Symptomatic, Genetic, and Mechanistic Overlaps between Autism and Alzheimer’s Disease
Abstract
:1. Introduction
2. Methodology
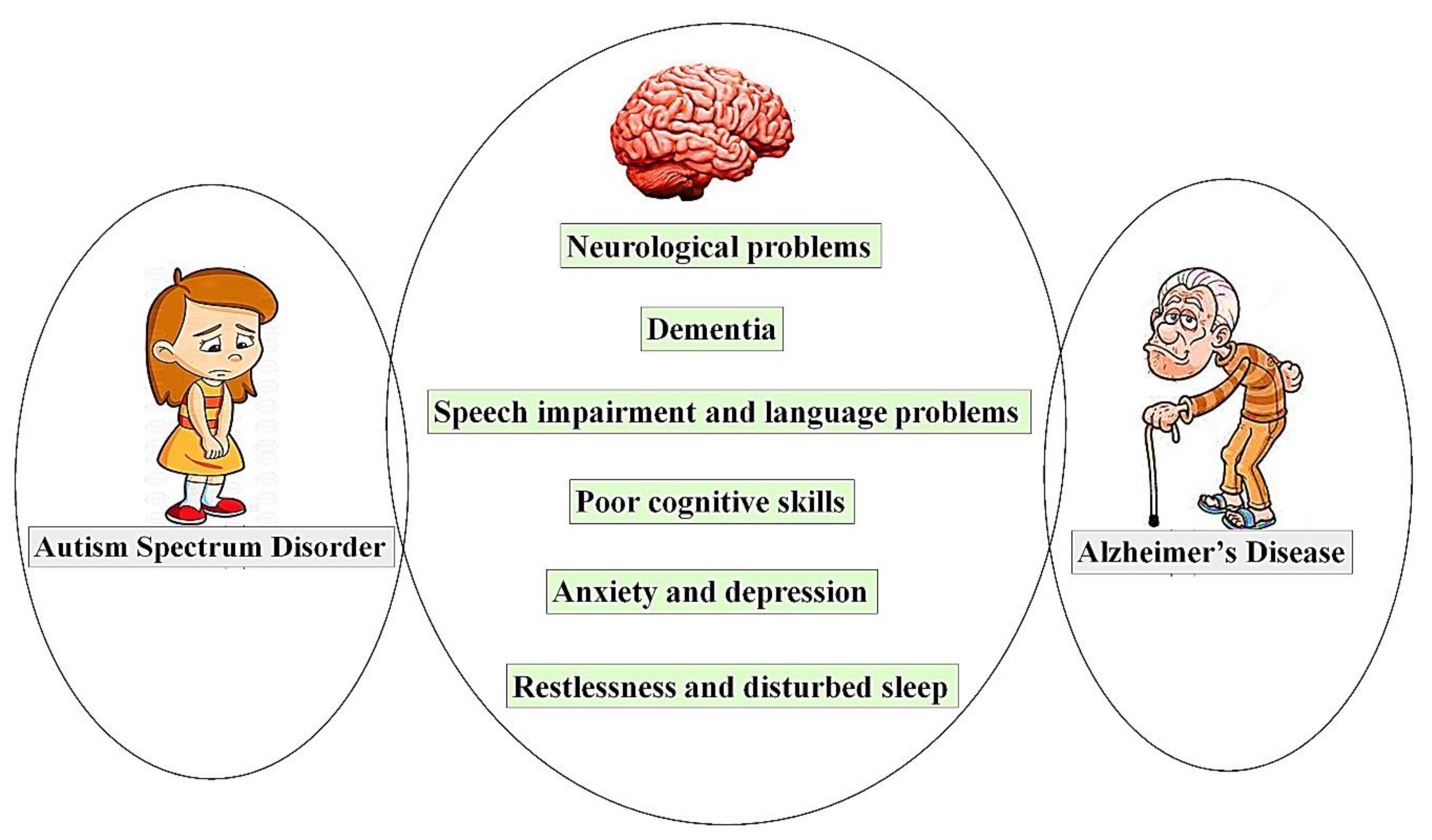
3. Shared Symptoms of ASD and AD
4. Genes and Genetics
5. Theories and Mechanism of Pathophysiology
5.1. Disrupted Neural Connectivity
5.2. Imbalanced Neurotransmitters
5.3. Overlapping Mechanisms of Pathogenesis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.D.; et al. Autism spectrum disorder: Neuropathology and animal models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef] [PubMed]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadeem, M.S.; Murtaza, B.N.; Al-Ghamdi, M.A.; Ali, A.; Zamzami, M.A.; Khan, J.A.; Ahmad, A.; Rehman, M.U.; Kazmi, I. Autism-A Comprehensive Array of Prominent Signs and Symptoms. Curr. Pharm. Des. 2021, 27, 1418–1433. [Google Scholar] [CrossRef]
- Dickinson, A.; Daniel, M.; Marin, A.; Gaonkar, B.; Dapretto, M.; McDonald, N.M.; Jeste, S. Multivariate neural connectivity patterns in early infancy predict later autism symptoms. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2021, 6, 59–69. [Google Scholar] [CrossRef]
- Nadeem, M.S.; Al-Abbasi, F.A.; Kazmi, I.; Murtaza, B.N.; Zamzami, M.A.; Kamal, M.A.; Arif, A.; Afzal, M.; Anwar, F. Multiple risk factors: A challenge in the management of Autism. Curr. Pharm. Des. 2020, 26, 743–754. [Google Scholar] [CrossRef]
- Katz, J.; Reichenberg, A.; Kolevzon, A. Prenatal and perinatal metabolic risk factors for autism: A review and integration of findings from population-based studies. Curr. Opin. Psychiatry 2021, 34, 94–104. [Google Scholar] [CrossRef]
- Soysal, P.; Tan, S.G. The prevalence and co-incidence of geriatric syndromes in older patients with early-stage Alzheimer’s disease and dementia with Lewy bodies. Aging Clin. Exp. Res. 2021, 33, 2599–2603. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.B.; Weuve, J.; Barnes, L.L.; McAninch, E.A.; Wilson, R.S.; Evans, D.A. Population estimate of people with clinical Alzheimer’s disease and mild cognitive impairment in the United States (2020–2060). Alzheimers Dement. 2021. [Google Scholar] [CrossRef]
- Lei, P.; Ayton, S.; Bush, A.I. The essential elements of Alzheimer’s disease. J. Biol. Chem. 2021, 296, 100105. [Google Scholar] [CrossRef] [PubMed]
- Mintun, M.A.; Lo, A.C.; Duggan Evans, C.; Wessels, A.M.; Ardayfio, P.A.; Andersen, S.W.; Shcherbinin, S.; Sparks, J.; Sims, J.R.; Brys, M.; et al. Donanemab in early Alzheimer’s disease. N. Engl. J. Med. 2021, 384, 1691–1704. [Google Scholar] [CrossRef]
- Arnsten, A.F.; Datta, D.; Del Tredici, K.; Braak, H. Hypothesis: Tau pathology is an initiating factor in sporadic Alzheimer’s disease. Alzheimers Dement. 2021, 17, 115–124. [Google Scholar] [CrossRef]
- Pereira, J.B.; Janelidze, S.; Smith, R.; Mattsson-Carlgren, N.; Palmqvist, S.; Teunissenm, C.E.; Zetterberg, H.; Stomrud, E.; Ashton, N.J.; Blennow, K.; et al. Plasma GFAP is an early marker of amyloid-β but not tau pathology in Alzheimer’s disease. Brain 2021, 14. [Google Scholar] [CrossRef]
- Johansson, M.; Stomrud, E.; Insel, P.S.; Leuzy, A.; Johansson, P.M.; Smith, R.; Ismail, Z.; Janelidze, S.; Palmqvist, S.; van Westen, D.; et al. Mild behavioral impairment and its relation to tau pathology in preclinical Alzheimer’s disease. Transl. Psychiatry 2021, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.D.; Hermann, B.; Mecollari, J.; Turkstra, L.S. Connected speech and language in mild cognitive impairment and Alzheimer’s disease: A review of picture description tasks. J. Clin. Exp. Neuropsychol. 2018, 40, 917–939. [Google Scholar] [CrossRef] [PubMed]
- Rhodus, E.K.; Barber, J.; Abner, E.L.; Duff, D.; Bardach, S.H.; Caban-Holt, A.; Lightner, D.; Rowles, G.D.; Schmitt, F.A.; Jicha, G.A. Behaviors characteristic of autism spectrum disorder in a geriatric cohort with mild cognitive impairment or early dementia. Alzheimer Dis. Assoc. Disord. 2020, 34, 66–71. [Google Scholar] [CrossRef]
- Reeves, S.; Williams, V.; Costela, F.M.; Palumbo, R.; Umoren, O.; Christopher, M.M.; Blacker, D.; Woods, R.L. Narrative video scene description task discriminates between levels of cognitive impairment in Alzheimer’s disease. Neuropsychology 2020, 34, 437. [Google Scholar] [CrossRef]
- Gevezova, M.; Sarafian, V.; Anderson, G.; Maes, M. Inflammation and mitochondrial dysfunction in autism spectrum disorder. CNS Neurol. Disord.-Drug Targets 2020, 19, 320–333. [Google Scholar] [CrossRef]
- Yoo, S.M.; Park, J.; Kim, S.H.; Jung, Y.K. Emerging perspectives on mitochondrial dysfunction and inflammation in Alzheimer’s disease. BMB Rep. 2020, 53, 35. [Google Scholar] [CrossRef]
- Zeidán-Chuliá, F.; de Oliveira, B.H.; Salmina, A. Altered expression of Alzheimer’s disease-related genes in the cerebellum of autistic patients: A model for disrupted brain connectome and therapy. Cell Death Dis. 2014, 5, e1250. [Google Scholar] [CrossRef]
- Sragovich, S.; Merenlender-Wagner, A.; Gozes, I. ADNP plays a key role in autophagy: From autism to schizophrenia and Alzheimer’s disease. Bioessays 2017, 39, 1700054. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Maloney, B.; Wang, R.; Sokol, D.K.; Rogers, J.T.; Westmark, C.J. How autism and Alzheimer’s disease are TrAPPed. Mol. Psychiatry 2021, 26, 26–29. [Google Scholar] [CrossRef]
- de Freitas Oliveira, L.; Camargos, E.F.; Martini, L.L.; Machado, F.V.; Novaes, M.R. Use of psychotropic agents to treat agitation and aggression in Brazilian patients with Alzheimer’s disease: A naturalistic and multicenter study. Psychiatry Res. 2021, 295, 113591. [Google Scholar] [CrossRef]
- Im, D.S. Treatment of aggression in adults with autism spectrum disorder: A review. Harv. Rev. Psychiatry 2021, 29, 35. [Google Scholar] [CrossRef]
- D’Alò, G.L.; De Crescenzo, F.; Amato, L.; Cruciani, F.; Davoli, M.; Fulceri, F.; Minozzi, S.; Mitrova, Z.; Morgano, G.P.; Nardocci, F.; et al. Impact of antipsychotics in children and adolescents with autism spectrum disorder: A systematic review and meta-analysis. Health Qual. Life Outcomes 2021, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Nanjappa, M.S.; Voyiaziakis, E.; Pradhan, B.; Thippaiah, S.M. Use of selective serotonin and norepinephrine reuptake inhibitors (SNRIs) in the treatment of autism spectrum disorder (ASD), comorbid psychiatric disorders and ASD-associated symptoms: A clinical review. CNS Spectr. 2020, 1–8. [Google Scholar] [CrossRef]
- Leshem, R.; Bar-Oz, B.; Diav-Citrin, O.; Gbaly, S.; Soliman, J.; Renoux, C.; Matok, I. Selective Serotonin Reuptake Inhibitors (SSRIs) and Serotonin Norepinephrine Reuptake Inhibitors (SNRIs) During Pregnancy and the Risk for Autism spectrum disorder (ASD) and Attention deficit hyperactivity disorder (ADHD) in the Offspring: A True Effect or a Bias? A Systematic Review & Meta-Analysis. Curr. Neuropharmacol. 2021, 19, 896–906. [Google Scholar] [CrossRef]
- Gunata, M.; Parlakpinar, H.A.; Acet, H.A. Melatonin: A review of its potential functions and effects on neurological diseases. Rev. Neurol. 2020, 176, 148–165. [Google Scholar] [CrossRef]
- Lalanne, S.; Fougerou-Leurent, C.; Anderson, G.M.; Schroder, C.M.; Nir, T.; Chokron, S.; Delormem, R.; Claustrat, B.; Bellissant, E.; Kermarrec, S.; et al. Melatonin: From Pharmacokinetics to Clinical Use in Autism Spectrum Disorder. Int. J. Mol. Sci. 2021, 22, 1490. [Google Scholar] [CrossRef]
