Role of CD36 in Palmitic Acid Lipotoxicity in Neuro-2a Neuroblastoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture
2.3. Establishment of N2a Cells Stably Transfected with CD36 shRNA
2.4. BODIPYTM FL C12 Uptake Assay by Flow Cytometry
2.5. BODIPYTM FL C12 Uptake Assay by Confocal Microscopy
2.6. RNA Isolation
2.7. Semi-Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) Assay
2.8. MTT (3-[4,5-Dimethylthiazol-2-yl]-2,5 Diphenyl Tetrazolium Bromide) Assay
2.9. Cell Cycle Analysis
2.10. Detection of Cell Surface Expression of CD36 by Immunostaining and Flow Cytometry
2.11. Bicinchoninic acid (BCA) Protein Assay
2.12. Western Blot Analysis
2.13. Statistical Analysis
3. Results
3.1. Effects of PA Treatment on CD36 Expression
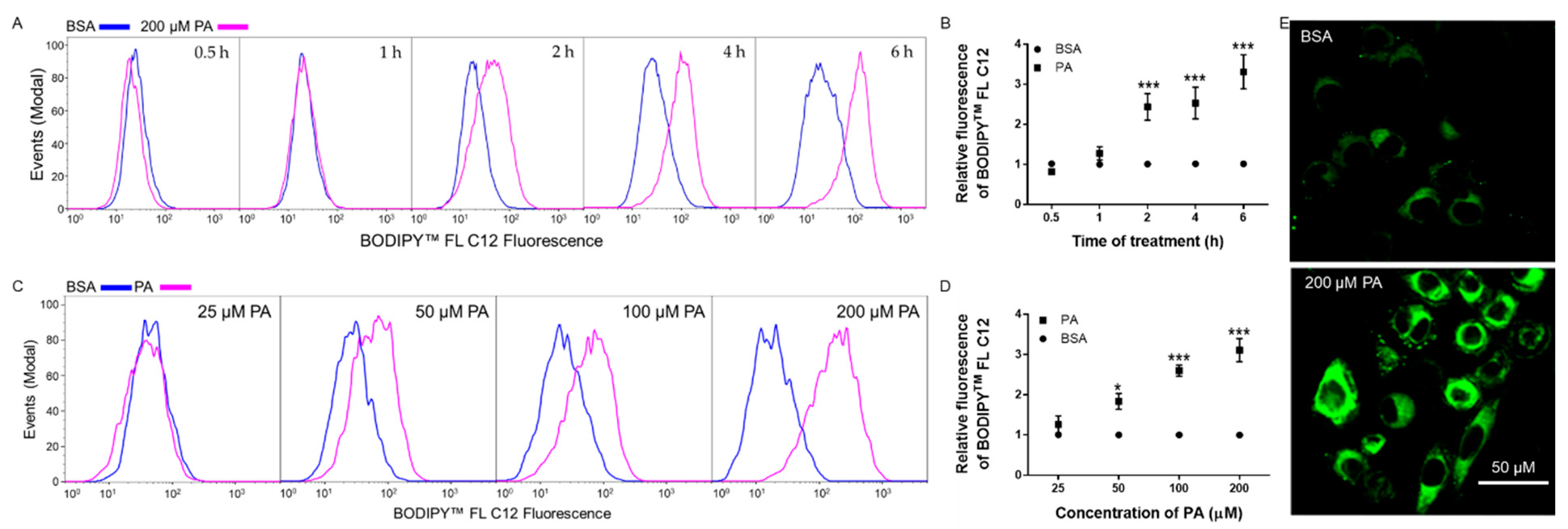
3.2. Time- and Dose-Dependent Effects of PA on BODIPYTM FL C12 Uptake in N2a Cells
3.3. Sulfo-N-Succinimidyl Oleate (SSO) Decreased BODIPYTM FL C12 Uptake in N2a Cells
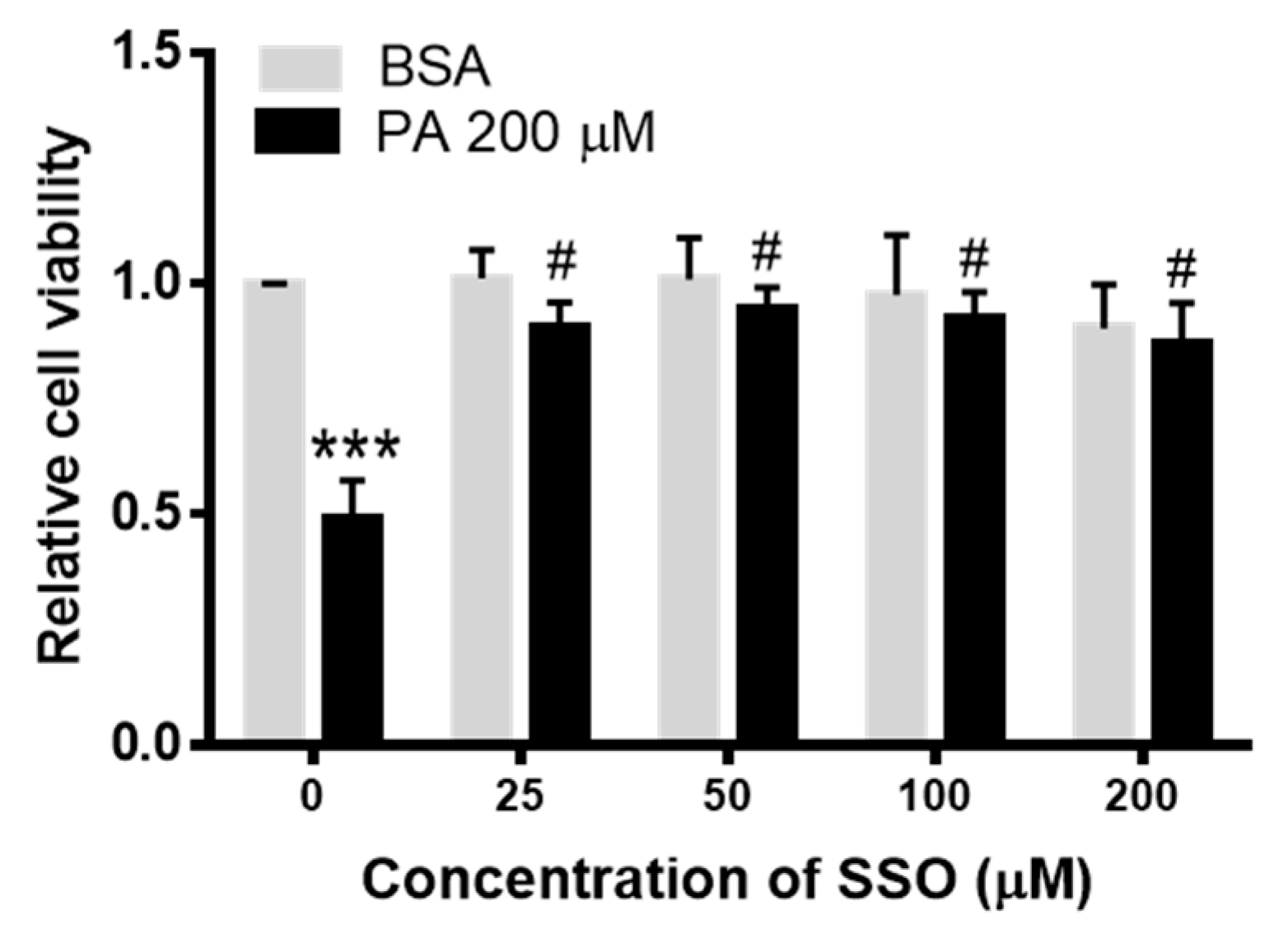
3.4. SSO Attenuated PA-Induced Decrease in N2a Cell Viability
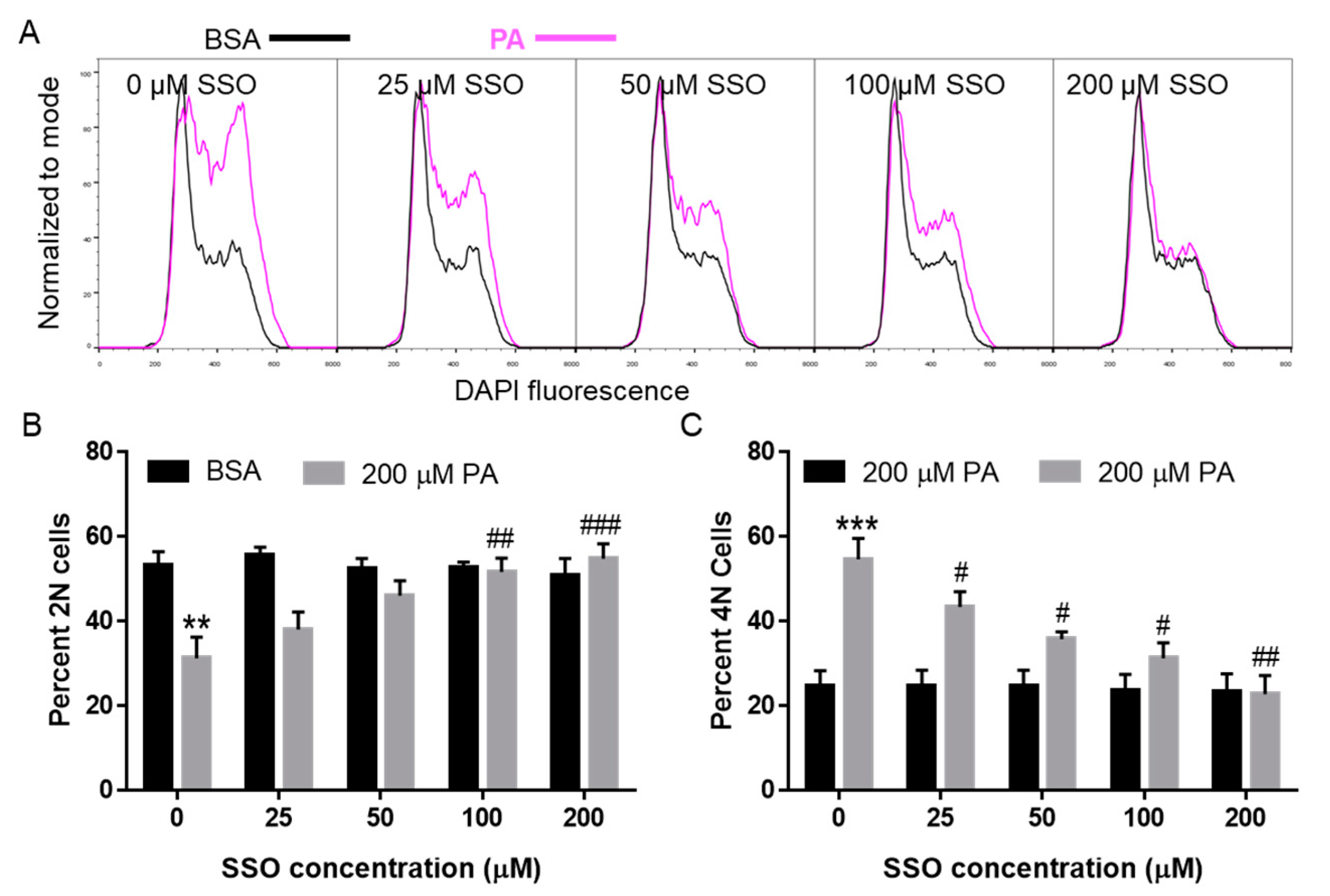
3.5. SSO Attenuated PA-Induced Cell Cycle Defects in N2a Cells
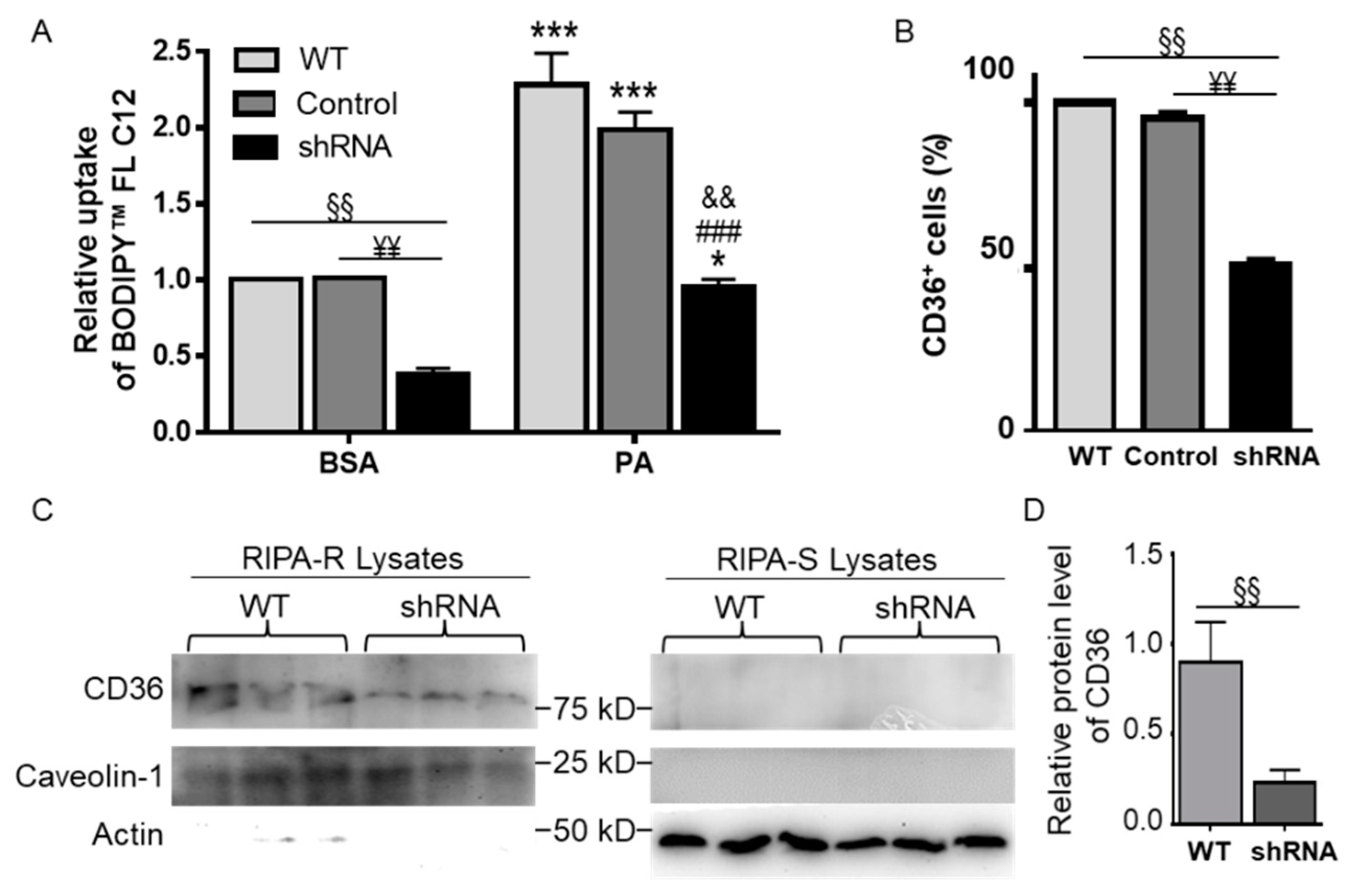
3.6. CD36 Knockdown Attenuated BODIPYTM FL C12 Uptake
3.7. CD36 Knockdown Attenuated PA-Induced Cell Cycle Defects
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arner, P.; Ryden, M. Fatty Acids, Obesity and Insulin Resistance. Obes. Facts 2015, 8, 147–155. [Google Scholar] [CrossRef]
- Rodriguez-Pacheco, F.; Gutierrez-Repiso, C.; Garcia-Serrano, S.; Alaminos-Castillo, M.A.; Ho-Plagaro, A.; Valdes, S.; Garcia-Arnes, J.; Gonzalo, M.; Andrade, R.J.; Moreno-Ruiz, F.J.; et al. The pro-/anti-inflammatory effects of different fatty acids on visceral adipocytes are partially mediated by GPR120. Eur. J. Nutr. 2016, 56, 1743–1752. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Y.; Xu, C.; Hong, Y.; Lu, H.; Wu, J.; Chen, Y. Association between serum free fatty acid levels and nonalcoholic fatty liver disease: A cross-sectional study. Sci. Rep. 2014, 4, 5832. [Google Scholar] [CrossRef] [Green Version]
