k3 > k4 or k3 < k4 | ↑ | ↑ | ↑ |
| ↑Km1, ↑Km2 | k3 > k4 or k3 < k4 | ↓ | ↓ | ↓ |
| ↑k1, ↑k21 | k3 > k4 or k3 < k4 | – | – | – |
| ↓k1, ↓k21 | k3 > k4 or k3 < k4 | – | – | – |
| ↑k2, ↑k2c | k3 > k4 | ↑ | ↑ | ↑ |
| ↑k2, ↑k2c | k3 < k4 | – | – | – |
| R vs. k0 | Rp vs. k0 | R + Rp vs. k0 | ||
| ↓k4 | k3 > k4 | – | – | – |
| ↑k4 | k3 < k4 | ↑ | – | ↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumbale, C.M.; Voit, E.O.; Zhang, Q. Emergence and Enhancement of Ultrasensitivity through Posttranslational Modulation of Protein Stability. Biomolecules 2021, 11, 1741. https://doi.org/10.3390/biom11111741
Kumbale CM, Voit EO, Zhang Q. Emergence and Enhancement of Ultrasensitivity through Posttranslational Modulation of Protein Stability. Biomolecules. 2021; 11(11):1741. https://doi.org/10.3390/biom11111741
Chicago/Turabian StyleKumbale, Carla M., Eberhard O. Voit, and Qiang Zhang. 2021. "Emergence and Enhancement of Ultrasensitivity through Posttranslational Modulation of Protein Stability" Biomolecules 11, no. 11: 1741. https://doi.org/10.3390/biom11111741
APA StyleKumbale, C. M., Voit, E. O., & Zhang, Q. (2021). Emergence and Enhancement of Ultrasensitivity through Posttranslational Modulation of Protein Stability. Biomolecules, 11(11), 1741. https://doi.org/10.3390/biom11111741






