Identification of miRNA-Target Gene Pairs Responsive to Fusarium Wilt of Cucumber via an Integrated Analysis of miRNA and Transcriptome Profiles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Foc Treatment
2.2. Total RNA Isolation and Library Preparation for sRNA Sequencing
2.3. miRNA-Seq Analysis
2.4. MicroRNA Family Identification and Target Prediction
2.5. Quantitative Real-Time RT-PCR (qRT-PCR) Assay of the miRNAs
2.6. Identification and Validation of Target Differentially Expressed Target Genes
3. Results
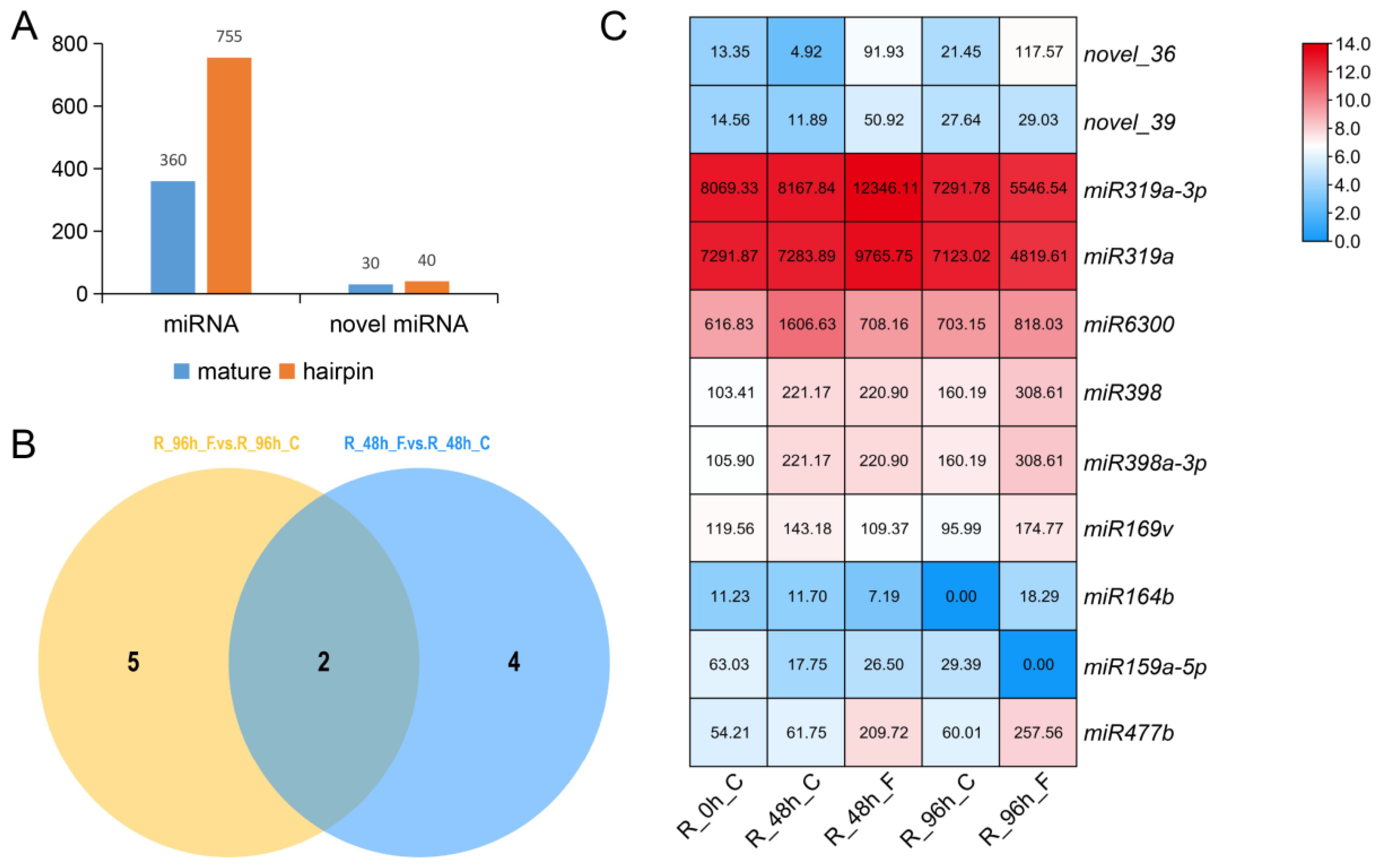
3.1. Identification of miRNAs Responsive to Foc
3.2. Validation of miRNA Expression by qRT-PCR
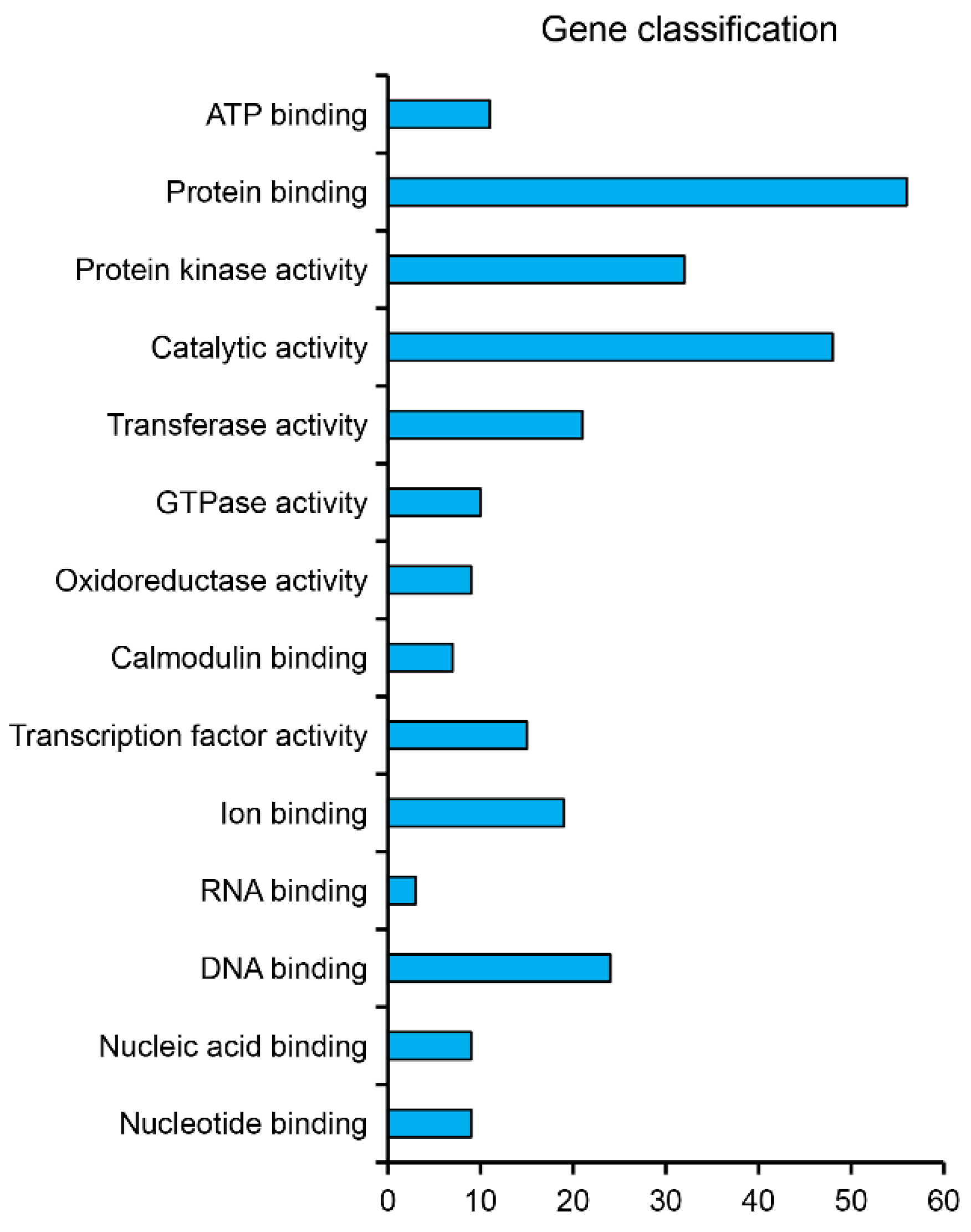
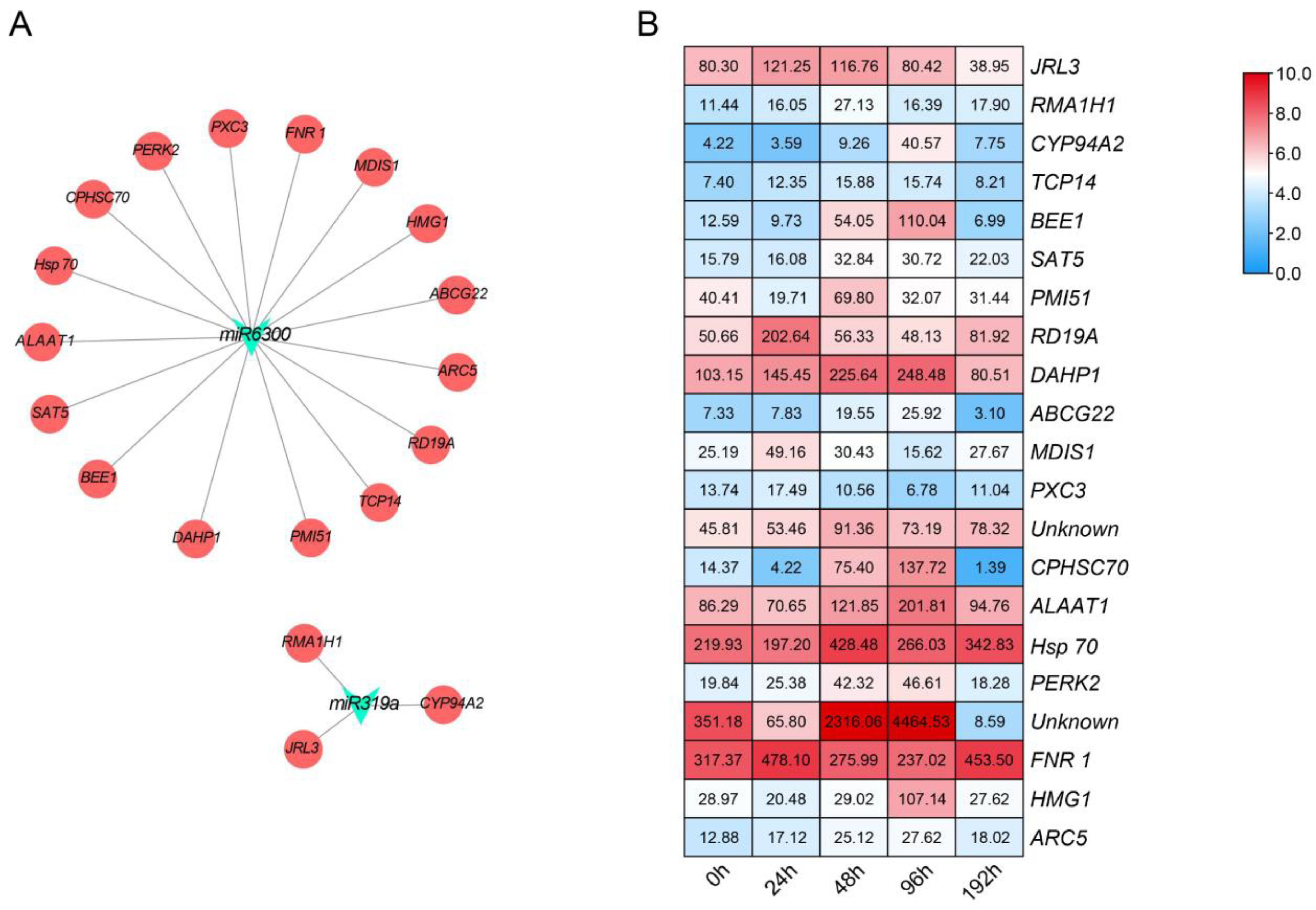
3.3. Identification of miRNA Target Genes and Profile Analyses
3.4. Verification of the Expression of Target Genes in Cucumber Infected with Foc
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FW | Fusarium wilt |
| Foc | Fusarium oxysporum f. sp. cucumerinum |
| DEM | differentially expressed miRNA |
| miRNA | microRNA |
| sRNA | small non-coding RNA |
| FPKM | fragments per kilobase of exon model per million mapped fragments |
| NCBI | National Center for Biotechnology Information |
| GO | Gene Ontology |
| qPCR | quantitative real-time polymerase chain reaction |
References
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, P.; Chung, H.S.; Lee, Y.H. Vegetative compatibility groups and pathogenicity among isolates of Fusarium oxysporum f. sp. cucumerinum. Plant Dis. 1998, 82, 244–246. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Shi, D.; Sun, F.; Lu, H.; Liu, J.; Wu, W. Efficacy of sludge and manure compost amendments against Fusarium wilt of cucumber. Environ. Sci. Pollut. Res. Int. 2012, 19, 3895–3905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.P.; Miao, H.; Yang, Y.H.; Xie, B.Y.; Wang, Y.; Gu, X.F. A major quantitative trait locus conferring resistance to Fusarium wilt was detected in cucumber by using recombinant inbred lines. Mol. Breed. 2014, 34, 1805–1815. [Google Scholar] [CrossRef]