- Frye, R.E.; Rossignol, D.A. Mitochondrial dysfunction can connect the diverse medical symptoms associated with autism spectrum disorders. Pediatric Res. 2011, 69, 41–47. [Google Scholar] [CrossRef]
- Luigetti, M.; Sauchelli, D.; Primiano, G. Peripheral neuropathy is a common manifestation of mitochondrial diseases: A single-centre experience. Eur. J. Neurol. 2016, 23, 1020–1027. [Google Scholar] [CrossRef]
- Luigetti, M.; Primiano, G.; Cuccagna, C. Small fibre neuropathy in mitochondrial diseases explored with sudoscan. Clin. Neurophysiol. 2018, 129, 1618–1623. [Google Scholar] [CrossRef]
- Morotti, H.; Mastel, S.; Keller, K.; Barnard, R.A.; Hall, T.; O’Roak, B.J.; Fombonne, E. Autism and attention-deficit/hyperactivity disorders and symptoms in children with neurofibromatosis type 1. Dev. Med. Child Neurol. 2021, 63, 226–232. [Google Scholar] [CrossRef]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef]
- Kumar, D.; Sharma, A.; Sharma, L. A comprehensive review of Alzheimer’s association with related proteins: Pathological role and therapeutic significance. Curr. Neuropharmacol. 2020, 18, 674–695. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Ikeda, M.; Brown, J.; Holland, A.J.; Fukuhara, R.; Hodges, J.R. Changes in appetite, food preference, and eating habits in frontotemporal dementia and Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry 2002, 73, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Shigenobu, K.; Ikeda, M.; Fukuhara, R.; Maki, N.; Hokoichi, K.; Nebu, A. The stereotypy rating inventory for frontotemporal lobar degeneration. Psychiatry Res. 2002, 110, 175–187. [Google Scholar] [CrossRef]
- Sakuta, S.; Hashimoto, M.; Ikeda, M.; Koyama, A.; Takasaki, A.; Hotta, M.; Fukuhara, R.; Ishikawa, T.; Yuki, S.; Miyagawa, Y.; et al. Clinical features of behavioral symptoms in patients with semantic dementia: Does semantic dementia cause autistic traits? PLoS ONE 2021, 18, e0247184. [Google Scholar] [CrossRef]
- Aschwanden, D.; Strickhouser, J.E.; Luchetti, M.; Stephan, Y.; Sutin, A.R.; Terracciano, A. Is personality associated with dementia risk? A meta-analytic investigation. Ageing Res. Rev. 2021, 6, 101269. [Google Scholar] [CrossRef]
- Sotiropoulos, I.; Catania, C.; Pinto, L.G.; Silva, R.; Pollerberg, G.E.; Takashima, A.; Sousa, N.; Almeida, O.F. Stress acts cumulatively to precipitate Alzheimer’s disease-like tau pathology and cognitive deficits. J. Neurosci. 2011, 25, 7840–7847. [Google Scholar] [CrossRef] [Green Version]
- Castillo, M.A.; Urdaneta, K.E.; Semprún-Hernández, N.; Brigida, A.L.; Antonucci, N.; Schultz, S.; Siniscalco, D. Speech-stimulating substances in autism spectrum disorders. Behav. Sci. 2019, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Tager-Flusberg, H.; Kasari, C. Minimally verbal school-aged children with autism spectrum disorder: The neglected end of the spectrum. Autism Res. 2013, 6, 468–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reindal, L.; Nærland, T.; Weidle, B.; Lydersen, S.; Andreassen, O.A.; Sund, A.M. Structural and Pragmatic Language Impairments in Children Evaluated for Autism Spectrum Disorder (ASD). J. Autism Dev. Dis. 2021, 1–9. [Google Scholar] [CrossRef]
- Matias-Guiu, J.A.; García-Ramos, R. Primary progressive aphasia: From syndrome to disease. Neurología 2013, 28, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, S.; Wicklund, A.H.; Salmon, D.P. The neuropsychological profile of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006171. [Google Scholar] [CrossRef]
- Laske, C.; Sohrabi, H.R.; Frost, S.M.; López-de-Ipiña, K.; Garrard, P.; Buscema, M.; Dauwels, J.; Soekadar, S.R.; Mueller, S.; Linnemann, C.; et al. Innovative diagnostic tools for early detection of Alzheimer’s disease. Alzheimers Dement. 2015, 11, 561–578. [Google Scholar] [CrossRef]
- Meilan, J.J.; Martinez-Sanchez, F.; Carro, J.; Carcavilla, N.; Ivanova, O. Voice markers of lexical access in mild cognitive impairment and Alzheimer’s disease. Curr. Alzheimer Res. 2018, 15, 111–119. [Google Scholar] [CrossRef]
- Pastoriza-Dominguez, P.; Torre, I.G.; Dieguez-Vide, F.; Gomez-Ruiz, I.; Gelado, S.; Bello-Lopez, J.; Avila-Rivera, A.; Matias-Guiu, J.; Pytel, V.; Hernandez-Fernandez, A. Speech pause distribution as an early marker for Alzheimer’s disease. medRxiv 2021. [Google Scholar] [CrossRef]
- Campbell, E.L.; Mesía, R.Y.; Docío-Fernández, L.; García-Mateo, C. Paralinguistic and linguistic fluency features for Alzheimer’s disease detection. Comput. Speech Lang. 2021, 68, 101198. [Google Scholar] [CrossRef]
- Nyrenius, J.; Billstedt, E. The functional impact of cognition in adults with autism spectrum disorders. Nordic J. Psychiatry 2019, 74, 220–225. [Google Scholar] [CrossRef]
- Kenny, L.; Cribb, S.J.; Pellicano, E. Childhood Executive Function Predicts Later Autistic Features and Adaptive Behavior in Young Autistic People: A 12-Year Prospective Study. J. Abnorm. Child Psychol. 2019, 47, 1089–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertelli, M.O. ASD and Intellectual Disability. In Psychopathology in Adolescents and Adults with Autism Spectrum Disorders; Keller, R., Ed.; Springer: Cham, Switzerland, 2019; pp. 111–130. [Google Scholar] [CrossRef]
- Livingston, L.A.; Happé, F. Conceptualising compensation in neurodevelopmental disorders: Reflections from autism spectrum disorder. Neurosci. Biobehav. Rev. 2017, 80, 729–742. [Google Scholar] [CrossRef] [Green Version]
- Mansour, R.; Ward, A.R.; Lane, D.M.; Loveland, K.A.; Aman, M.G.; Jerger, S.; Schachar, R.J.; Pearson, D.A. ADHD severity as a predictor of cognitive task performance in children with Autism Spectrum Disorder (ASD). Res. Dev. Disabil. 2021, 111, 103882. [Google Scholar] [CrossRef]
- Cornelis, M.C.; Wang, Y.; Holland, T.; Agarwal, P.; Weintraub, S.; Morris, M.C. Age and cognitive decline in the UK Biobank. PLoS ONE 2019, 14, e013948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malpetti, M.; Kievit, R.A.; Passamonti, L.; Jones, P.S.; Tsvetanov, K.A.; Rittman, T.; Mak, E.; Nicastro, N.; Bevan-Jones, W.R.; Su, L.; et al. Microglial activation and tau burden predict cognitive decline in Alzheimer’s disease. Brain 2020, 143, 1588–1602. [Google Scholar] [CrossRef]
- Marshall, G.A.; Rentz, D.M.; Frey, M.T.; Locascio, J.J.; Johnson, K.A.; Sperling, R.A. Alzheimer’s Disease Neuroimaging Initiative. Executive function and instrumental activities of daily living in mild cognitive impairment and Alzheimer’s disease. Alzheimers Dement. 2011, 7, 300–308. [Google Scholar] [CrossRef] [Green Version]
- Alzheimer Association. Early Signs and Symptoms of Alzheimer’s Alzheimer’s and Dementia; Alzheimer Association: Washington, DC, USA, 2019; pp. 1–88. [Google Scholar]
- Chang, C.H.; Lin, C.H.; Liu, C.Y.; Huang, C.S.; Chen, S.J.; Lin, W.C.; Yang, H.T.; Lane, H.Y. Plasma d-glutamate levels for detecting mild cognitive impairment and Alzheimer’s disease: Machine learning approaches. J. Psychopharmacol. 2021, 35, 265–272. [Google Scholar] [CrossRef]
- Wijnhoven, L.A.; Creemers, D.H.; Vermulst, A.A. Prevalence and risk factors of anxiety in a clinical Dutch sample of children with an autism spectrum disorder. Front Psychiatry 2018, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- McVey, A.J. The neurobiological presentation of anxiety in autism spectrum disorder: A systematic review. Autism Res. 2019, 12, 346–369. [Google Scholar] [CrossRef]
