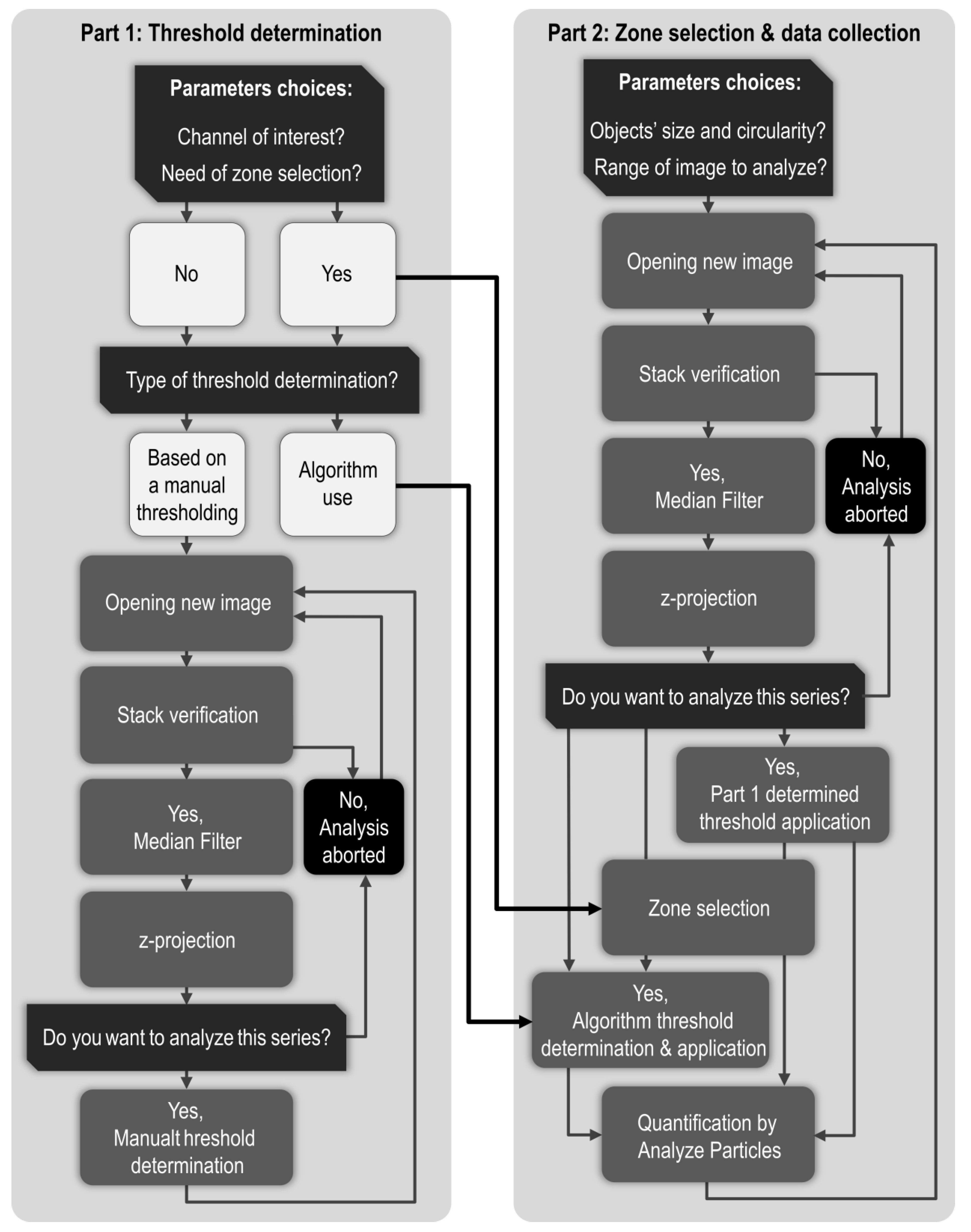
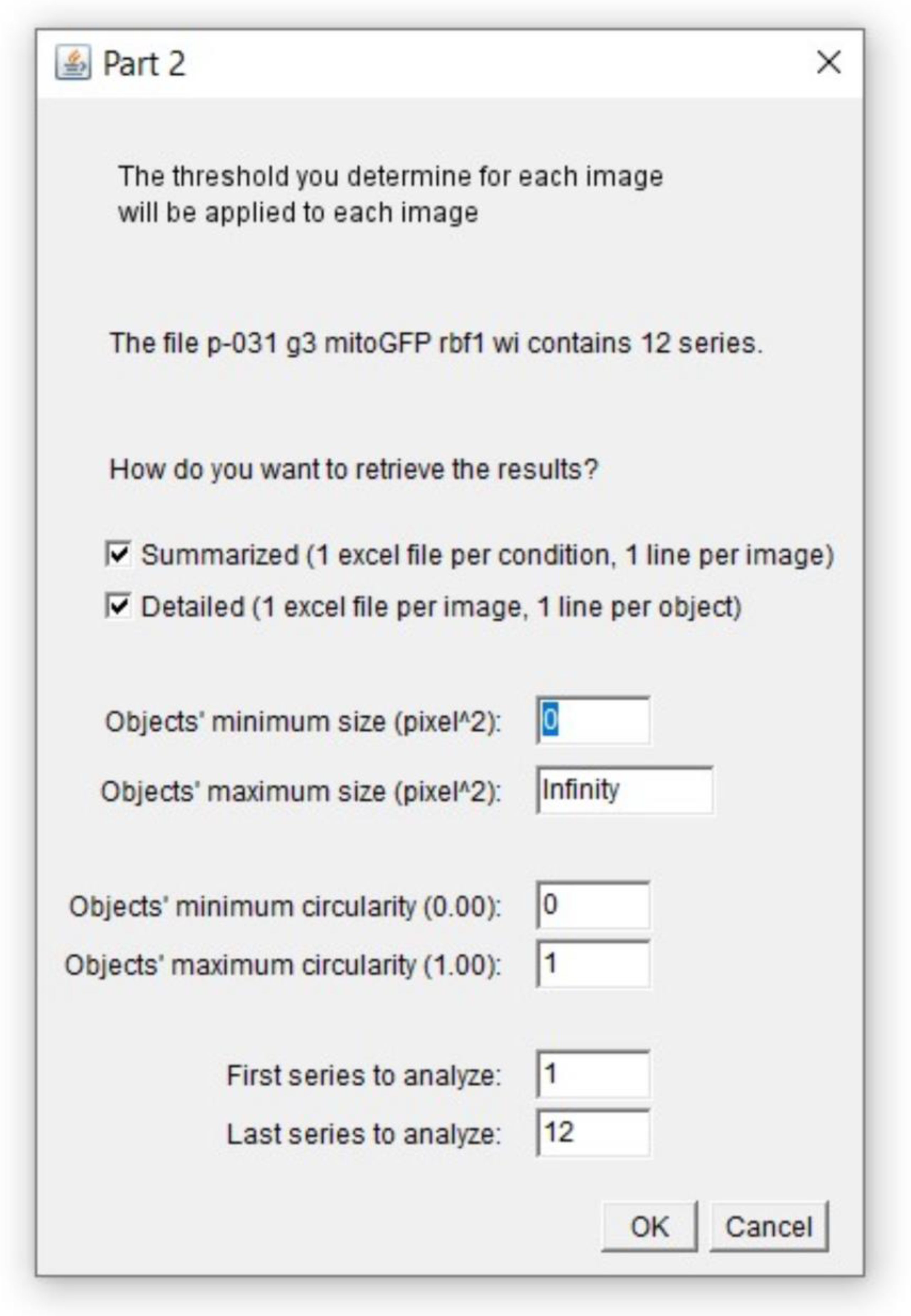
Steps of image processing directly depend on the chosen readout. In order to get both the number of objects and the stained area, our image processing protocol is based on three major steps: 1. Background noise reduction, 2. Compression of our 3D images into 2D by a z-axis projection, 3. Thresholding. Those steps allow appropriate segmentation required for relevant quantification by the “Analyze Particles” function. Importantly, on ImageJ/Fiji, there are many ways to minimize background signal, several ways of compressing a 3D image in 2D and multiple ways to determine a threshold, resulting in countless combinations of possible image processing. In this study, we investigated the weight of these parameters on signal segmentation to end up with an optimized and unbiased protocol for apoptosis quantification.