- Feng, R.; Luo, C.; Li, C.; Du, S.; Okekunle, A.P.; Li, Y.; Chen, Y.; Zi, T.; Niu, Y. Free fatty acids profile among lean, overweight and obese non-alcoholic fatty liver disease patients: A case-control study. Lipids Health Dis. 2017, 16, 165. [Google Scholar] [CrossRef] [Green Version]
- Capel, F.; Acquaviva, C.; Pitois, E.; Laillet, B.; Rigaudiere, J.P.; Jouve, C.; Pouyet, C.; Gladine, C.; Comte, B.; Vianey Saban, C.; et al. DHA at nutritional doses restores insulin sensitivity in skeletal muscle by preventing lipotoxicity and inflammation. J. Nutr. Biochem. 2015, 26, 949–959. [Google Scholar] [CrossRef]
- Cheon, H.G.; Cho, Y.S. Protection of palmitic acid-mediated lipotoxicity by arachidonic acid via channeling of palmitic acid into triglycerides in C2C12. J. Biomed. Sci. 2014, 21, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, L.; Liu, X.; Luo, R.; Liao, G.; Liu, J.; Cheng, J.; Lu, Y.; Chen, Y. Oleic acid protects saturated fatty acid mediated lipotoxicity in hepatocytes and rat of non-alcoholic steatohepatitis. Life Sci. 2018, 203, 291–304. [Google Scholar] [CrossRef]
- Kwon, B.; Lee, H.K.; Querfurth, H.W. Oleate prevents palmitate-induced mitochondrial dysfunction, insulin resistance and inflammatory signaling in neuronal cells. Biochim. Biophys. Acta 2014, 1843, 1402–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urso, C.J.; Zhou, H. Differential Effects of Unsaturated Fatty Acids and Saturated Fatty Acids in Lipotoxicity and Neutral Lipid Accumulation in Neuro-2a Cells. Biomed. J. Sci. Tech. Res. 2021, 37, 29516–29524. [Google Scholar] [CrossRef]
- Urso, C.J.; Zhou, H. Palmitic Acid Lipotoxicity in Microglia Cells Is Ameliorated by Unsaturated Fatty Acids. Int. J. Mol. Sci. 2021, 22, 9093. [Google Scholar] [CrossRef]