- Kerns, C.M.; Kendall, P.C. The presentation and classification of anxiety in autism spectrum disorder. Clin. Psychol. 2012, 19, 323–347. [Google Scholar] [CrossRef]
- Cervantes, P.; Matson, J.L.; Tureck, K. The relationship of comorbid anxiety symptom severity and challenging behaviors in infants and toddlers with autism spectrum disorder. Res. Autism Spectr. Disord. 2013, 7, 1528–1534. [Google Scholar] [CrossRef]
- Vasa, R.A.; Carroll, L.M.; Nozzolillo, A.A. A systematic review of treatments for anxiety in youth with autism spectrum disorders. J. Autism Dev. Disord. 2014, 44, 3215–3229. [Google Scholar] [CrossRef]
- Davis III, T.E.; Hess, J.A.; Moree, B.N. Anxiety symptoms across the lifespan in people diagnosed with autistic disorder. Res. Autism Spectr. Disord. 2011, 5, 112–118. [Google Scholar] [CrossRef]
- Uljarević, M.; Hedley, D.; Rose-Foley, K.; Magiati, I.; Cai, R.Y.; Dissanayake, C.; Richdale, A.; Trollor, J. Anxiety and depression from adolescence to old age in autism spectrum disorder. J. Autism Dev. Dis. 2019, 49, 559–572. [Google Scholar] [CrossRef]
- Salazar, F.; Baird, G.; Chandler, S. Co-occurring psychiatric disorders in preschool and elementary school-aged children with autism spectrum disorder. J. Autism Dev. Disord. 2015, 45, 2283–2294. [Google Scholar] [CrossRef] [Green Version]
- Hull, L.; Levy, L.; Lai, M.C.; Petrides, K.V.; Baron-Cohen, S.; Allison, C.; Smith, P.; Mandy, W. Is social camouflaging associated with anxiety and depression in autistic adults? Mol. Autism. 2021, 12, 13. [Google Scholar] [CrossRef]
- Agüera-, L.; García-Ramos, R.; Grandas, F.J.; López-Álvarez, J.; Rodríguez, J.M.M.; Rodríguez, F.J.O.; Olivera Pueyo, J.; Pelegrín Valero, C.; Porta-Etessam, J. Depression in Alzheimer’s disease: A Delphi consensus on etiology, risk factors, and clinical management. Front. Psychiatry 2021, 12, 141. [Google Scholar] [CrossRef]
- Asmer, M.S.; Kirkham, J.; Newton, H.; Ismail, Z.; Elbayoumi, H.; Leung, R.H.; Seitz, D.P. Meta-analysis of the prevalence of major depressive disorder among older adults with dementia. J. Clin. Psychiatry 2018, 79, 17r11772. [Google Scholar] [CrossRef] [PubMed]
- Starkstein, S.E.; Jorge, R.; Mizrahi, R.; Robinson, R.G. The construct of minor and major depression in Alzheimer’s disease. Am. J. Psychiatry 2005, 162, 2086–2093. [Google Scholar] [CrossRef]
- Kotagal, S.; Broomall, E. Sleep in children with autism spectrum disorder. Pediatric Neurol. 2012, 47, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Mannion, A.; Leader, G. Sleep problems in autism spectrum disorder: A literature review. J. Autism Dev. Disord. 2014, 1, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, A.; Hayashi, W.; Nishio, T.; Hanawa, Y.; Aoyagi, K.; Okajima, Y.; Iwanami, A. Similarity of subjective symptoms between autism spectrum disorder and attention-deficit/hyperactivity disorder in adults: Preliminary findings. Neuropsychopharmacol. Rep. 2021, 41, 237–241. [Google Scholar] [CrossRef]
- Outen, J.; Spira, A.; Wanigatunga, S.; Zipunnikov, V.; Wu, M.; Rosenberg, P. Circadian Rhythm Disturbance in Agitation of Alzheimer’s disease. Am. J. Geriatr. Psychiatry 2021, 29, S111–S113. [Google Scholar] [CrossRef]
- Cheng, W.; Rolls, E.; Gong, W.; Du, J.; Zhang, J.; Zhang, X.Y.; Li, F.; Feng, J. Sleep duration, brain structure, and psychiatric and cognitive problems in children. Mol. Psychiatry 2020, 26, 3992–4003. [Google Scholar] [CrossRef]
- Lee, S.B.; Park, J.; Kwak, Y.; Park, Y.U.; Nhung, T.T.; Suh, B.K.; Woo, Y.; Suh, Y.; Cho, E.; Cho, S.; et al. Disrupted-in-schizophrenia 1 enhances the quality of circadian rhythm by stabilizing BMAL1. Transl. Psychiatry 2021, 11, 110. [Google Scholar] [CrossRef]
- Kent, B.A.; Feldman, H.H.; Nygaard, H.B. Sleep and its regulation: An emerging pathogenic and treatment frontier in Alzheimer’s disease. Prog. Neurobiol. 2021, 197, 101902. [Google Scholar] [CrossRef]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Dekosky, S.T.; Barberger-Gateau, P.; Cummings, J. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007, 6, 734–746. [Google Scholar] [CrossRef]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Hampel, H.; Molinuevo, J.L.; Blennow, K.; DeKosky, S.T.; Gauthier, S.; Selkoe, D.; Bateman, R.; et al. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014, 13, 614–629. [Google Scholar] [CrossRef]
- Alexiou, A.; Mantzavinos, V.D.; Greig, N.H.; Kamal, M.A. A Bayesian Model for the Prediction and Early Diagnosis of Alzheimer’s disease. Front. Aging Neurosci. 2017, 9, 77. [Google Scholar] [CrossRef]
- Xiong, J.; Chen, S.; Pang, N.; Deng, X.; Yang, L.; He, F.; Wu, L.; Chen, C.; Yin, F.; Peng, J. Neurological diseases with autism spectrum disorder: Role of ASD risk genes. Front. Neurosci. 2019, 13, 349. [Google Scholar] [CrossRef]
- Santos, J.X.; Rasga, C.; Marques, A.R.; Martiniano, H.F.; Asif, M.; Vilela, J.; Oliveira, G.; Vicente, A.M. A role for gene-environment interactions in Autism Spectrum Disorder is suggested by variants in genes regulating exposure to environmental factors. bioRxiv 2019, 520544. [Google Scholar] [CrossRef]
- Hoffmann, A.; Spengler, D. Chromatin Remodeler CHD8 in Autism and Brain Development. J. Clin. Med. 2021, 10, 366. [Google Scholar] [CrossRef]
- Li, W.; Pozzo-Miller, L. Dysfunction of the corticostriatal pathway in autism spectrum disorders. J. Neurosci Res. 2020, 98, 2130–2147. [Google Scholar] [CrossRef]
- Chen, J.; Yu, S.; Fu, Y.; Li, X. Synaptic proteins and receptors defects in autism spectrum disorders. Front. Cell. Neurosci. 2014, 8, 276. [Google Scholar] [CrossRef] [Green Version]
- De Rubeis, S.; Buxbaum, J.D. Genetics and genomics of autism spectrum disorder: Embracing complexity. Hum. Mol. Genet. 2015, 24, R24–R31. [Google Scholar] [CrossRef]
- Puffenberger, E.G.; Jinks, R.N.; Wang, H.; Xin, B.; Fiorentini, C.; Sherman, E.A.; Degrazio, D.; Shaw, C.; Sougnez, C.; Cibulskis, K.; et al. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat. 2012, 33, 1639–1646. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Guo, H. Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders. Mol. Psychiatry 2017, 22, 1282–1290. [Google Scholar] [CrossRef]
- Peng, J.; Zhou, Y.; Wang, K. Multiplex gene and phenotype network to characterize shared genetic pathways of epilepsy and autism. Sci. Rep. 2021, 11, 952. [Google Scholar] [CrossRef]
- Rahbar, M.H.; Samms-Vaughan, M.; Saroukhani, S.; Bressler, J.; Hessabi, M.; Grove, M.L.; Shakspeare-Pellington, S.; Loveland, K.A.; Beecher, C.; McLaughlin, W. Associations of Metabolic Genes (GSTT1, GSTP1, GSTM1) and Blood Mercury Concentrations Differ in Jamaican Children with and without Autism Spectrum Disorder. Int. J. Environ. Res. Public Health 2021, 18, 1377. [Google Scholar] [CrossRef]
- Guo, H.; Peng, Y.; Hu, Z.; Li, Y.; Xun, G.; Ou, J.; Sun, L.; Xiong, Z.; Liu, Y.; Wang, T.; et al. Genome-wide copy number variation analysis in a Chinese autism spectrum disorder cohort. Sci. Rep. 2017, 7, 44155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascano, M.; Mukherjee, N.; Bandaru, P.; Miller, J.B.; Nusbaum, J.D.; Corcoran, D.L.; Langlois, C.; Munschauer, M.; Dewell, S.; Hafner, M.; et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 2012, 492, 382. [Google Scholar] [CrossRef]