- Wang, M.; Sun, Y.M.; Sun, G.M.; Liu, X.; Zhai, L.; Shen, Q.; Guo, S. Water balance altered in cucumber plants infected with Fusarium oxysporum f. sp. cucumerinum. Sci. Rep. 2015, 5, 7722. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Pu, X.; Feng, B.; Yang, Q.; Shen, H.; Zhang, J.; Lin, B. FocVel1 influences asexual production, filamentous growth, biofilm formation, and virulence in Fusarium oxysporum f. sp. cucumerinum. Front. Plant Sci. 2015, 6, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGovern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Snyder, W.C.; Hansen, H.N. The species concept in Fusarium. Am. J. Bot. 1940, 27, 64–67. [Google Scholar] [CrossRef]
- Martínez, R.; Aguilar, M.I.; Guirado, M.L.; Alvarez, A.; Gomez, J. First report of Fusarium wilt of cucumber caused by Fusarium oxysporum in Spain. Plant Pathol. 2003, 52, 410. [Google Scholar] [CrossRef]
- Chen, L.H.; Huang, X.Q.; Zhang, F.G.; Zhao, D.K.; Yang, X.M.; Shen, Q.R. Application of Trichoderma harzianum SQR-T037 bioorganic fertiliser significantly controls Fusarium wilt and affects the microbial communities of continuously cropped soil of cucumber. J. Sci. Food Agric. 2012, 92, 2465–2470. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Reinhart, B.J.; Bartel, D.P. A biochemical framework for RNA silencing in plants. Gene 2003, 17, 49–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.A. MicroRNA as translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Sun, C.; Zhao, Q.; Liu, D.D. Ectopic expression of the apple Md-miRNA156h gene regulates flower and fruit development in Arabidopsis. Plant Cell Tissue 2013, 112, 343–351. [Google Scholar] [CrossRef]
- Tang, F.; Wei, H.R.; Zhao, S.T. Identification of microRNAs involved in regeneration of the secondary vascular system in Populus tomentosa Carr. Front. Plant Sci. 2016, 7, 724–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.H.; Bao, M.L.; Sun, Y.Z.; Yang, Y.J.; Xu, X.H.; Wang, J.H.; Han, N.; Bian, H.W.; Zhu, M.Y. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 2011, 77, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sanuy, F.; Peris-Peris, C.; Tomiyama, S.; Okada, K.; Hsing, Y.I.; San Segundo, B.; Campo, S. Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus. BMC Plant Biol. 2019, 19, 563. [Google Scholar] [CrossRef] [Green Version]
