Nucleic Acids Analytical Methods for Viral Infection Diagnosis: State-of-the-Art and Future Perspectives
Abstract
1. Introduction
2. Conventional Methods for Virus Detection
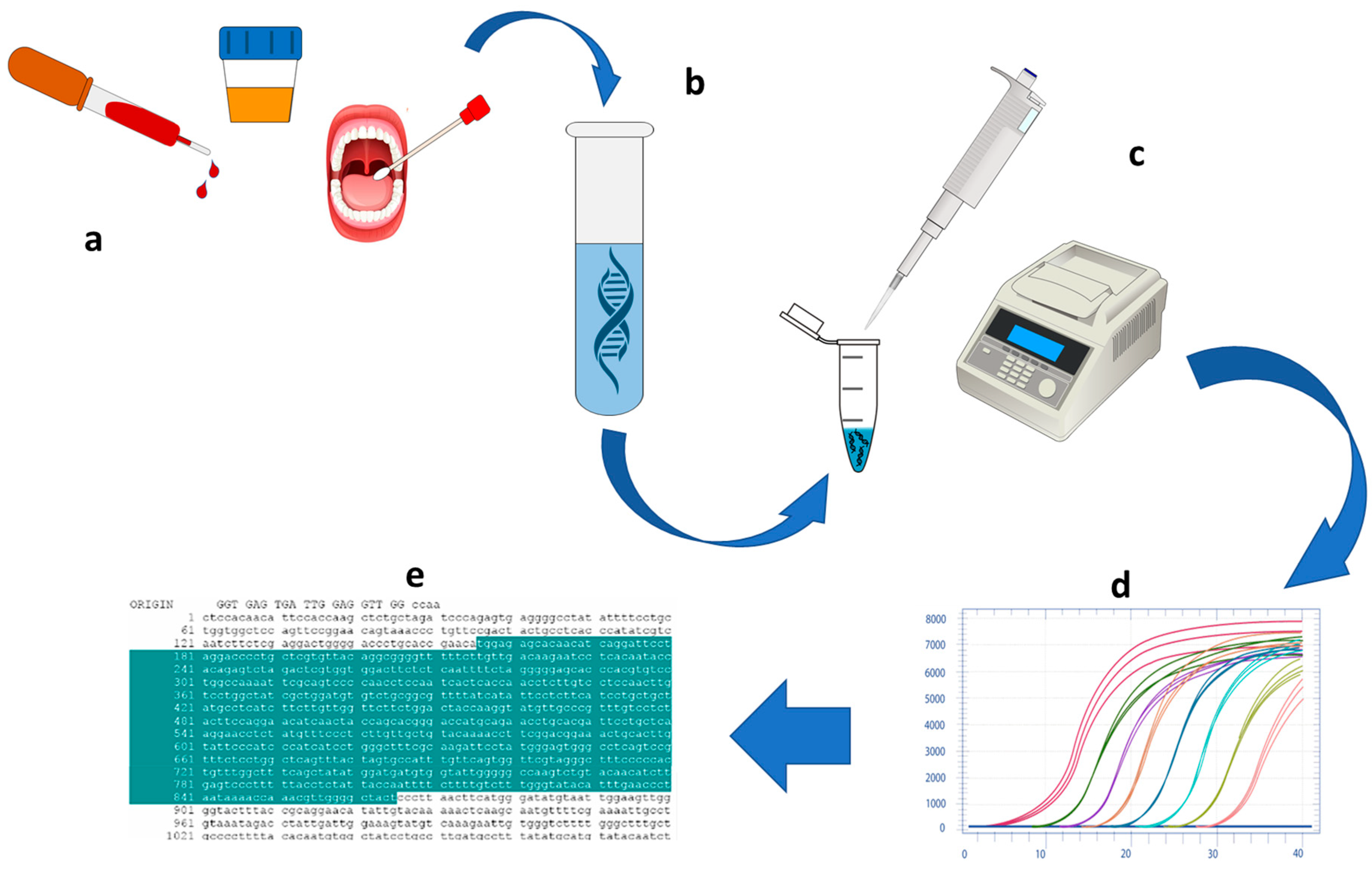
3. NATs Improvements towards PoC Applications
3.1. NATs Evolution: NA Extraction
3.2. NATs Evolution: Redox PCR Probes
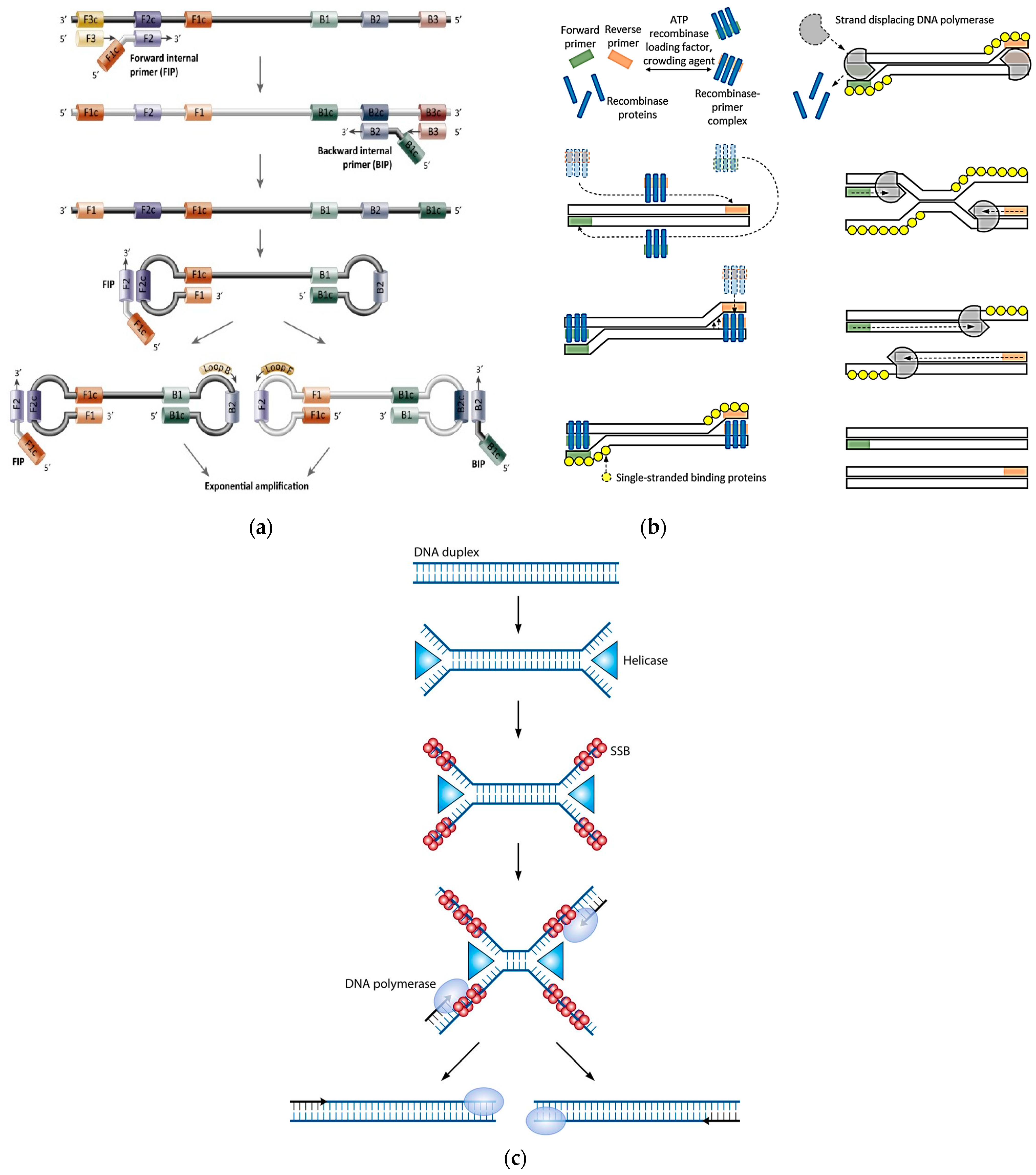
3.3. NATs Evolution: Isothermal PCR
4. Genetic PoC Systems for Viral Infection Diagnosis
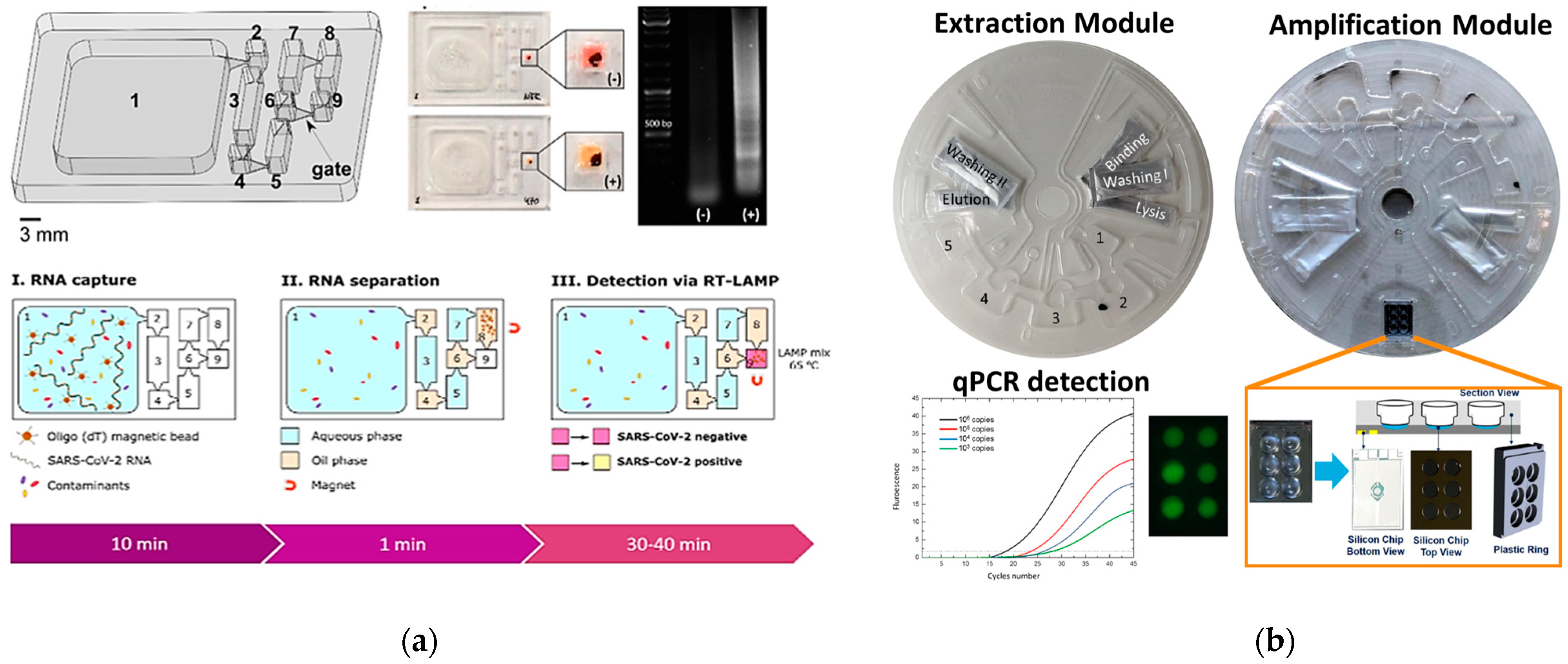
4.1. PCR-Based Genetic PoC
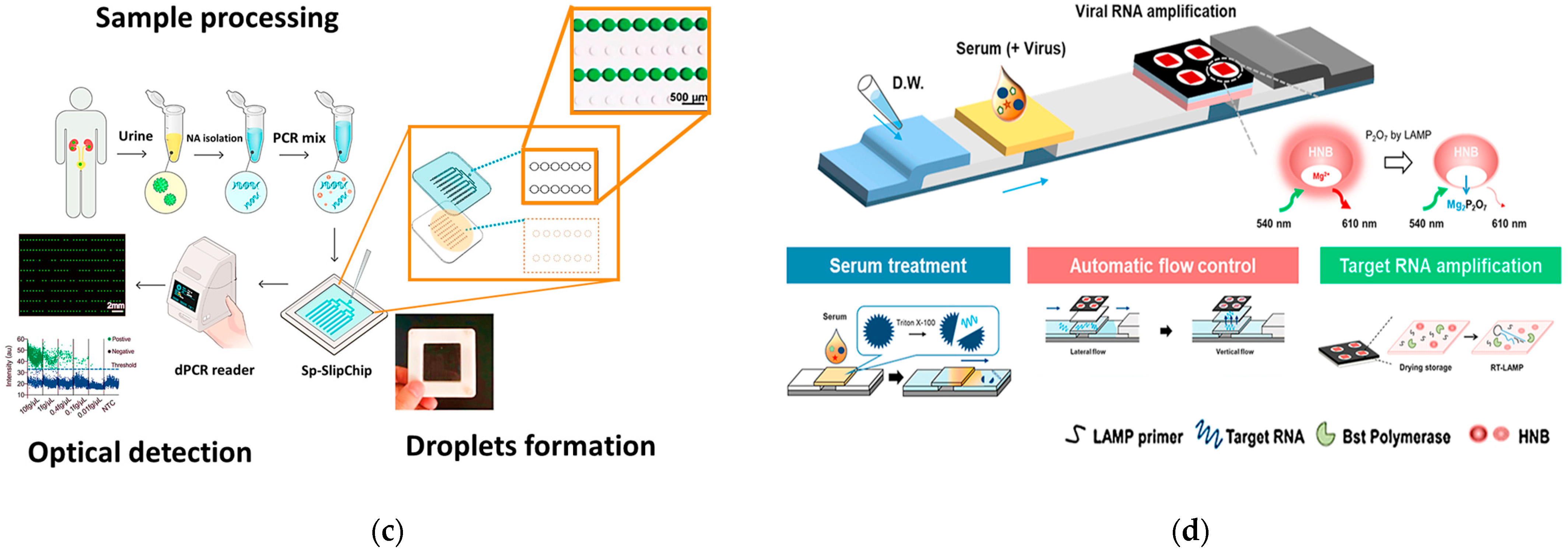
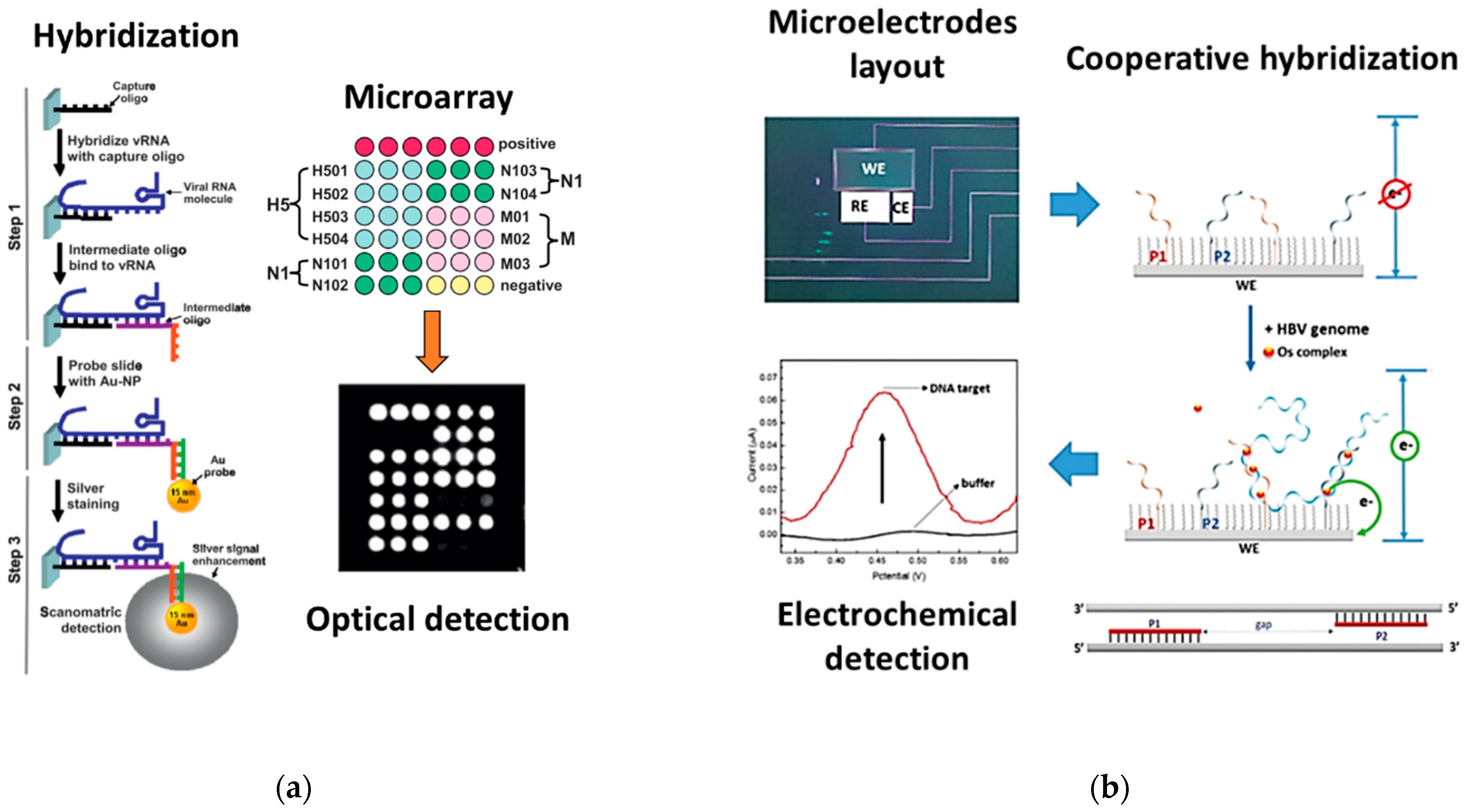
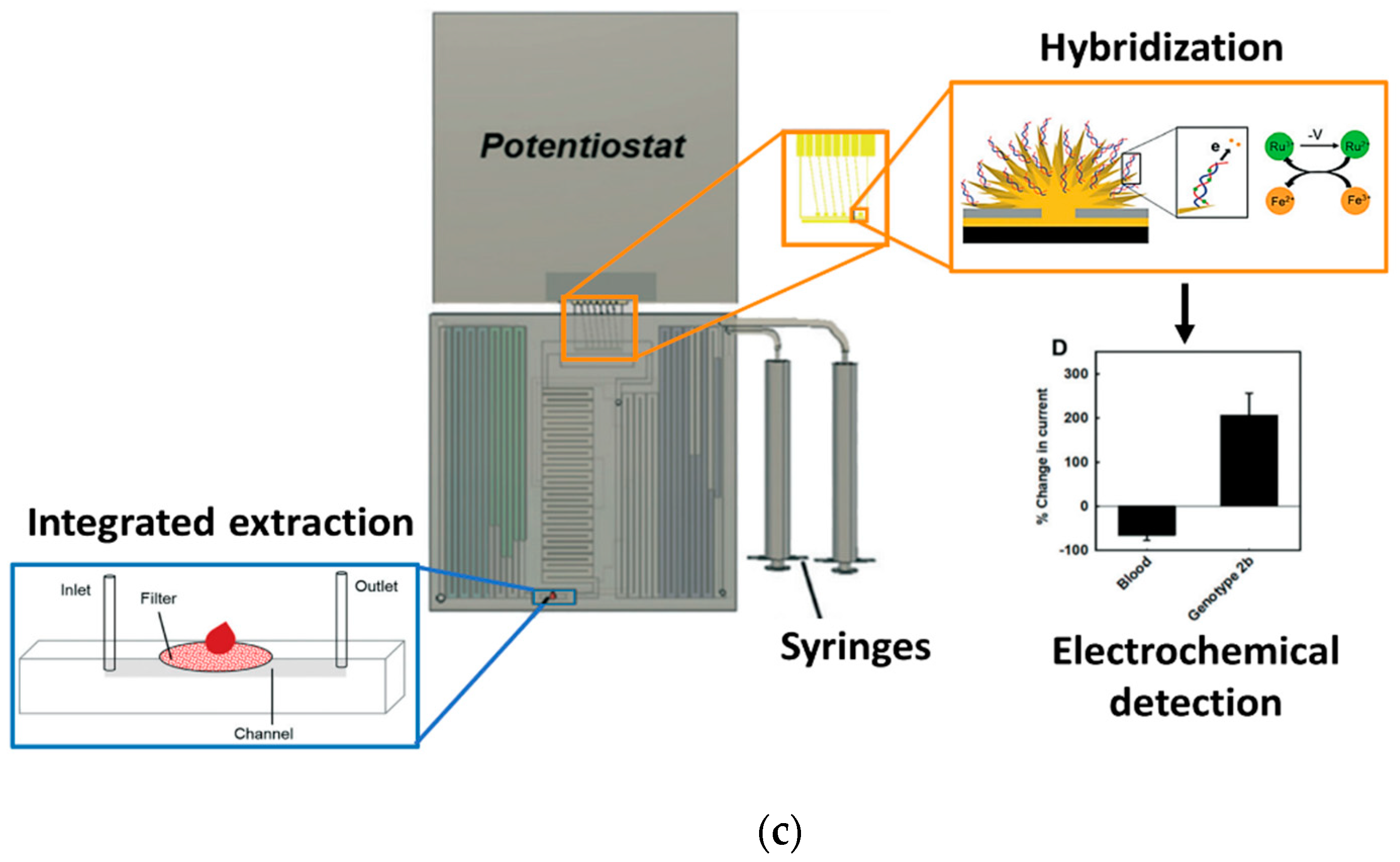
4.2. PCR-Free Genetic PoC
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef]