- Garrido, N.; Cruz, F.; Egea, R.R.; Simon, C.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; Maamar, M.B.; Skinner, M.K. Sperm DNA methylation epimutation biomarker for paternal offspring autism susceptibility. Clin. Epigenet. 2021, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Takumi, T. Genomic and genetic aspects of autism spectrum disorder. Biochem. Biophys. Res. Commun. 2014, 452, 244–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niego, A.; Benítez-Burraco, A. Autism and Williams syndrome: Dissimilar socio-cognitive profiles with similar patterns of abnormal gene expression in the blood. Autism 2021, 25, 464–489. [Google Scholar] [CrossRef]
- Cacace, R.; Sleegers, K.; Van Broeckhoven, C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016, 12, 733–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoogmartens, J.; Hens, E.; Engelborghs, S.; De Deyn, P.P.; van der Zee, J.; Van Broeckhoven, C.; Cacace, R. Investigation of the role of matrix metalloproteinases in the genetic etiology of Alzheimer’s disease. Neurobiol. Aging 2021, 104, 105.e1–105.e6. [Google Scholar] [CrossRef]
- Lanoiselée, H.M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.H.; Hulette, C.; Saunders, A.M.; Einstein, G.; Pericak-Vance, M.; Strittmatter, W.J.; Roses, A.D.; Schmechel, D.E. Apolipoprotein E is present in hippocampal neurons without neurofibrillary tangles in Alzheimer’s disease and in age-matched controls. Exp. Neurol. 1994, 128, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Wolters, F.J.; Beiser, A.; Haan, M.; Ikram, M.A.; Karlawish, J.; Langbaum, J.B.; Neuhaus, J.M.; Reiman, E.M.; Roberts, J.S.; et al. APOE-related risk of mild cognitive impairment and dementia for prevention trials: An analysis of four cohorts. PLoS Med. 2017, 14, e1002254. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.; Cheng, T.L.; Li, G.Z.; Sun, S.B.; Yu, S.Y.; Zhang, Y.; Du, Y.S.; Qiu, Z. Identification of autism-related MECP2 mutations by whole-exome sequencing and functional validation. Mol. Autism 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Quan, W.; Wang, Z.; Chen, Y.; Zhang, H.; Zhou, Y. AD7c-NTP Impairs Adult Striatal Neurogenesis by Affecting the Biological Function of MeCP2 in APP/PSl Transgenic Mouse Model of Alzheimer’s Disease. Front. Aging Neurosci. 2021, 12, 478. [Google Scholar] [CrossRef]
- Ivashko-Pachima, Y.; Hadar, A.; Grigg, I.; Korenková, V.; Kapitansky, O.; Karmon, G.; Gershovits, M.; Sayas, C.L.; Kooy, R.F.; Attems, J.; et al. Discovery of autism/intellectual disability somatic mutations in Alzheimer’s brains: Mutated ADNP cytoskeletal impairments and repair as a case study. Mol. Psychiatry 2019, 26, 1619–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreoli, V.; De Marco, E.V.; Trecroci, F.; Cittadella, R.; Di Palma, G.; Gambardella, A. Potential involvement of GRIN2B encoding the NMDA receptor subunit NR2B in the spectrum of Alzheimer’s disease. J. Neural Transm. 2014, 121, 533–542. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, J.; Guo, H.; Ou, J.; Peng, Y.; Liu, Q.; Shen, Y.; Shi, L.; Liu, Y.; Xiong, Z.; et al. Association of genetic variants of GRIN2B with autism. Sci. Rep. 2015, 5, 8296. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Chen, W.; Myers, S.J.; Yuan, H.; Traynelis, S.F. Human GRIN2B variants in neurodevelopmental disorders. J. Pharmacol. Sci. 2016, 132, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Ilyas, M.; Salpietro, V.; Efthymiou, S.; Bourinaris, T.; Tariq, A.; Imdad, M.; Ahmad, A.; Ahmad, H.; Houlden, H. Identification of common genetic markers of paroxysmal neurological disorders using a network analysis approach. Neurol. Sci. 2019, 41, 851–857. [Google Scholar] [CrossRef]
- Spratt, P.W.; Ben-Shalom, R.; Keeshen, C.M.; Burke, K.J., Jr.; Clarkson, R.L.; Sanders, S.J.; Bender, K.J. The autism-associated gene Scn2a contributes to dendritic excitability and synaptic function in the prefrontal cortex. Neuron 2019, 103, 673–685. [Google Scholar] [CrossRef]
- Tristán-Clavijo, E.; Camacho-Garcia, R.J.; Robles-Lanuza, E.; Ruiz, A.; van der Zee, J.; Van Broeckhoven, C.; Hernandez, I.; Martinez-Mir, A.; Scholl, F.G. A truncating mutation in Alzheimer’s disease inactivates neuroligin-1 synaptic function. Neurobiol. Aging 2015, 36, 3171–3175. [Google Scholar] [CrossRef]
- Jamain, S.; Quach, H.; Betancur, C.; Råstam, M.; Colineaux, C.; Gillberg, I.C.; Soderstrom, H.; Giros, B.; Leboyer, M.; Gillberg, C.; et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003, 34, 27–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sindi, I.A.; Dodd, P.R. New insights into Alzheimer’s disease pathogenesis: The involvement of neuroligins in synaptic malfunction. Neurodegener. Dis. Manag. 2015, 5, 137–145. [Google Scholar] [CrossRef]
- Alarcón, M.; Abrahams, B.S.; Stone, J.L.; Duvall, J.A.; Perederiy, J.V.; Bomar, J.M.; Sebat, J.; Wigler, M.; Martin, C.L.; Ledbetter, D.H.; et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008, 82, 150–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñagarikano, O.; Geschwind, D.H. What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol. Med. 2012, 18, 156–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, A.; Ohara, T.; Takahashi, A.; Aoki, M.; Fuyuno, Y.; Ashikawa, K.; Morihara, T.; Takeda, M.; Kamino, K.; Oshima, E.; et al. A genome-wide association study of late-onset Alzheimer’s disease in a Japanese population. Psychiatry Genet. 2015, 25, 139–146. [Google Scholar] [CrossRef]
- Tábuas-Pereira, M.; Santana, I.; Guerreiro, R.; Brás, J. Alzheimer’s disease Genetics: Review of Novel Loci Associated with Disease. Curr. Genet. Med. Rep. 2020, 8, 1–16. [Google Scholar] [CrossRef]
- Louwersheimer, E.; Cohn-Hokke, P.E.; Pijnenburg, Y.A.; Weiss, M.M.; Sistermans, E.A.; Rozemuller, A.J.; Hulsman, M.; van Swieten, J.C.; van Duijn, C.M.; Barkhof, F.; et al. Rare Genetic Variant in SORL1 May Increase Penetrance of Alzheimer’s Disease in a Family with Several Generations of APOE-ɛ4 Homozygosity. J. Alzheimers Dis. 2017, 56, 63–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caubit, X.; Gubellini, P.; Andrieux, J.; Roubertoux, P.L.; Metwaly, M.; Jacq, B.; Fatmi, A.; Had-Aissouni, L.; Kwan, K.Y.; Salin, P.; et al. TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons. Nat. Genet. 2016, 48, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Chabbert, D.; Caubit, X.; Roubertoux, P.L.; Carlier, M.; Habermann, B.; Jacq, B.; Salin, P.; Metwaly, M.; Frahm, C.; Fatmi, A.; et al. Postnatal Tshz3 deletion drives altered corticostriatal function and autism spectrum disorder–like behavior. Biol. Psychiatry 2019, 86, 274–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchino, S.; Waga, C. SHANK3 as an autism spectrum disorder-associated gene. Brain Dev. 2013, 35, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Mossa, A.; Giona, F.; Pagano, J.; Sala, C.; Verpelli, C. SHANK genes in autism: Defining therapeutic targets. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 84, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Durand, C.M.; Betancur, C.; Boeckers, T.M.; Bockmann, J.; Chaste, P.; Fauchereau, F.; Nygren, G.; Rastam, M.; Gillberg, I.C.; Anckarsäter, H.; et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007, 39, 25–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Jaber, V.R.; LeBeauf, A.; Sharfman, N.M.; Lukiw, W.J. microRNA-34a (miRNA-34a) mediated down-regulation of the post-synaptic cytoskeletal element SHANK3 in sporadic Alzheimer’s disease (AD). Front. Neurol. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Matas, E.; Maisterrena, A.; Thabault, M.; Balado, E.; Francheteau, M.; Balbous, A.; Galvan, L.; Jaber, M. Major motor and gait deficits with sexual dimorphism in a Shank3 mutant mouse model. Mol. Autism 2021, 12, 2. [Google Scholar] [CrossRef]