- Zhou, H.; Urso, C.J.; Jadeja, V. Saturated Fatty Acids in Obesity-Associated Inflammation. J. Inflamm. Res. 2020, 13, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Almaguel, F.G.; Liu, J.W.; Pacheco, F.J.; Casiano, C.A.; De Leon, M. Activation and reversal of lipotoxicity in PC12 and rat cortical cells following exposure to palmitic acid. J. Neurosci. Res. 2009, 87, 1207–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Li, L.; Zhang, C.; Cheng, X.; Zhang, Y.; Guo, Y.; Long, M.; Yang, S.; He, J. Palmitic Acid and beta-Hydroxybutyrate Induce Inflammatory Responses in Bovine Endometrial Cells by Activating Oxidative Stress-Mediated NF-kappaB Signaling. Molecules 2019, 24, 2421. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Guan, G.; Lei, L.; Liu, J.; Cao, L.; Wang, X. Oxidative and endoplasmic reticulum stresses are involved in palmitic acid-induced H9c2 cell apoptosis. Biosci. Rep. 2019, 39, BSR20190225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuzefovych, L.; Wilson, G.; Rachek, L. Different effects of oleate vs. palmitate on mitochondrial function, apoptosis, and insulin signaling in L6 skeletal muscle cells: Role of oxidative stress. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E1096–E1105. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, E.; Matsuda, T.; Kawamoto, T.; Takahashi, H.; Mieda, Y.; Matsuura, Y.; Takai, T.; Kanno, A.; Koyanagi-Kimura, M.; Asahara, S.I.; et al. Docosahexaenoic Acid Reduces Palmitic Acid-Induced Endoplasmic Reticulum Stress in Pancreatic Beta Cells. Kobe J. Med. Sci. 2018, 64, E43–E55. [Google Scholar]
- Mayer, C.M.; Belsham, D.D. Palmitate attenuates insulin signaling and induces endoplasmic reticulum stress and apoptosis in hypothalamic neurons: Rescue of resistance and apoptosis through adenosine 5’ monophosphate-activated protein kinase activation. Endocrinology 2010, 151, 576–585. [Google Scholar] [CrossRef]
- Kamp, F.; Hamilton, J.A. How fatty acids of different chain length enter and leave cells by free diffusion. Prostaglandins Leukot. Essent. Fatty Acids 2006, 75, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Jay, A.G.; Hamilton, J.A. The enigmatic membrane fatty acid transporter CD36: New insights into fatty acid binding and their effects on uptake of oxidized LDL. Prostaglandins Leukot. Essent. Fatty Acids 2018, 138, 64–70. [Google Scholar] [CrossRef]