- Kotra, L.P. Infectious Diseases. xPharm Compr. Pharmacol. Ref. 2007, 1–2. [Google Scholar] [CrossRef]
- Leland, D.S.; Ginocchio, C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. 2007, 20, 49–78. [Google Scholar] [CrossRef] [PubMed]
- Gleaves, C.A.; Smith, T.F.; Shuster, E.A.; Pearson, G.R. Rapid detection of cytomegalovirus in MRC-5 cells inoculated with urine specimens by using low-speed centrifugation and monoclonal antibody to an early antigen. J. Clin. Microbiol. 1984, 19, 917–919. [Google Scholar] [CrossRef]
- Hematian, A.; Sadeghifard, N.; Mohebi, R.; Taherikalani, M.; Nasrolahi, A.; Amraei, M.; Ghafourian, S. Traditional and modern cell culture in virus diagnosis. Osong Public Health Res. Perspect. 2016, 7, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; O’Kennedy, R. The structure of natural and recombinantantibodies. Methods Mol. Biol. 2015, 1348, 7–11. [Google Scholar]
- Usuda, S.; Okamoto, H.; Tanaka, T.; Kidd-Ljunggren, K.; Holland, P.V.; Miyakawa, Y.; Mayumi, M. Differentiation of hepatitis B virus genotypes D and E by ELISA using monoclonal antibodies to epitopes on the preS2-region product. J. Virol. Methods 2016, 87, 81–89. [Google Scholar] [CrossRef]
- Yu, C.; Wu, J.; Zhou, X. Detection and subgrouping of Cucumber mosaic virus isolates by TAS-ELISA and immunocapture RT-PCR. J. Virol. Methods 2005, 123, 155–161. [Google Scholar] [CrossRef]