- Zhou, J.; Parada, L.F. PTEN signaling in autism spectrum disorders. Curr. Opin. Neurobiol. 2012, 22, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Knafo, S.; Sánchez-Puelles, C.; Palomer, E.; Delgado, I.; Draffin, J.E.; Mingo, J.; Wahle, T.; Kaleka, K.; Mou, L.; Pereda-Perez, I.; et al. PTEN recruitment controls synaptic and cognitive function in Alzheimer’s models. Nat. Neurosci. 2016, 19, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Frere, S.; Slutsky, I. Targeting PTEN interactions for Alzheimer’s disease. Nat. Neurosci. 2016, 9, 416–418. [Google Scholar] [CrossRef]
- Matsuda, S.; Nakagawa, Y.; Tsuji, A.; Kitagishi, Y.; Nakanishi, A.; Murai, T. Implications of PI3K/AKT/PTEN signaling on superoxide dismutases expression and in the pathogenesis of Alzheimer’s disease. Diseases 2018, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, O.H.; Cho, H.J.; Han, E.; Hong, T.I.; Ariyasiri, K.; Choi, J.H.; Hwang, K.S.; Jeong, Y.M.; Yang, S.Y.; Yu, K.; et al. Zebrafish knockout of Down syndrome gene, DYRK1A, shows social impairments relevant to autism. Mol. Autism 2017, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- van Bon, B.W.; Coe, B.P.; Bernier, R.; Green, C.; Gerdts, J.; Witherspoon, K.; Kleefstra, T.; Willemsen, M.H.; Kumar, R.; Bosco, P.; et al. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry 2016, 21, 126–132. [Google Scholar] [CrossRef] [PubMed]
- García-Cerro, S.; Rueda, N.; Vidal, V.; Lantigua, S.; Martínez-Cué, C. Normalizing the gene dosage of Dyrk1A in a mouse model of Down syndrome rescues several Alzheimer’s disease phenotypes. Neurobiol. Dis. 2017, 106, 76–88. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.; Medda, F.; Gokhale, V.; Dunckley, T.; Hulme, C. Recent advances in the design, synthesis, and biological evaluation of selective DYRK1A inhibitors: A new avenue for a disease modifying treatment of Alzheimer’s? ACS Chem. Neurosci. 2012, 3, 857–872. [Google Scholar] [CrossRef] [Green Version]
- Fehér, Á.; Juhász, A.; Pákáski, M.; Kálmán, J.; Janka, Z. Genetic analysis of the RELN gene: Gender specific association with Alzheimer’s disease. Psychiatry Res. 2015, 230, 716–718. [Google Scholar] [CrossRef]
- Seripa, D.; Matera, M.G.; Franceschi, M.; Daniele, A.; Bizzarro, A.; Rinaldi, M.; Panza, F.; Fazio, V.M.; Gravina, C.; D’Onofrio, G.; et al. The RELN locus in Alzheimer’s disease. J. Alzheimers Dis. 2008, 14, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Lammert, D.B.; Howell, B.W. RELN mutations in autism spectrum disorder. Front. Cell. Neurosci. 2016, 10, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, W.H.; Gau, S.F.; Chen, C.H.; Tsai, W.C.; Wu, Y.Y.; Chen, P.H.; Shang, C.Y.; Chen, C.H. Increased gene expression of FOXP1 in patients with autism spectrum disorders. Mol. Autism 2013, 4, 23. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, K.; Friedman, B.A.; Etxeberria, A.; Huntley, M.A.; Van Der Brug, M.P.; Foreman, O.; Paw, J.S.; Modrusan, Z.; Beach, T.G.; Serrano, G.E.; et al. Alzheimer’s patient brain myeloid cells exhibit enhanced aging and unique transcriptional activation. BioRxiv 2019, 1, 610345. [Google Scholar] [CrossRef]
- Garcia-Oscos, F.; Koch, T.M.; Pancholi, H.; Trusel, M.; Daliparthi, V.; Park, S.E.; Ayhan, F.; Alam, D.H.; Holdway, J.E.; Konopka, G.; et al. Autism-linked gene FoxP1 selectively regulates the cultural transmission of learned vocalizations. Sci. Adv. 2021, 7, eabd2827. [Google Scholar] [CrossRef] [PubMed]
- Al-Erjan, A.M.; Kadhim, S.; Ahmed, M.A.; Kadham, M.J. Determine Some Mutations in the Foxp1 Gene in Autistic Patients in Baghdad Governorate. Ann. Rom. Soc. Cell Biol. 2021, 11, 2757–2762. [Google Scholar]
- Seyfried, N.T.; Dammer, E.B.; Swarup, V.; Nandakumar, D.; Duong, D.M.; Yin, L.; Deng, Q.; Nguyen, T.; Hales, C.M.; Wingo, T.; et al. A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s disease. Cell Syst. 2017, 4, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Rolland, T.; Cliquet, F.; Anney, R.J.; Traut, N.; Mathieu, A.; Huguet, G.; Leblond, C.S.; Douard, E.; Amsellem, F.; Malesys, S.; et al. Towards a gene-level map of resilience to genetic variants associated with autism. medRxiv 2021. [Google Scholar] [CrossRef]
- Lin, G.; Ji, K.; Li, S.; Ma, W.; Pan, X. The Genetics Analysis of Molecular Pathogenesis for Alzheimer’s Disease. Eur. Neurol. 2020, 83, 458–467. [Google Scholar] [CrossRef]
- Mahdavi, M.; Kheirollahi, M.; Riahi, R.; Khorvash, F.; Khorrami, M.; Mirsafaie, M. Meta-analysis of the association between GABA receptor polymorphisms and autism spectrum disorder (ASD). J. Mol. Neurosci. 2018, 65, 1–9. [Google Scholar] [CrossRef]
- Singh, B.K.; Vatsa, N.; Kumar, V.; Shekhar, S.; Sharma, A.; Jana, N.R. Ube3a deficiency inhibits amyloid plaque formation in APPswe/PS1δE9 mouse model of Alzheimer’s disease. Hum. Mol. Genet. 2017, 26, 4042–4054. [Google Scholar] [CrossRef] [PubMed]
- Jason, J.Y.; Paranjape, S.R.; Walker, M.P.; Choudhury, R.; Wolter, J.M.; Fragola, G.; Emanuele, M.J.; Major, M.B.; Zylka, M.J. The autism-linked UBE3A T485A mutant E3 ubiquitin ligase activates the Wnt/β-catenin pathway by inhibiting the proteasome. J. Biol. Chem. 2017, 292, 12503–12515. [Google Scholar] [CrossRef] [Green Version]
- Olabarria, M.; Pasini, S.; Corona, C.; Robador, P.; Song, C.; Patel, H.; Lefort, R. Dysfunction of the ubiquitin ligase E3A Ube3A/E6-AP contributes to synaptic pathology in Alzheimer’s disease. Commun. Biol. 2019, 2, 111. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.S.; Mizuno, K.; Ghosh, A.; Aziz, W.; Troakes, C.; Daoud, J.; Golash, V.; Noble, W.; Hortobágyi, T.; Giese, K.P. Alzheimer-related decrease in CYFIP2 links amyloid production to tau hyperphosphorylation and memory loss. Brain 2016, 139, 2751–2765. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tao, Y.; Song, F.; Sun, Y.; Ott, J.; Saffen, D. Common regulatory variants of CYFIP1 contribute to susceptibility for autism spectrum disorder (ASD) and classical autism. Ann. Hum. Genet. 2015, 79, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.J.; Li, W.X.; Liu, J.Q.; Guo, Y.C.; Wang, Q.; Li, G.H.; Dai, S.X.; Huang, J.F. Low expression of aging-related NRXN3 is associated with Alzheimer disease: A systematic review and meta-analysis. Medicine 2018, 97, 28. [Google Scholar] [CrossRef]
- Tromp, A.; Mowry, B.; Giacomotto, J. Neurexins in autism and schizophrenia-a review of patient mutations, mouse models and potential future directions. Mol. Psychiatry 2020, 26, 747–760. [Google Scholar] [CrossRef]
- Costa, A.S.; Guerini, F.R.; Arosio, B.; Galimberti, D.; Zanzottera, M.; Bianchi, A.; Nemni, R.; Clerici, M. SNARE complex polymorphisms associate with alterations of visual selective attention in Alzheimer’s Disease. J. Alzheimers Dis. 2019, 69, 179–188. [Google Scholar] [CrossRef]