3.2.1. Median Filter and Size Limitation Efficiently Reduce Artefacts
When quantification is automatized, definition of the signal of interest by segmentation is even more critical. Indeed, wrong segmentation can lead to quantification of unreal objects and thus give useless results. Thus, the signal of interest boundaries has to be better defined while background noise has to be decreased. General background noise can be minimized in many ways depending on the kind of images, the desired readout, and the defaults faced. In our case, mandatory use of a threshold would blacken every low intensity pixel constitutive of the general background noise. However, if binarization of the image efficiently removes diffuse low intensity background noise, it is not sufficient to erase artifactual pixels with aberrant high intensity, i.e., whose intensity is higher than the threshold value. Fortunately, isolated aberrant high pixels can be dealt with filters. Filters are matrix operations that re-calculate a pixel intensity value based on itself and its neighbors. The two mainly used filters are “Mean Filter” and “Median Filter” (
Figure 2A).
A “Mean Filter” with a radius of 1 gives a pixel an intensity value that corresponds to the mean of its value and those of its direct neighbors (
Figure 2A(b)). Hence, the value of an isolated pixel with aberrant high intensity is attenuated by the intensity value of its neighbors. However, as extreme values impact mean calculation, every neighboring pixel is affected by the isolated aberrant high pixel and their intensity is artificially increased (
Figure 2A(b)). By contrast, a “Median Filter” with a radius of 1 gives to a pixel an intensity value corresponding to the median of its value and those of its direct neighbors (
Figure 2A(c)), which is expected to be much closer to local intensity value. Besides, as extreme values effect on median calculation is low, high intensity isolated pixel impact on its neighbors is negligible. Overall, the “Mean Filter” tends to spread an aberrant high value whereas a “Median Filter” tends to confine it. This effect is illustrated in
Figure 2B, where the “1” arrows of the “No filter” panel show typical isolated aberrant high pixels that are efficiently erased by a “Median Filter” with a radius of 1 (
Figure 2B compare (b) and (c)). Aside from this benefit, the area pointed by the “2” arrow exemplifies the median filter ability to preserve edges of an object. Indeed, on the illustration, human eyes easily detect that the “2” arrow targets a marked cell (
Figure 2B(a)). However, this object is heterogeneous: in a restricted space, it contains few pixels of high intensity and many pixels of low intensity (i.e., below the chosen threshold). Without any filter, only high intensity pixels are kept after the thresholding step thereby fragmenting this object in several small groups of pixels (
Figure 2B(b)). Thus, with no further treatment, multiple objects will be counted in this area, which does not reflect reality. However, as these high intensity pixels are close to each other, the “Median Filter” with a radius of 1 homogenizes intensity values within this object. This allows its reconstruction and gives a segmentation consistent with reality (
Figure 2B(c)). Thus, when the “Median Filter” with a radius of 1 is applied, the “number of objects” decreases only to be closer to what a human eye would count.
Although the “Median Filter” with a radius of 1 efficiently reduces the number of artifactual objects by erasing isolated high pixels, the issue of groups of pixels with an intensity higher than the threshold value remains. A way to eliminate most of those artifacts is to limit our analysis to objects with a size consistent with the smallest biological object of interest. In our case, this smallest biological object is a TUNEL-labeled nucleus, we assessed their size on a few random images and thus set a size limit at 2 µm. Importantly, this “>2 µm size limitation” fits our data but should not be taken as a default value and must be adapted for other kinds of signals or cell types. Moreover, as the “Analyze Particles” function records the size of every object, this filtering can be carried out after quantification. In
Figure 2B, the “>2 µm limitation” eliminates artifactual objects pointed out by the “3” arrow as well as individual pixels such as those pointed by the “1” arrows. It thus appears very powerful in order to “clean” the image. However, as efficient as the size limitation may be, it cannot replace “Median filter”. Indeed, as already explained, in the absence of a “Median Filter”, the cell indicated by the “2” arrow in
Figure 2B becomes fragmented in several small groups of pixels, each one being smaller than 2 µm (
Figure 2B(b)). Thus, without the “Median Filter”, these pixels are eliminated by the “>2 µm limitation” and the actually labeled cell indicated by the “2” arrow is not included in the quantification of the apoptotic signal. Here, reconstruction of the object by the “Median Filter” prevents its elimination by the “>2 µm limitation” (
Figure 2B(c)). In the end, combination of a “Median Filter” with a radius of 1 and “>2 µm limitation” allows a better segmentation and a more accurate quantification.