- Denner, J. Sensitive detection systems for infectious agents in xenotransplantation. Xenotransplantation 2020, e12594. [Google Scholar] [CrossRef] [PubMed]
- Hema, M.; Charan, K.N. Recent developments in detection and diagnosis of plant viruses. In Recent Developments in Applied Microbiology and Biochemistry; Viswanath, B., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 2, pp. 163–180. [Google Scholar]
- Petralia, S.; Conoci, S. PCR technologies for Point of Care testing: Progresses and Perspectives. ACS Sens. 2017, 2, 876–891. [Google Scholar] [CrossRef]
- Almassian, D.R.; Cockrell, L.M.; Nelson, W.M. Portable nucleic acid thermocyclers. Chem. Soc. Rev. 2013, 42, 8769–8798. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Diagnostics for the developing world. Nat. Rev. Microbiol 2004, 2, 231–240. [Google Scholar] [CrossRef]
- Hennion, M.C. Solid-phase extraction: Method development, sorbents, and coupling with liquid chromatography. J. Chromatogr. A 1999, 856, 3–5. [Google Scholar] [CrossRef]
- Chatfield, L. Forensic DNA Typing: Biology and Technology behind STR Markers. Heredity 2002, 89, 327. [Google Scholar] [CrossRef][Green Version]
- MagaZorb(R) DNA Mini-Prep Kit Technical Bulletin, TB376. Available online: https://ita.promega.com/-/media/files/resources/protocols/technical-bulletins/101/magazorb-dna-mini-prep-kit-protocol.pdf?rev=e050e0de33ff4b1c8dd9247d2c2717c8&la=en (accessed on 17 September 2021).