- Tasaki, S.; Gaiteri, C.; Mostafavi, S.; De Jager, P.L.; Bennett, D.A. The molecular and neuropathological consequences of genetic risk for Alzheimer’s dementia. Front. Neurosci. 2018, 12, 699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartier, E.; Hamilton, P.J.; Belovich, A.N.; Shekar, A.; Campbell, N.G.; Saunders, C.; Andreassen, T.F.; Gether, U.; Veenstra-Vanderweele, J.; Sutcliffe, J.S.; et al. Rare autism-associated variants implicate syntaxin 1 (STX1 R26Q) phosphorylation and the dopamine transporter (hDAT R51W) in dopamine neurotransmission and behaviors. EBioMedicine 2015, 2, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Coley, A.A.; Gao, W.J. PSD95: A synaptic protein implicated in schizophrenia or autism? Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 82, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Qian, H.; Wang, L. Identification of Key Genes Related with Alzheimer’s Disease Treatment Through Bioinformatics Analysis. J. Biol. Life Sci. 2018, 9, 78–90. [Google Scholar]
- Quan, X.; Liang, H.; Chen, Y.A.; Qin, Q.; Wei, Y.; Liang, Z. Related network and differential expression analyses identify nuclear genes and pathways in the hippocampus of Alzheimer disease. Medical science monitor: Int. Med. J. Exp. Clin. Res. 2020, 26, e919311-1–e919311-11. [Google Scholar] [CrossRef] [PubMed]
- Ikezu, T.; Chen, C.; DeLeo, A.M.; Zeldich, E.; Fallin, M.D.; Kanaan, N.M.; Lunetta, K.L.; Abraham, C.R.; Logue, M.W.; Farrer, L.A. Tau phosphorylation is impacted by rare AKAP9 mutations associated with Alzheimer disease in African Americans. J. Neuroimmune Pharmacol. 2018, 13, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Poelmans, G.; Franke, B.; Pauls, D.L.; Glennon, J.C.; Buitelaar, J.K. AKAPs integrate genetic findings for autism spectrum disorders. Transl. Psychiatry 2013, 3, e270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villela, D.; Suemoto, C.K.; Pasqualucci, C.A.; Grinberg, L.T.; Rosenberg, C. Do copy number changes in CACNA2D2, CACNA2D3, and CACNA1D constitute a predisposing risk factor for Alzheimer’s disease? Front. Genet. 2016, 7, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willsey, A.J.; State, M.W. Autism spectrum disorders: From genes to neurobiology. Curr. Opin. Neurobiol. 2015, 30, 92–99. [Google Scholar] [CrossRef] [Green Version]
- Richard, A.E.; Scheffer, I.E.; Wilson, S.J. Features of the broader autism phenotype in people with epilepsy support shared mechanisms between epilepsy and autism spectrum disorder. Neurosci. Biobehav. Rev. 2017, 75, 203–233. [Google Scholar] [CrossRef] [Green Version]
- Oshodi, Y.; Ojewunmi, O.; Oshodi, T.A.; Ijarogbe, G.T.; Ogun, O.C.; Aina, O.F.; Lesi, F.E. Oxidative stress markers and genetic polymorphisms of glutathione S-transferase T1, M1, and P1 in a subset of children with autism spectrum disorder in Lagos, Nigeria. Niger. J. Clin. Pract. 2017, 20, 1161–1167. [Google Scholar] [CrossRef] [Green Version]
- Jafarian, Z.; Saliminejad, K.; Kamali, K.; Ohadi, M.; Kowsari, A.; Nasehi, L.; Khorshid, K.H.R. Association of glutathione S-transferases M1, P1 and T1 variations and risk of late-onset Alzheimer’s disease. Neurol. Res. 2018, 40, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.; Au, J.; Hagerman, P. FMR1 premutation and full mutation molecular mechanisms related to autism. J. Neurodev. Disord. 2011, 3, 211–224. [Google Scholar] [CrossRef] [Green Version]
- Renoux, A.J.; Carducci, N.M.; Ahmady, A.A.; Todd, P.K. Fragile X mental retardation protein expression in Alzheimer’s disease. Front. Genet. 2014, 5, 360. [Google Scholar] [CrossRef] [PubMed]
- Klusek, J.; Thurman, A.J.; Abbeduto, L. Maternal Pragmatic Language Difficulties in the FMR1 Premutation and the Broad Autism Phenotype: Associations with Individual and Family Outcomes. J. Autism Dev. Disord. 2021, 4, 1–7. [Google Scholar] [CrossRef]
- Ashley-Koch, A.E.; Jaworski, J.; Mei, H.; Ritchie, M.D.; Skaar, D.A.; Delong, G.R.; Worley, G.; Abramson, R.K.; Wright, H.H.; Cuccaro, M.L.; et al. Investigation of potential gene–gene interactions between APOE and RELN contributing to autism risk. Psychiatr. Genet. 2007, 17, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.G.; Tulloch, J.; Chen, S.; Leong, L.; Saxton, A.D.; Kraemer, B.; Darvas, M.; Keene, C.D.; Shutes-David, A.; Todd, K.; et al. Redefining transcriptional regulation of the APOE gene and its association with Alzheimer’s disease. PLoS ONE 2020, 15, e0227667. [Google Scholar] [CrossRef]
- Husain, M.A.; Laurent, B.; Plourde, M. APOE and Alzheimer’s disease: From lipid transport to physiopathology and therapeutics. Front. Neurosci. 2021, 15, 85. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, Y.; Zhao, Y.; Yu, H.; Ying, X.; Zhou, D.; Zhong, J.; Zheng, Z.; Liu, J.; Pan, R.; et al. APOE hypermethylation is associated with autism spectrum disorder in a Chinese population. Exp. Ther. Med. 2018, 15, 4749–4754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westmark, C.J.; Sokol, D.K.; Maloney, B.; Lahiri, D.K. Novel roles of amyloid-beta precursor protein metabolites in fragile X syndrome and autism. Mol. Psychiatry 2016, 21, 1333–1341. [Google Scholar] [CrossRef] [Green Version]
- Jonsson, T.; Atwal, J.K.; Steinberg, S.; Snaedal, J.; Jonsson, P.V.; Bjornsson, S.; Stefansson, H.; Sulem, P.; Gudbjartsson, D.; Maloney, J.; et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 2012, 488, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.D.; Sun, S.J.; Yang, J.; Chu, R.X.; Tu, W.; Liu, Q. Elevated serum brain-derived neurotrophic factor (BDNF) but not BDNF gene Val66Met polymorphism is associated with autism spectrum disorders. Mol. Neurobiol. 2017, 54, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Bryn, V.; Halvorsen, B.; Ueland, T.; Isaksen, J.; Kolkova, K.; Ravn, K.; Skjeldal, O.H. Brain derived neurotrophic factor (BDNF) and autism spectrum disorders (ASD) in childhood. Eur. J. Paediatr. Neurol. 2015, 19, 411–444. [Google Scholar] [CrossRef] [PubMed]
- Li, G.D.; Bi, R.; Zhang, D.F.; Xu, M.; Luo, R.; Wang, D.; Fang, Y.; Li, T.; Zhang, C.; Yao, Y.G.; et al. Female-specific effect of the BDNF gene on Alzheimer’s disease. Neurobiol. Aging 2017, 53, 192-e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF: Neurobiology and treatment of Alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Huang, L.; Li, X.; Li, H.; Zhou, Y.; Zhu, H.; Pan, T.; Kendrick, K.M.; Xu, W. Immunological cytokine profiling identifies TNF-α as a key molecule dysregulated in autistic children. Oncotarget 2017, 8, 82390. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Zhou, X.W.; Wang, J.Z. The dual roles of cytokines in Alzheimer’s disease: Update on interleukins, TNF-α, TGF-β and IFN-γ. Transl. Neurodegener. 2016, 5, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrò, M.; Mandelli, L.; Crisafulli, C.; Porcelli, S.; Albani, D.; Politis, A.; Papadimitriou, G.N.; Di Nicola, M.; Janiri, L.; Colombo, R.; et al. Psychiatric disorders and SLC6A4 gene variants: Possible effects on alcohol dependence and alzheimer’s disease. Mol. Biol. Rep. 2020, 47, 191–200. [Google Scholar] [CrossRef]