- Campo, S.; Sánchez-Sanuy, F.; Camargo-Ramírez, R.; Gómez-Ariza, J.; Baldrich, P.; Campos-Soriano, L.; Soto-Suárez, M.; San Segundo, B. A novel transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 2021, 19, 1798–1811. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Lei, Y.; Liu, J.; Hao, M.; Zhang, Z.; Tang, Y.; Chen, A.; Wu, J. The ghr-miR164 and GhNAC100 modulate cotton plant resistance against Verticillium dahlia. Plant Sci. 2020, 293, 110438. [Google Scholar] [CrossRef]
- Jiang, N.; Cui, J.; Shi, Y.; Yang, G.; Zhou, X.; Hou, X.; Meng, J.; Luan, Y. Tomato lncRNA23468 functions as a competing endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic. Res. 2019, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.P.; Qi, X.H.; Xu, Q.; Chen, X.H. Segregation and identification of Fusarium oxysporum f. sp. cucumerinum and analysis of cucumber varieties′ resistance difference. Mol. Plant Breed. 2017, 15, 3648–3653. [Google Scholar]
- Dong, J.P.; Wang, Y.A.; Xian, Q.Q.; Chen, X.; Xu, J. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant Biol. 2020, 20, 334. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Gu, L.; Li, P.; Song, X.; Cao, X. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica). Front. Biol. 2010, 5, 67–90. [Google Scholar] [CrossRef]
- Wu, H.J.; Ma, Y.K.; Chen, T.; Wang, M.; Wang, X.J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Howell, M.D.; Kasschau, K.D.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Law, T.F.; Grant, S.R.; Dangl, J.L.; et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2007, 2, e219. [Google Scholar] [CrossRef]
- Gheysarzadeh, A.; Sadeghifard, N.; Afraidooni, L.; Pooyan, F.; Mofid, M.R.; Valadbeigi, H.; Bakhtiari, H.; Keikhavani, S. Serum-based microRNA biomarkers for major depression: MiR-16, miR-135a, and miR-1202. J. Res. Med. Sci. 2018, 23, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, X.; Wang, X.; Zhang, S.; Guo, X. ghr-miR5272a-mediated regulation of GhMKK6 gene transcription contributes to the immune response in cotton. J. Exp. Bot. 2017, 68, 