3.2.2. Max Intensity Z-Projection Improves Contrast
Confocal microscopy gives the possibility to capture objects in 3D. However, image processing often requires transforming volumes into 2D images by compressing the z-axis. In our case, the “Analyze Particles” function used to quantify the signal of interest requires 2D images. Flattening a 3D volume can seem counterproductive, as separate objects on the same z-axis will be reduced to a single one on the final 2D image. In our case, this is unlikely to happen since imaginal disc cells are organized in a monolayer with only limited folding and this can also be true for tissue sections as long as they are thin enough. In the case of a multilayer tissue, z-projection can still be used if the apoptosis rate is low enough to prevent the occurrence of having multiple apoptotic cells in the same z-axis.
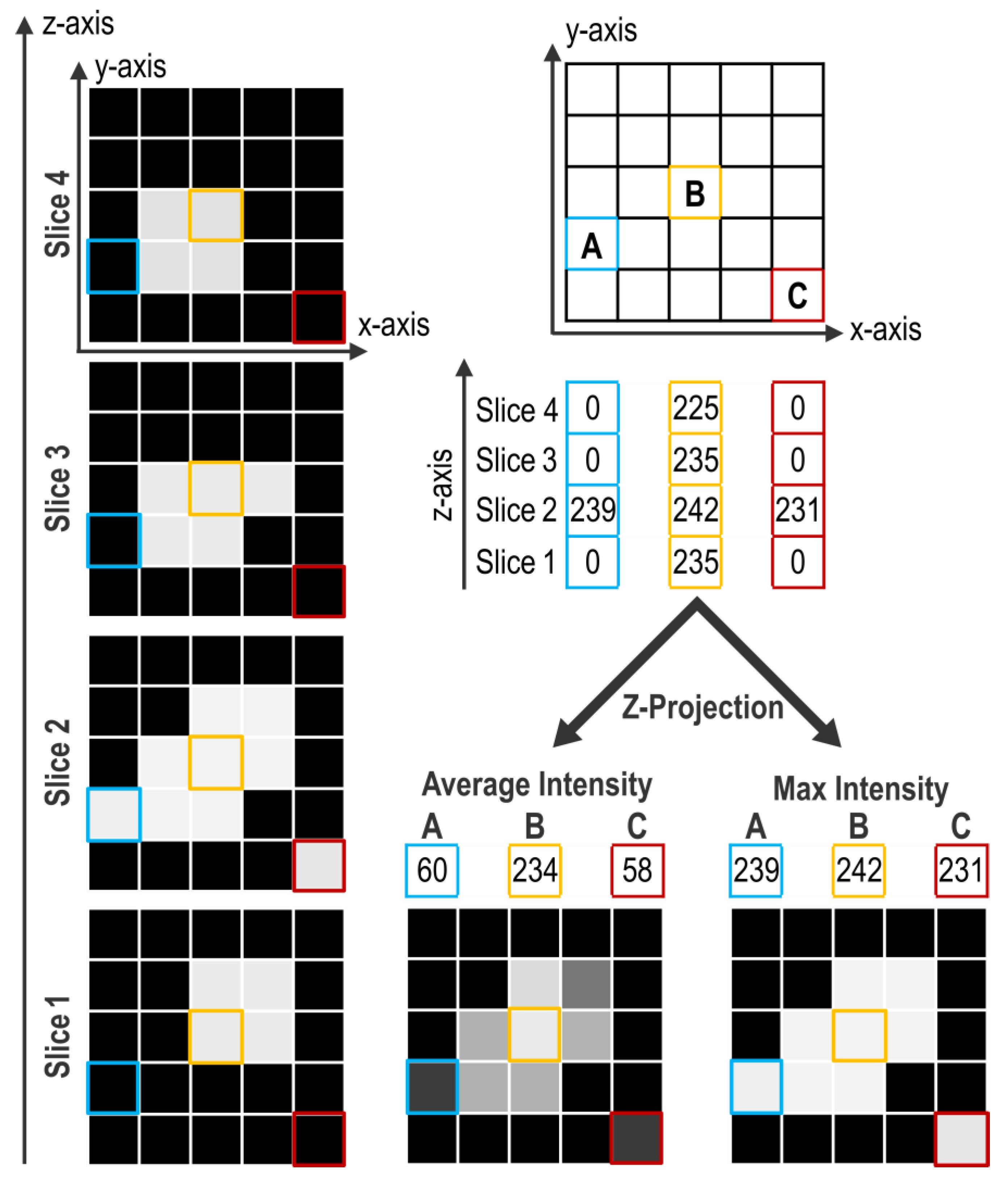
Projection consists of compressing the signal contained in every pixel of a z-axis into a single one. With ImageJ/Fiji, projection can be performed in several ways but two of them are mainly used. The first method, Average Intensity (AI), calculates the average intensity of all the pixels of a z-axis. The second, Max Intensity (MI), only retains the maximal intensity value along the z-axis.