- Schwenk, R.W.; Holloway, G.P.; Luiken, J.J.; Bonen, A.; Glatz, J.F. Fatty acid transport across the cell membrane: Regulation by fatty acid transporters. Prostaglandins Leukot. Essent. Fatty Acids 2010, 82, 149–154. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M. CD36, a scavenger receptor implicated in atherosclerosis. Exp. Mol. Med. 2014, 46, e99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverstein, R.L.; Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Ueno, M.; Kusaka, T.; Miki, T.; Nagai, Y.; Nakagawa, T.; Kanenishi, K.; Hosomi, N.; Sakamoto, H. CD36 expression in the brains of SAMP8. Arch. Gerontol. Geriatr. 2013, 56, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Verpoorten, S.; Sfyri, P.; Scully, D.; Mitchell, R.; Tzimou, A.; Mougios, V.; Patel, K.; Matsakas, A. Loss of CD36 protects against diet-induced obesity but results in impaired muscle stem cell function, delayed muscle regeneration and hepatic steatosis. Acta Physiol. 2020, 228, e13395. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Peng, Y.; Hang, W.; Nie, J.; Zhou, N.; Wang, D.W. The role of CD36 in cardiovascular disease. Cardiovasc. Res. 2020, cvaa319. [Google Scholar] [CrossRef] [PubMed]
- Pepino, M.Y.; Kuda, O.; Samovski, D.; Abumrad, N.A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014, 34, 281–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baylin, A.; Kabagambe, E.K.; Siles, X.; Campos, H. Adipose tissue biomarkers of fatty acid intake. Am. J. Clin. Nutr. 2002, 76, 750–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlivan, V.H.; Wilson, M.H.; Ruzicka, J.; Farber, S.A. An HPLC-CAD/fluorescence lipidomics platform using fluorescent fatty acids as metabolic tracers. J. Lipid Res. 2017, 58, 1008–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surriga, O.; Ortega, A.; Jadeja, V.; Bellafronte, A.; Lasala, N.; Zhou, H. Altered hepatic inflammatory response in the offspring following prenatal LPS exposure. Immunol. Lett. 2009, 123, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Varlamov, O.; Chu, M.P.; McGee, W.K.; Cameron, J.L.; O’Rourke, R.W.; Meyer, K.A.; Bishop, C.V.; Stouffer, R.L.; Roberts, C.T., Jr. Ovarian