- QIAprep Spin Miniprep Columns. Available online: https://www.qiagen.com/gb/shop/sample-technologies/dna/qiaprep-spin-miniprep-columns (accessed on 17 September 2021).
- Thatcher, S.A. DNA/RNA Preparation for Molecular Detection. Clin. Chem. 2015, 61, 89–99. [Google Scholar] [CrossRef]
- Sciuto, E.L.; Bongiorno, C.; Scandurra, A.; Petralia, S.; Cosentino, T.; Conoci, S.; Sinatra, F.; Libertino, S. Functionalization of Bulk SiO2 Surface with Biomolecules for Sensing Applications: Structural and Functional Characterizations. Chemosensors 2018, 6, 59. [Google Scholar] [CrossRef]
- Nakagawa, T.; Tanaka, T.; Niwa, D.; Osaka, T.; Takeyama, H.; Matsunaga, T. Fabrication of amino silane-coated microchip for DNA extraction from whole blood. J. Biotechnol. 2005, 116, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P.; Dutta, J.; Tripathi, V. Chitin and chitosan: Chemistry, properties and applications. J. Sci. Ind. Res. 2003, 63, 20–31. [Google Scholar]
- Li, C.; Chen, J.; Chen, Y.; Wang, J.; Ping, H.; Lu, A. Graphene-Derivatized Silica Composite as Solid-Phase Extraction Sorbent Combined with GC-MS/MS for the Determination of Polycyclic Musks in Aqueous Samples. Molecules 2018, 23, 318. [Google Scholar] [CrossRef]
- Sortino, S.; Petralia, S.; Condorelli, G.G.; Conoci, S.; Condorelli, G. Novel photoactive self-assembled monolayer for immobilization and cleavage of DNA. Langmuir 2003, 19, 536–539. [Google Scholar] [CrossRef]
- Kim, J.; Johnson, M.; Hill, P.; Gale, B.K. Microfluidic sample preparation: Cell lysis and nucleic acid purification. Integr. Biol. 2009, 1, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Hu, Q.; Amamoto, T.; Refinetti, P.; Mimori, K.; Funatsu, T.; Kato, M. Extraction of cell-free DNA from urine, using polylysine-coated silica particles. Anal. Bioanal. Chem. 2017, 409, 4021–4025. [Google Scholar] [CrossRef] [PubMed]
- Birch, C.; Esfahani, M.M.N.; Shaw, K.J.; Kemp, C.; Haswell, S.J.; Dyer, C. The preparation of microfluidic architecture with monolithic materials using a dual porous silica structure. Electrophoresis 2017, 38, 2996–3002. [Google Scholar] [CrossRef]
- Petralia, S.; Sciuto, E.L.; Conoci, S. A novel miniaturized biofilter based on silicon micropillars for nucleic acid extraction. Analyst 2016, 142, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Conoci, S.; Mascali, A.; Pappalardo, F. Synthesis, DNA binding properties and electrochemistry towards an electrode-bound DNA of a novel anthracene–viologen conjugate. RSC Adv. 2014, 4, 2845–2850. [Google Scholar] [CrossRef]
- Santangelo, M.F.; Pagano, R.; Lombardo, S.; Sciuto, E.; Sanfilippo, D.; Fallica, G.; Sinatra, F.; Libertino, S. Silicon photomultipliers applications to biosensors. In Proceedings of the Silicon Photonics IX, San Francisco, CA, USA, 3–5 February 2014. [Google Scholar]
- Martin, A.; Bouffier, L.; Grant, K.B.; Limoges, B.; Marchal, D. Real-time electrochemical LAMP: A rational comparative study of different DNA intercalating and nonintercalating redox probes. Analyst 2016, 141, 4196–4203. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.H.; Wei, M.Y.; Chen, H. Multiple DNA Binding Modes of a Metallointercalator Revealed by DNA Film Voltammetry. J. Phys. Chem. B 2006, 110, 20568–20571. [Google Scholar] [CrossRef]
- Sciuto, E.L.; Santangelo, M.F.; Villaggio, G.; Fulvia, S.; Bongiorno, C.; Nicotra, G.; Libertino, S. Photo-physical characterization of fluorophore Ru(bpy)32+ for optical biosensing applications. Sens. Bio-Sens. Res. 2015, 6, 67–71. [Google Scholar] [CrossRef]
- Sciuto, E.L.; Villaggio, G.; Santangelo, M.F.; Laudani, S.; Federico, C.; Saccone, S.; Sinatra, F.; Libertino, S. Study of a Miniaturizable System for Optical Sensing Application to Human Cells. App. Sci. 2019, 9, 975. [Google Scholar] [CrossRef]
- Defever, T.; Druet, M.; Evrard, D.; Marchal, D.; Limoges, B. Real-Time Electrochemical PCR with a DNA Intercalating Redox Probe. Anal. Chem. 2011, 83, 1815–1821. [Google Scholar] [CrossRef]
- Petralia, S.; Castagna, M.E.; Cappello, E.; Puntoriero, F.; Trovato, E.; Gagliano, A.; Conoci, S. A miniaturized silicon based device for nucleic acids electrochemical detection. Sens. Bio-Sens. Res. 2015, 6, 90–94. [Google Scholar] [CrossRef][Green Version]