- Ma, D.Q.; Rabionet, R.; Konidari, I.; Jaworski, J.; Cukier, H.N.; Wright, H.H.; Abramson, R.K.; Gilbert, J.R.; Cuccaro, M.L.; Pericak-Vance, M.A.; et al. Association and gene–gene interaction of SLC6A4 and ITGB3 in autism. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2010, 153, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courchesne, E.; Mouton, P.R.; Calhoun, M.E.; Semendeferi, K.; Ahrens-Barbeau, C.; Hallet, M.J.; Barnes, C.C.; Pierce, K. Neuron number and size in prefrontal cortex of children with autism. JAMA 2011, 306, 2001–2010. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, E.; Pierce, K.; Schumann, C.M.; Redcay, E.; Buckwalter, J.A.; Kennedy, D.P.; Morgan, J. Mapping early brain development in autism. Neuron 2007, 56, 399–413. [Google Scholar] [CrossRef] [Green Version]
- Marchetto, M.C.; Belinson, H.; Tian, Y.; Freitas, B.C.; Fu, C.; Vadodaria, K.; Beltrao-Braga, P.C.; Trujillo, C.A.; Mendes, A.P.; Padmanabhan, K.; et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 2017, 22, 820–835. [Google Scholar] [CrossRef]
- Segawa, M. Early motor disturbances in Rett syndrome and its pathophysiological importance. Brain Dev. 2005, 27, S54–S58. [Google Scholar] [CrossRef]
- Xu, X.; Miller, E.C.; Pozzo-Miller, L. Dendritic spine dysgenesis in Rett syndrome. Front. Neuroanat. 2014, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangasamy, S.; Olfers, S.; Gerald, B.; Hilbert, A.; Svejda, S.; Narayanan, V. Reduced neuronal size and mTOR pathway activity in the Mecp2 A140V Rett syndrome mouse model. F1000Research 2016, 5, 2269. [Google Scholar] [CrossRef] [PubMed]
- Winden, K.D.; Ebrahimi-Fakhari, D.; Sahin, M. Abnormal mTOR Activation in Autism. Ann. Rev. Neurosci. 2018, 41, 1–23. [Google Scholar] [CrossRef]
- Onore, C.; Yang, H.; Van de Water, J.; Ashwood, P. Dynamic Akt/mTOR signaling in children with autism spectrum disorder. Front. Pediatrics 2017, 5, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busche, M.A.; Konnerth, A. Impairments of neural circuit function in Alzheimer’s disease. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150429. [Google Scholar] [CrossRef]
- Dziewczapolski, G.; Glogowski, C.M.; Masliah, E.; Heinemann, S.F. Deletion of the α7 nicotinic acetylcholine receptor gene improves cognitive deficits and synaptic pathology in a mouse model of Alzheimer’s disease. J. Neurosci. 2009, 29, 8805–8815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriele, S.; Sacco, R.; Persico, A.M. Blood serotonin levels in autism spectrum disorder: A systematic review and meta-analysis. Eur. Neuropsychopharmacol. 2014, 24, 919–929. [Google Scholar] [CrossRef]
- Murphy, D.L.; Lesch, K.P. Targeting the murine serotonin transporter: Insights into human neurobiology. Nat. Rev. Neurosci. 2008, 9, 85–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnin, A.; Goeden, N.; Chen, K.; Wilson, M.L.; King, J.; Shih, J.C.; Blakely, R.D.; Deneris, E.S.; Levitt, P. A transient placental source of serotonin for the fetal forebrain. Nature 2011, 472, 347–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanari, A.; Amenta, F.; Silvestrelli, G.; Tomassoni, D.; Parnetti, L. Neurotransmitter deficits in behavioural and psychological symptoms of Alzheimer’s disease. Mech. Ageing Dev. 2006, 127, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Previc, F.H. The role of the extrapersonal brain systems in religious activity. Conscious. Cogn. 2006, 15, 500–539. [Google Scholar] [CrossRef]
- Delaveau, P.; Salgado-Pineda, P.; Wicker, B.; Micallef-Roll, J.; Blinm, O. Effect of levodopa on healthy volunteers’ facial emotion perception: An FMRI study. Clin. Neuropharmacol. 2005, 28, 255–261. [Google Scholar] [CrossRef]
- Bachevalier, J.; Loveland, K.A. The orbitofrontal-amygdala circuit and self-regulation of social–emotional behavior in autism. Neurosci. Biobehav. Rev. 2006, 30, 97–117. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Kaminga, A.C.; Wen, S.W.; Wu, X.; Acheampong, K.; Liu, A. Dopamine and dopamine receptors in Alzheimer’s disease: A systematic review and network meta-analysis. Front. Aging Neurosci. 2019, 11, 175. [Google Scholar] [CrossRef] [Green Version]
- Nam, E.; Derrick, J.S.; Lee, S.; Kang, J.; Han, J.; Lee, S.J.; Chung, S.W.; Lim, M.H. Regulatory activities of dopamine and its derivatives toward metal-free and metal-induced amyloid-β aggregation, oxidative stress, and inflammation in Alzheimer’s disease. ACS Chem. Neurosci. 2018, 9, 2655–2666. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Reddy, P.H. Role of glutamate and NMDA receptors in Alzheimer’s disease. J. Alzheimers Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef] [Green Version]
- Eltokhi, A.; Santuy, A.; Merchan-Perez, A.; Sprengel, R. Glutamatergic dysfunction and synaptic ultrastructural alterations in schizophrenia and autism spectrum disorder: Evidence from human and rodent studies. Int. J. Mol. Sci. 2021, 22, 59. [Google Scholar] [CrossRef]
- Srivastava, A.; Das, B.; Yao, A.Y.; Yan, R. Metabotropic glutamate receptors in Alzheimer’s disease synaptic dysfunction: Therapeutic opportunities and hope for the future. J. Alzheimers Dis. 2020, 78, 1345–1361. [Google Scholar] [CrossRef]
- Khlebodarova, T.M.; Kogai, V.V.; Trifonova, E.A.; Likhoshvai, V.A. Dynamic landscape of the local translation at activated synapses. Mol. Psychiatry 2018, 23, 107–114. [Google Scholar] [CrossRef]
- Nordahl, C.W.; Scholz, R.; Yang, X.; Buonocore, M.H.; Simon, T.; Rogers, S.; Amaral, D.G. Increased rate of amygdala growth in children aged 2 to 4 years with autism spectrum disorders: A longitudinal study. Arch. Gen. Psychiatry 2012, 69, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Courchesne, E. Abnormal early brain development in autism. Mol. Psychiatry 2002, 7, S21–S23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexiou, A.; Soursou, G.; Yarla, N.S.; Ashraf, G.M. Proteins commonly linked to autism spectrum disorder and Alzheimer’s disease. Curr. Protein Pept. Sci. 2018, 19, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Mameza, M.G.; Dvoretskova, E.; Bamann, M.; Hönck, H.H.; Güler, T.; Boeckers, T.M.; Schoen, M.; Verpelli, C.; Sala, C.; Barsukov, I.; et al. SHANK3 gene mutations associated with autism facilitate ligand binding to the Shank3 ankyrin repeat region. J. Biol. Chem. 2013, 288, 26697–26708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucher, M.; Niebling, S.; Han, Y.; Molodenskiy, D.; Kreienkamp, H.J.; Svergun, D.; Kim, E.; Kostyukova, A.S.; Kreutz, M.R.; Mikhaylova, M. Autism associated SHANK3 missense point mutations impact conformational fluctuations and protein turnover at synapses. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jaber, V.; Zhao, Y.; Lukiw, W.J. Alterations in micro RNA-messenger RNA (miRNA-mRNA) coupled signaling networks in sporadic Alzheimer’s disease (AD) hippocampal CA1. J. Alzheimers Dis. Parkinsonism 2017, 7, 312. [Google Scholar] [CrossRef]
- Sokol, D.K.; Maloney, B.; Long, J.M.; Ray, B.; Lahiri, D.K. Autism, Alzheimer disease, and fragile X: APP, FMRP, and mGluR5 are molecular links. Neurology 2011, 76, 1344–1352. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, J.; Webberm, C. The roles of FMRP-regulated genes in autism spectrum disorder: Single-and multiple-hit genetic etiologies. Am. J. Hum. Genet. 2013, 93, 825–839. [Google Scholar] [CrossRef] [Green Version]
- Borreca, A.; Gironi, K.; Amadoro, G.; Ammassari-Teule, M. Opposite dysregulation of fragile-X mental retardation protein and heteronuclear ribonucleoprotein C protein associates with enhanced APP translation in Alzheimer disease. Mol. Neurobiol. 2016, 53, 3227–3234. [Google Scholar] [CrossRef]