cycle-specific regulation of adipose tissue lipid storage by testosterone in female nonhuman primates. Endocrinology 2013, 154, 4126–4135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salameh, A.; Daquinag, A.C.; Staquicini, D.I.; An, Z.; Hajjar, K.A.; Pasqualini, R.; Arap, W.; Kolonin, M.G. Prohibitin/annexin 2 interaction regulates fatty acid transport in adipose tissue. JCI Insight 2016, 1, e86351. [Google Scholar] [CrossRef] [PubMed]
- Ahowesso, C.; Black, P.N.; Saini, N.; Montefusco, D.; Chekal, J.; Malosh, C.; Lindsley, C.W.; Stauffer, S.R.; DiRusso, C.C. Chemical inhibition of fatty acid absorption and cellular uptake limits lipotoxic cell death. Biochem. Pharmacol. 2015, 98, 167–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wei, E.; Quiroga, A.D.; Sun, X.; Touret, N.; Lehner, R. Altered lipid droplet dynamics in hepatocytes lacking triacylglycerol hydrolase expression. Mol. Biol. Cell 2010, 21, 1991–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolahi, K.; Louey, S.; Varlamov, O.; Thornburg, K. Real-Time Tracking of BODIPY-C12 Long-Chain Fatty Acid in Human Term Placenta Reveals Unique Lipid Dynamics in Cytotrophoblast Cells. PLoS ONE 2016, 11, e0153522. [Google Scholar] [CrossRef] [Green Version]
- Coburn, C.T.; Knapp, F.F., Jr.; Febbraio, M.; Beets, A.L.; Silverstein, R.L.; Abumrad, N.A. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 2000, 275, 32523–32529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintana-Castro, R.; Aguirre-Maldonado, I.; Soto-Rodriguez, I.; Deschamps-Lago, R.A.; Gruber-Pagola, P.; Urbina de Larrea, Y.K.; Juarez-Rivera, V.E.; Ramos-Manuel, L.E.; Alexander-Aguilera, A. Cd36 gene expression in adipose and hepatic tissue mediates the lipids accumulation in liver of obese rats with sucrose-induced hepatic steatosis. Prostaglandins Other Lipid Mediat. 2020, 147, 106404. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.W.; Edmundson, C.L.; Miller, D.W.; Hatch, G.M. On the mechanism of oleate transport across human brain microvessel endothelial cells. J. Neurochem. 