5895–5906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Liu, D.; Chen, D.; Cheng, Y.; Zhang, X.; Song, L.; Hu, M.; Dong, J.; Shen, F. MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 2019, 16, 362–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Hao, M.; Wang, L.; Liu, J.; Zhang, Z.; Tang, Y.; Peng, Q.; Yang, Z.; Wu, J. The cotton miR477-CBP60A module participates in plant defence against Verticillium dahliae. Mol. Plant Microbe Interact. 2019, 33, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mu, X.Y.; Liu, C.; Cai, J.; Shi, K.; Zhu, W.; Yang, Q. Over expression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Soto-Suárez, M.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; San Segundo, B. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci. Rep. 2017, 7, 44898. [Google Scholar] [CrossRef] [Green Version]
- Boccara, M.; Sarazin, A.; Thiebeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.J.; Lee, H.J.; Kwak, K.J.; Lee, K.; Hong, S.W.; Kang, H. MicroRNA400-guided cleavage of Pentatricopeptide repeat protein mRNAs Renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 2014, 55, 1660–1668. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Park, Y.J.; Kwak, K.J.; Kim, D.; Park, J.H.; Lim, J.Y.; Shin, C.; Yang, K.Y.; Kang, H. MicroRNA844-Guided Down-regulation of Cytidinephosphate Diacylglycerol Synthase3 (CDS3) mRNA Affects the Response of Arabidopsis thaliana to Bacteria and Fungi. Mol. Plant Microbe Interact. 2015, 28, 892–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camargo-Ramírez, R.; Val-Torregrosa, B.; San Segundo, B. MiR858-Mediated Regulation of Flavonoid-Specific MYB Transcription Factor Genes Controls Resistance to Pathogen Infection in Arabidopsis. Plant Cell Physiol. 2018, 59, 190–204. [Google Scholar] [CrossRef]
- Salvador-Guirao, R.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; San Segundo, B. The MicroRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Mol. Plant Microbe Interact. 2018, 31, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandran, V.; Wang, H.; Gao, F.; Cao, X.L.; Chen, Y.P.; Li, G.B.; Zhu, Y.; Yang, X.M.; Zhang, L.L.; Zhao, Z.X.; et al. miR396-OsGRFs module balances growth and rice blast disease-resistance. Front. Plant Sci. 2019, 9, 1999. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Hong, G.; Li, L.; Zhang, X.; Kong, Y.; Sun, Z.; Li, J.; Chen, J.; He, Y. miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 2019, 109, 632–642. [Google Scholar]