- Giamblanco, N.; Conoci, S.; Russo, D.; Marletta, G. Single-step label-free hepatitis B virus detection by a piezoelectric biosensor. RSC Adv. 2015, 5, 38152–38158. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Mason, M.G.; Botella, J.R. Evaluation and improvement of isothermal amplification methods for point-of-need plant disease diagnostics. PLoS ONE 2020, 15, e0235216. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, 63e. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Alhassan, A.; Li, Z.; Poole, C.B.; Carlow, C.K. Expanding the MDx toolbox for filarial diagnosis and surveillance. Trends Parasitol. 2015, 31, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Buchan, B.W.; Ledeboer, N.A. Emerging technologies for the clinical microbiology laboratory. Clin. Microbiol. Rev. 2014, 27, 783–822. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S.; Petralia, S.; Vicario, N.; Cirillo, D.; Conoci, S. An innovative silicon-chip for sensitive real time PCR improvement in pathogen detection. Analyst 2019, 144, 2353–2358. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.S.; Scott, L.; Noble, L.; Gous, N.; Dheda, K. Impact of the GeneXpert MTB/RIF technology on tuberculosis control. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Xpert® Ebola. Available online: https://p.widencdn.net/pesypn/Cepheid-Xpert-Ebola-Brochure-CE-IVD-3058-English (accessed on 12 October 2021).
- Andersson, M.E.; Olofsson, S.; Lindh, M. Comparison of the filmarray assay and in-house real-time PCR for detection of respiratory infection. Scand. J. Infect. Dis. 2014, 46, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mateos, P.; Ngamsom, B.; Walter, C.; Dyer, C.E.; Gitaka, J.; Iles, A.; Pamme, N. A lab-on-a-chip platform for integrated extraction and detection of SARS-CoV-2 RNA in resource-limited settings. Anal. Chim. Acta 2021, 1177, 338758. [Google Scholar] [CrossRef] [PubMed]
- Sciuto, E.L.; Petralia, S.; Calabrese, G.; Conoci, S. Integrated Biosensor Platform for Nucleic Acids Extraction and Detection. Biotechnol. Bioeng. 2020, 117, 1554–1561. [Google Scholar] [CrossRef]
- Xu, L.; Qu, H.; Alonso, D.G.; Yu, Z.; Yu, Y.; Shi, Y.; Hu, C.; Zhu, T.; Wu, N.; Shen, F. Portable integrated digital PCR system for the point-of-care quantification of BK virus from urine samples. Biosens. Bioelectron. 2021, 175, 112908. [Google Scholar] [CrossRef]
- Seok, Y.; Batule, B.S.; Kim, M.G. Lab-on-paper for all-in-one molecular diagnostics (LAMDA) of zika, dengue, and chikungunya virus from human serum. Biosens. Bioelectron. 2020, 165, 112400. [Google Scholar] [CrossRef]
- Sun, F.; Ganguli, A.; Nguyen, J.; Brisbin, R.; Shanmugam, K.; Hirschberg, D.L.; Wheeler, M.B.; Bashir, R.; Nash, D.M.; Cunningham, B.T. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip 2020, 20, 1621–1627. [Google Scholar] [CrossRef]
- Tanriverdi, S.; Chen, L.J.; Chen, S.Q. A rapid and automated sample-to-result HIV load test for near-patient application. J. Infect. Dis. 2010, 201, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, L.; Bergeron, M.G. The GenePOC platform, a rational solution for extreme point-of-care testing. Micromachines 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, Y.; Fang, X.; Liu, Y.; Du, M.; Lu, X.; Li, Q.; Sun, Y.; Ma, J.; Lan, T. Microfluidic-RT-LAMP chip for the point-of-care detection of emerging and re-emerging enteric coronaviruses in swine. Anal. Chim. Acta 2020, 1125, 57–65. [Google Scholar] [CrossRef]
- Shin, D.J.; Wang, T.-H. Magnetic Droplet Manipulation Platforms for Nucleic Acid Detection at the Point of Care. Ann. Biomed. Eng. 2014, 42, 2289–2302. [Google Scholar] [CrossRef]
- Lehmann, U.; Vandevyver, C.; Parashar, V.K.; Gijs, M.A.M. Droplet-Based DNA Purification in a Magnetic Lab-on-a-Chip. Angew. Chem. Int. Ed. 2006, 45, 3062–3067. [Google Scholar] [CrossRef]
- Pipper, J.; Inoue, M.; Ng, L.F.; Neuzil, P.; Zhang, Y.; Novak, L. Catching bird flu in a droplet. Nat. Med. 2007, 13, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Introduction to Digital PCR. Available online: https://www.bio-rad.com/it-it/applications-technologies/introduction-digital-pcr?ID=MDV300E8Z (accessed on 12 October 2021).