2009, 110, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Ioghen, O.; Chitoiu, L.; Gherghiceanu, M.; Ceafalan, L.C.; Hinescu, M.E. CD36—A novel molecular target in the neurovascular unit. Eur. J. Neurosci. 2021, 53, 2500–2510. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, J.W.; Wang, X.; Guo, H.; Sun, H.H.; Lai, X.Y.; Liu, L.Y.; Zhu, M.; Wang, H.Y.; Li, Y.F.; et al. DHHC4 and DHHC5 Facilitate Fatty Acid Uptake by Palmitoylating and Targeting CD36 to the Plasma Membrane. Cell Rep. 2019, 26, 209–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Li, M.Y.; Lin, Q.; Liang, Z.; Xin, Q.; Wang, M.; He, Z.; Wang, X.; Wu, X.; Chen, G.G.; et al. Lipid oversupply induces CD36 sarcolemmal translocation via dual modulation of PKCzeta and TBC1D1: An early event prior to insulin resistance. Theranostics 2020, 10, 1332–1354. [Google Scholar] [CrossRef]
- Glatz, J.F.C.; Luiken, J.; Nabben, M. CD36 (SR-B2) as a Target to Treat Lipid Overload-Induced Cardiac Dysfunction. J. Lipid Atheroscler. 2020, 9, 66–78. [Google Scholar] [CrossRef]
- Chabowski, A.; Gorski, J.; Luiken, J.J.; Glatz, J.F.; Bonen, A. Evidence for concerted action of FAT/CD36 and FABPpm to increase fatty acid transport across the plasma membrane. Prostaglandins Leukot. Essent. Fatty Acids 2007, 77, 345–353. [Google Scholar] [CrossRef]
- Luiken, J.J.; Chanda, D.; Nabben, M.; Neumann, D.; Glatz, J.F. Post-translational modifications of CD36 (SR-B2): Implications for regulation of myocellular fatty acid uptake. Biochim. Biophys. Acta 2016, 1862, 2253–2258. [Google Scholar] [CrossRef]
- Ehehalt, R.; Sparla, R.; Kulaksiz, H.; Herrmann, T.; Fullekrug, J.; Stremmel, W. Uptake of long chain fatty acids is regulated by dynamic interaction of FAT/CD36 with cholesterol/sphingolipid enriched microdomains (lipid rafts). BMC Cell Biol. 2008, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.G.; Jacobson, K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 2002, 296, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Ventura, R.; Mordec, K.; Waszczuk, J.; Wang, Z.; Lai, J.; Fridlib, M.; Buckley, D.; Kemble, G.; Heuer, T.S. Inhibition of de novo Palmitate Synthesis by Fatty Acid Synthase Induces Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting Signaling Pathways, and Reprogramming Gene Expression. EBioMedicine 2015, 2, 808–824. [Google Scholar] [CrossRef] [Green Version]
- Moulle, V.S.; Le Foll, C.; Philippe, E.; Kassis, N.; Rouch, C.; Marsollier, N.; Bui, L.C.; Guissard, C.; Dairou, J.; Lorsignol, A.; et al. Fatty acid transporter CD36 mediates hypothalamic effect of fatty acids on food intake in rats. PLoS ONE 2013, 8, e74021. [Google Scholar] [CrossRef]
- Enciso-Ramirez, M.; Reyes-Castillo, Z.; Llamas-Covarrubias, M.A.; Guerrero, L.; Lopez-Espinoza, A.; Valdes-Miramontes, E.H. CD36 gene polymorphism -31118 G > A (rs1761667) is associated with overweight and obesity but not with fat preferences in Mexican children. Int. J. Vitam. Nutr. Res. 2020, 91, 1664–2821. [Google Scholar] [CrossRef] [PubMed]
- Gharib, M.; Tao, H.; Fungwe, T.V.; Hajri, T. Cluster Differentiating 36 (CD36) Deficiency Attenuates Obesity-Associated Oxidative Stress in the Heart. PLoS ONE 2016, 11, e0155611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricciarelli, R.; D’Abramo, C.; Zingg, J.M.; Giliberto, L.; Markesbery, W.; Azzi, A.; Marinari, U.M.; Pronzato, M.A.; Tabaton, M. CD36 overexpression in human brain correlates with beta-amyloid deposition but not with Alzheimer’s disease. Free. Radic. Biol. Med. 2004, 36, 1018–1024. [Google Scholar] [CrossRef] [PubMed]
- Sery, O.; Janoutova, J.; Ewerlingova, L.; Halova, A.; Lochman, J.; Janout, V.; Khan, N.A.; Balcar, V.J. CD36 gene polymorphism is associated with Alzheimer’s disease. Biochimie 2017, 135, 46–53. [Google Scholar] [CrossRef]
- Grajchen, E.; Wouters, E.; van de Haterd, B.; Haidar, M.; Hardonniere, K.; Dierckx, T.; Van Broeckhoven, J.; Erens, C.; Hendrix, S.; Kerdine-Romer, S.; et al. CD36-mediated uptake of myelin debris by macrophages and microglia reduces neuroinflammation. J. Neuroinflamm. 2020, 17, 224. [Google Scholar] [CrossRef]