- Barone, R.; Fichera, M.; De Grandi, M.; Battaglia, M.; Lo Faro, V.; Mattina, T.; Rizzo, R. Familial 18q12. 2 deletion supports the role of RNA-binding protein CELF4 in autism spectrum disorders. Am. J. Med. Genet. 2017, 173, 1649–1655. [Google Scholar] [CrossRef]
- McLane, R.D.; Schmitt, L.M.; Pedapati, E.V.; Shaffer, R.C.; Dominick, K.C.; Horn, P.S.; Gross, C.; Erickson, C.A. Peripheral amyloid precursor protein derivative expression in fragile X syndrome. Fronti. Integr. Neurosci. 2019, 13, 49. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Hoang, D.; Miller, N.; Ansaloni, S.; Huang, Q.; Rogers, J.T.; Lee, J.C.; Saunders, A.J. MicroRNAs can regulate human APP levels. Mol. Neurodegener. 2008, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, N.; Wang, R.; Maloney, B.; Nho, K.; Beck, J.S.; Pourshafie, N.; Niculescu, A.; Saykin, A.J.; Rinaldi, C.; Counts, S.E.; et al. MicroRNA-298 reduces levels of human amyloid-β precursor protein (APP), β-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol. Psychiatry 2020, 15, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruberti, F.; Barbato, C.; Cogoni, C. Post-transcriptional regulation of amyloid precursor protein by microRNAs and RNA binding proteins. Commun. Integr. Biol. 2010, 3, 499–503. [Google Scholar] [CrossRef]
- Pantopoulos, K. Iron metabolism and the IRE/IRP regulatory system: An update. Ann. N. Y. Acad. Sci. 2004, 1012, 1–13. [Google Scholar] [CrossRef]
- Zhou, Z.D.; Tan, E.K. Iron regulatory protein (IRP)-iron responsive element (IRE) signaling pathway in human neurodegenerative diseases. Mol. Neurodegener. 2017, 12, 75. [Google Scholar] [CrossRef]
- Lee, E.K.; Kim, H.H.; Kuwano, Y.; Abdelmohsen, K.; Srikantan, S.; Subaran, S.S. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010, 17, 732–739. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.J.; Shi, H.; Zhu, A.C.; Lu, Z.; Miller, N.; Edens, B.M.; Ma, Y.C.; He, C. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine–containing mRNAs. J. Biol. Chem. 2019, 294, 19889–19895. [Google Scholar] [CrossRef]
- Schepens, B.; Tinton, S.A.; Bruynooghe, Y.; Parthoens, E.; Haegman, M.; Beyaert, R. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007, 26, 158–169. [Google Scholar] [CrossRef] [Green Version]
- Westmark, C.J.; Maloney, B.; Alisch, R.S.; Sokol, D.K.; Lahiri, D.K. FMRP Regulates the Nuclear Export of Adam9 and Psen1 mRNAs: Secondary Analysis of an N 6-Methyladenosine Dataset. Sci. Rep. 2020, 10, 10781. [Google Scholar] [CrossRef]
- Ray, B.; Maloney, B.; Sambamurti, K.; Karnati, H.K.; Nelson, P.T.; Greig, N.H. Rivastigmine modifies the α-secretase pathway and potentially early Alzheimer’s disease. Transl. Psychiatry 2020, 10, 47. [Google Scholar] [CrossRef] [Green Version]
- Rice, H.C.; De Malmazet, D.; Schreurs, A.; Frere, S.; Van Molle, I.; Volkov, A.N.; Creemers, E.; Vertkin, I.; Nys, J.; Ranaivoson, F.M.; et al. Secreted amyloid-β precursor protein functions as a GABABR1a ligand to modulate synaptic transmission. Science 2019, 363, eaao4827. [Google Scholar] [CrossRef]
- Tang, B.L. Amyloid precursor protein (APP) and GABAergic neurotransmission. Cells 2019, 8, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, B.; Long, J.M.; Sokol, D.K.; Lahiri, D.K. Increased secreted amyloid precursor protein-α (sAPPα) in severe autism: Proposal of a specific, anabolic pathway and putative biomarker. PLoS ONE 2011, 22, e20405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, K.; Kang, J.; Ouyang, G.; Li, J.; Han, J.; Wang, Y.; Sokhadze, E.M.; Casanova, M.F.; Li, X. Disrupted brain network in children with autism spectrum disorder. Sci. Rep. 2017, 7, 16253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, B.; Sokol, D.K.; Maloney, B.; Lahiri, D.K. Finding novel distinctions between the sAPPα-mediated anabolic biochemical pathways in Autism Spectrum Disorder and Fragile X Syndrome plasma and brain tissue. Sci Rep. 2016, 6, 26052. [Google Scholar] [CrossRef]
- Sokol, D.K.; Maloney, B.; Westmark, C.J.; Lahiri, D.K. Novel contribution of secreted amyloid-β precursor protein to white matter brain enlargement in autism spectrum disorder. Front. Ppsychiatry 2019, 10, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Sr. No. | Genes Associated with ASD and AD | Reference |
|---|---|---|
| 1 | MECP2 (methyl-CpG binding protein 2) | [102,103]. |
| 2 | ADNP (activity-dependent neuroprotective protein) | [20,104]. |
| 3 | GRIN2B (glutamate ionotropic receptor NMDA type subunit 2B) | [105,106,107]. |
| 4 | SCN2A (sodium voltage-gated channel alpha subunit 2) | [108,109]. |
| 5 | NLGN (neuroligin) | [110,111,112]. |
| 6 | CNTNAP2 (contactin-associated protein 2) | [113,114,115,116]. |
| 7 | TSHZ3 (teashirt zinc finger homeobox 3) | [117,118,119]. |
| 8 | SHANK | [120,121,122,123,124]. |
| 9 | PTEN (phosphatase and tensin homolog) | [125,126,127,128]. |
| 10 | DYRK1A (dual-specificity tyrosine phosphorylation-regulated kinase 1A) | [129,130,131,132]. |
| 11 | RELN (reelin) | [133,134,135]. |
| 12 | FOXP1 (forkhead box protein P1) | [136,137,138,139]. |
| 13 | SYNGAP1 (synaptic Ras GTPase-activating protein 1) | [140,141]. |
| 14 | GABRA5 (gamma-aminobutyric acid type A receptor subunit alpha5) | [142,143]. |
| 15 | UBE3A (ubiquitin-protein ligase E3A) | [144,145,146]. |
| 16 | CYFIP1 (cytoplasmic FMR1-interacting protein 1) | [147,148]. |
| 17 | NRXN (neurexin) | [149,150]. |
| 18 | STX1A (syntaxin 1A) | [151,152,153]. |
| 19 | DLG4 (discs large MAGUK scaffold protein 4) | [154,155,156]. |
| 20 | AKAP9 (A-kinase anchoring protein 9) | [157,158]. |
| 21 | CACNA2D3 (calcium voltage-gated channel auxiliary subunit alpha2delta 3) | [159,160,161]. |
| 22 | GSTT1(glutathione S-transferase theta-1), GSTM1(glutathione S-transferase Mu 1), GSTP1(glutathione S-transferase pi 1) | [162,163]. |
| 23 | FMR1 (fragile X mental retardation 1) | [164,165,166]. |
| 24 | APOE (apolipoprotein E) | [167,168,169,170]. |
| 25 | APP (beta-amyloid precursor protein) | [171,172]. |
| 26 | BDNF (brain-derived neurotrophic factor) | [173,174,175,176]. |
| 27 | TNF (tumor necrosis factor) | [177,178]. |
| 28 | SLC6A4 (solute carrier family 6 (neurotransmitter transporter, serotonin), member 4) | [179,180]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nadeem, M.S.; Hosawi, S.; Alshehri, S.; Ghoneim, M.M.; Imam, S.S.; Murtaza, B.N.; Kazmi, I. Symptomatic, Genetic, and Mechanistic Overlaps between Autism and Alzheimer’s Disease. Biomolecules 2021, 11, 1635. https://doi.org/10.3390/biom11111635
Nadeem MS, Hosawi S, Alshehri S, Ghoneim MM, Imam SS, Murtaza BN, Kazmi I. Symptomatic, Genetic, and Mechanistic Overlaps between Autism and Alzheimer’s Disease. Biomolecules. 2021; 11(11):1635. https://doi.org/10.3390/biom11111635
Chicago/Turabian StyleNadeem, Muhammad Shahid, Salman Hosawi, Sultan Alshehri, Mohammed M. Ghoneim, Syed Sarim Imam, Bibi Nazia Murtaza, and Imran Kazmi. 2021. "Symptomatic, Genetic, and Mechanistic Overlaps between Autism and Alzheimer’s Disease" Biomolecules 11, no. 11: 1635. https://doi.org/10.3390/biom11111635
APA StyleNadeem, M. S., Hosawi, S., Alshehri, S., Ghoneim, M. M., Imam, S. S., Murtaza, B. N., & Kazmi, I. (2021). Symptomatic, Genetic, and Mechanistic Overlaps between Autism and Alzheimer’s Disease. Biomolecules, 11(11), 1635. https://doi.org/10.3390/biom11111635