- Yu, Z.; Lyu, W.; Yu, M.; Wang, Q.; Qu, H.; Ismagilov, R.F.; Han, X.; Lai, D.; Shen, F. Self-partitioning SlipChip for slip-induced droplet formation and human papillomavirus viral load quantification with digital LAMP. Biosens. Bioelectron. 2020, 155, 112107. [Google Scholar] [CrossRef]
- Rodriguez, N.M.; Wong, W.S.; Liu, L.; Dewar, R.; Klapperich, C.M. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 2016, 16, 753–763. [Google Scholar] [CrossRef]
- Ahn, H.; Batule, B.S.; Seok, Y.; Kim, M.G. Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal. Chem. 2018, 90, 10211–10216. [Google Scholar] [CrossRef]
- Seok, Y.; Jang, H.; Oh, J.; Joung, H.A.; Kim, M.G. A handheld lateral flow strip for rapid DNA extraction from staphylococcus aureus cell spiked in various samples. Biomed. Phys. Eng. Exp. 2019, 5, 035035. [Google Scholar] [CrossRef]
- Liu, F.; Choi, K.S.; Park, T.J.; Lee, S.Y.; Seo, T.S. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011, 5, 123–128. [Google Scholar] [CrossRef]
- Chen, Y.J.; Chiang, C.L.; Huang, J.T. Wireless Portable Graphene-FET Biosensor for Detecting H1N1 Virus. 2017. Available online: https://www.semanticscholar.org/paper/Wireless-Portable-Graphene-FET-Biosensor-for-H-1-N-Chen-Chiang/edd3649cfc53aa765de7193fbd4e86aceaa0c433 (accessed on 17 September 2021).
- Sengupta, J.; Hussain, C.M. Graphene-based field-effect transistor biosensors for the rapid detection and analysis of viruses: A perspective in view of COVID-19. Carbon Trends 2020, 2, 100011. [Google Scholar] [CrossRef]
- Townsend, M.B.; Dawson, E.D.; Mehlmann, M.; Smagala, J.A.; Dankbar, D.M.; Moore, C.L.; Smith, C.B.; Cox, N.J.; Kuchta, R.D.; Rowlen, K.L. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J. Clin. Microbiol. 2006, 44, 2863–2871. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tang, S.; Storhoff, J.; Marla, S.; Bao, Y.P.; Wang, X.; Wong, E.Y.; Ragupathy, V.; Ye, Z.; Hewlett, I.K. Multiplexed, rapid detection of H5N1 using a PCR-free nanoparticle-based genomic microarray assay. BMC Biotechnol. 2010, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Petralia, S.; Sciuto, E.L.; Di Pietro, M.L.; Zimbone, M.; Grimaldi, M.G.; Conoci, S. An innovative chemical strategy for PCR-free genetic detection of pathogens by an integrated electrochemical biosensor. Analyst 2017, 142, 2090–2093. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Das, J.; Mepham, A.H.; Nemr, C.R.; Sargent, E.H.; Kelley, S.O. A fully-integrated and automated testing device for PCR-free viral nucleic acid detection in whole blood. Lab Chip 2018, 18, 1928–1935. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sciuto, E.L.; Leonardi, A.A.; Calabrese, G.; Luca, G.D.; Coniglio, M.A.; Irrera, A.; Conoci, S. Nucleic Acids Analytical Methods for Viral Infection Diagnosis: State-of-the-Art and Future Perspectives. Biomolecules 2021, 11, 1585. https://doi.org/10.3390/biom11111585
Sciuto EL, Leonardi AA, Calabrese G, Luca GD, Coniglio MA, Irrera A, Conoci S. Nucleic Acids Analytical Methods for Viral Infection Diagnosis: State-of-the-Art and Future Perspectives. Biomolecules. 2021; 11(11):1585. https://doi.org/10.3390/biom11111585
Chicago/Turabian StyleSciuto, Emanuele Luigi, Antonio Alessio Leonardi, Giovanna Calabrese, Giovanna De Luca, Maria Anna Coniglio, Alessia Irrera, and Sabrina Conoci. 2021. "Nucleic Acids Analytical Methods for Viral Infection Diagnosis: State-of-the-Art and Future Perspectives" Biomolecules 11, no. 11: 1585. https://doi.org/10.3390/biom11111585
APA StyleSciuto, E. L., Leonardi, A. A., Calabrese, G., Luca, G. D., Coniglio, M. A., Irrera, A., & Conoci, S. (2021). Nucleic Acids Analytical Methods for Viral Infection Diagnosis: State-of-the-Art and Future Perspectives. Biomolecules, 11(11), 1585. https://doi.org/10.3390/biom11111585