- Hussain, G.; Anwar, H.; Rasul, A.; Imran, A.; Qasim, M.; Zafar, S.; Imran, M.; Kamran, S.K.S.; Aziz, N.; Razzaq, A.; et al. Lipids as biomarkers of brain disorders. Crit. Rev. Food Sci. Nutr. 2020, 60, 351–374. [Google Scholar] [CrossRef]
- Adibhatla, R.M.; Hatcher, J.F. Phospholipase A(2), reactive oxygen species, and lipid peroxidation in CNS pathologies. BMB Rep. 2008, 41, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Chew, H.; Solomon, V.A.; Fonteh, A.N. Involvement of Lipids in Alzheimer’s Disease Pathology and Potential Therapies. Front. Physiol. 2020, 11, 598. [Google Scholar] [CrossRef] [PubMed]
- Alecu, I.; Bennett, S.A.L. Dysregulated Lipid Metabolism and Its Role in alpha-Synucleinopathy in Parkinson’s Disease. Front. Neurosci. 2019, 13, 328. [Google Scholar] [CrossRef]
- Opazo-Rios, L.; Mas, S.; Marin-Royo, G.; Mezzano, S.; Gomez-Guerrero, C.; Moreno, J.A.; Egido, J. Lipotoxicity and Diabetic Nephropathy: Novel Mechanistic Insights and Therapeutic Opportunities. Int. J. Mol. Sci. 2020, 21, 2632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo-Lanussa, O.; Baez-Jurado, E.; Echeverria, V.; Ashraf, G.M.; Sahebkar, A.; Garcia-Segura, L.M.; Melcangi, R.C.; Barreto, G.E. Lipotoxicity, neuroinflammation, glial cells and oestrogenic compounds. J. Neuroendocrinol. 2020, 32, e12776. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urso, C.J.; Zhou, H. Role of CD36 in Palmitic Acid Lipotoxicity in Neuro-2a Neuroblastoma Cells. Biomolecules 2021, 11, 1567. https://doi.org/10.3390/biom11111567
Urso CJ, Zhou H. Role of CD36 in Palmitic Acid Lipotoxicity in Neuro-2a Neuroblastoma Cells. Biomolecules. 2021; 11(11):1567. https://doi.org/10.3390/biom11111567
Chicago/Turabian StyleUrso, C. J., and Heping Zhou. 2021. "Role of CD36 in Palmitic Acid Lipotoxicity in Neuro-2a Neuroblastoma Cells" Biomolecules 11, no. 11: 1567. https://doi.org/10.3390/biom11111567
APA StyleUrso, C. J., & Zhou, H. (2021). Role of CD36 in Palmitic Acid Lipotoxicity in Neuro-2a Neuroblastoma Cells. Biomolecules, 11(11), 1567. https://doi.org/10.3390/biom11111567