- Campo, S.; Peris-Peris, C.; Siré, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Y.G.; Shi, Y.; Wu, L.; Xu, Y.J.; Huang, F.; Guo, X.Y.; Zhang, Y.; Fan, J.; Zhao, J.Q.; et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhao, S.L.; Li, J.L.; Hu, X.H.; Wang, H.; Cao, X.L.; Xu, Y.J.; Zhao, Z.X.; Xiao, Z.Y.; Yang, N.; et al. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Cao, X.L.; Zhu, Y.; Yang, X.M.; Zhang, K.N.; Xiao, Z.Y.; Wang, H.; Zhao, J.H.; Zhang, L.L.; Li, G.B.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.P.; Ma, X.C.; Wang, H.; Zhu, Y.; Liu, X.X.; Li, T.T.; Zheng, Y.P.; Zhao, J.Q.; Zhang, J.W.; Huang, Y.Y.; et al. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and yield. Rice 2020, 13, 38. [Google Scholar] [CrossRef]
- Salvador-Guirao, R.; Hsing, Y.I.; San Segundo, B. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front. Plant Sci. 2018, 9, 337. [Google Scholar] [CrossRef]
- Zhang, L.L.; Li, Y.; Zheng, Y.P.; Wang, H.; Yang, X.; Chen, J.F.; Zhou, S.X.; Wang, L.F.; Li, X.P.; Ma, X.C.; et al. Expressing a target mimic of miR156fhl-3p enhances rice blast disease resistance without yield penalty by improving SPL14 expression. Front. Genet. 2020, 11, 327. [Google Scholar] [CrossRef]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae Induces the Expression of a MicroRNA to Suppress the Immune Response in Rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, Y.; Chern, M.; Zhu, Y.; Zhang, L.L.; Lu, J.H.; Li, X.P.; Dang, W.Q.; Ma, X.C.; Yang, Z.R.; et al. Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 2021, 7, 129–136. [Google Scholar] [CrossRef]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.X.; Feng, Q.; Cao, X.L.; Zhu, Y.; Wang, H.; Chandran, V.; Fan, J.; Zhao, J.Q.; Pu, M.; Li, Y.; et al. Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integr. Plant Biol. 2020, 62, 702–715. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Zhu, Y.; Wang, L.; Zheng, Y.P.; Chen, J.F.; Li, T.T.; Yang, X.M.; Wang, H.; Li, X.P.; Ma, X.C.; et al. Osa-miR1873 fine-tunes rice immunity against Magnaporthe oryzae and yield traits. J. Integr. Plant Biol. 2019, 62, 1213–1226. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Song, M.; Wei, Z.; Tong, J.; Zhang, L.; Xiao, L.; Ma, Z.; Wang, Y. A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 2011, 62, 5471–5483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Q.; Dong, L.; Gao, T.; Liu, T.; Li, N.; Wang, L.; Chang, X.; Wu, J.; Xu, P.; Zhang, S. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018, 69, 2527–2541. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Dong, X.N.; Ausubel, F.M.; Fink, G.R. Differential induction of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc. Natl. Acad. Sci. USA 1991, 88, 8821–8825. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Xian, Q.; Zhang, N.; Wang, K.; Zhou, X.; Li, Y.; Dong, J.; Chen, X. Identification of miRNA-Target Gene Pairs Responsive to Fusarium Wilt of Cucumber via an Integrated Analysis of miRNA and Transcriptome Profiles. Biomolecules 2021, 11, 1620. https://doi.org/10.3390/biom11111620
Xu J, Xian Q, Zhang N, Wang K, Zhou X, Li Y, Dong J, Chen X. Identification of miRNA-Target Gene Pairs Responsive to Fusarium Wilt of Cucumber via an Integrated Analysis of miRNA and Transcriptome Profiles. Biomolecules. 2021; 11(11):1620. https://doi.org/10.3390/biom11111620
Chicago/Turabian StyleXu, Jun, Qianqian Xian, Ningyuan Zhang, Ke Wang, Xin Zhou, Yansong Li, Jingping Dong, and Xuehao Chen. 2021. "Identification of miRNA-Target Gene Pairs Responsive to Fusarium Wilt of Cucumber via an Integrated Analysis of miRNA and Transcriptome Profiles" Biomolecules 11, no. 11: 1620. https://doi.org/10.3390/biom11111620
APA StyleXu, J., Xian, Q., Zhang, N., Wang, K., Zhou, X., Li, Y., Dong, J., & Chen, X. (2021). Identification of miRNA-Target Gene Pairs Responsive to Fusarium Wilt of Cucumber via an Integrated Analysis of miRNA and Transcriptome Profiles. Biomolecules, 11(11), 1620. https://doi.org/10.3390/biom11111620