Figure 3 presents examples of these projection methods on a virtual object without any other treatment (i.e., median filter).
The z-axis presented in “A” shows only one illuminated pixel on slice #2 and this pixel is included in the object. The “B” z-axis shows numerous pixels highly illuminated, comprised in the object. The “C” z-axis presents an artifactually illuminated pixel on slice #2 that is not comprised in the object. Comparison of the projection methods shows that AI projection decreases the importance of the artifactual pixel of the “C” z-axis, while the MI projection increases its weight. However, in the processing protocol, the preceding use of a “Median Filter” with a radius of 1 eliminates most of those artefacts that are thus not present anymore at the projection step. Conversely, the AI projection of the “A” z-axis leads to loss of signal even if it is part of the object. Furthermore, with the AI projection, the contrast between object and background is weak, therefore the range for the appropriate threshold value is limited (
Figure 3). After a MI projection, contrast is enhanced and thus threshold determination is easier for the experimenter, which helps limiting the experimenter bias. This is particularly important for signals with low contrast such as TUNEL. In our case, these two projection methods do not end up in drastically different results but, overall, MI projection presents more benefits than AI projection.
3.2.3. Use of Custom Manual Thresholds Gives the Best Segmentation for Relevant Quantification
The “Analyze Particles” function used for quantification requires the image to be binary. The transition from a greyscale image to a black and white image involves the setting of a threshold that defines an intensity value above which a pixel is turned to white and under which a pixel is turned to black. Ideally, this value should enable to get an image where white only corresponds to the signal of interest. Thresholding is the last step of segmentation and finally defines the objects of interest, which is critical for accurate quantification. Therefore, among the steps of image processing, it is the one that has the most dramatic effect on quantification accuracy, thus we dedicated particular attention to the threshold determination method.
Threshold can be automatically set by algorithms that analyze specific features of the image intensity histogram to determine a threshold value using either simple indicators such as the mean, maximal, or minimal intensity values, or more complex formulas. Hence, algorithms appear as an unbiased method to obtain a specific threshold value per image. We thus wondered whether any of the 16 thresholding algorithms currently available in the menu Image > Adjust > Threshold of ImageJ/Fiji could be used to determine a threshold that properly segments apoptotic signal in our images. Using some randomly chosen images, after application of a “Median filter” with a radius of 1 and a Max Intensity projection, we visually checked if these algorithms could provide a threshold value allowing a relevant segmentation (i.e., consistent with apoptotic signal). Most algorithms did not pass the visual inspection step as they yielded unrealistic segmentation either by ignoring a great portion of the signal or by including artifactual signal. However, two of them, Otsu [
15] and Moments [
16], seemed capable of discriminating actual apoptotic staining from background. We then performed a more detailed analysis of the threshold values obtained with these algorithms by comparing them to the ones obtained by experimenters. To this end, for the 28 images of the
vg >
rbf1 genotype, three experimenters determined the threshold to use for each staining (TUNEL or anti-cleaved Dcp-1) by eye and in triplicate (see
Supplementary Figure S1). The criteria for manual threshold determination were to set a threshold value that allows the positive signal to be consistent with the original image and located in the vestigial domain. Thresholds for these images were also determined using the 16 algorithms. As expected, algorithms inducing obvious unrealistic segmentation of the apoptotic signal yielded threshold values very far from the range of the ones determined by experimenters (
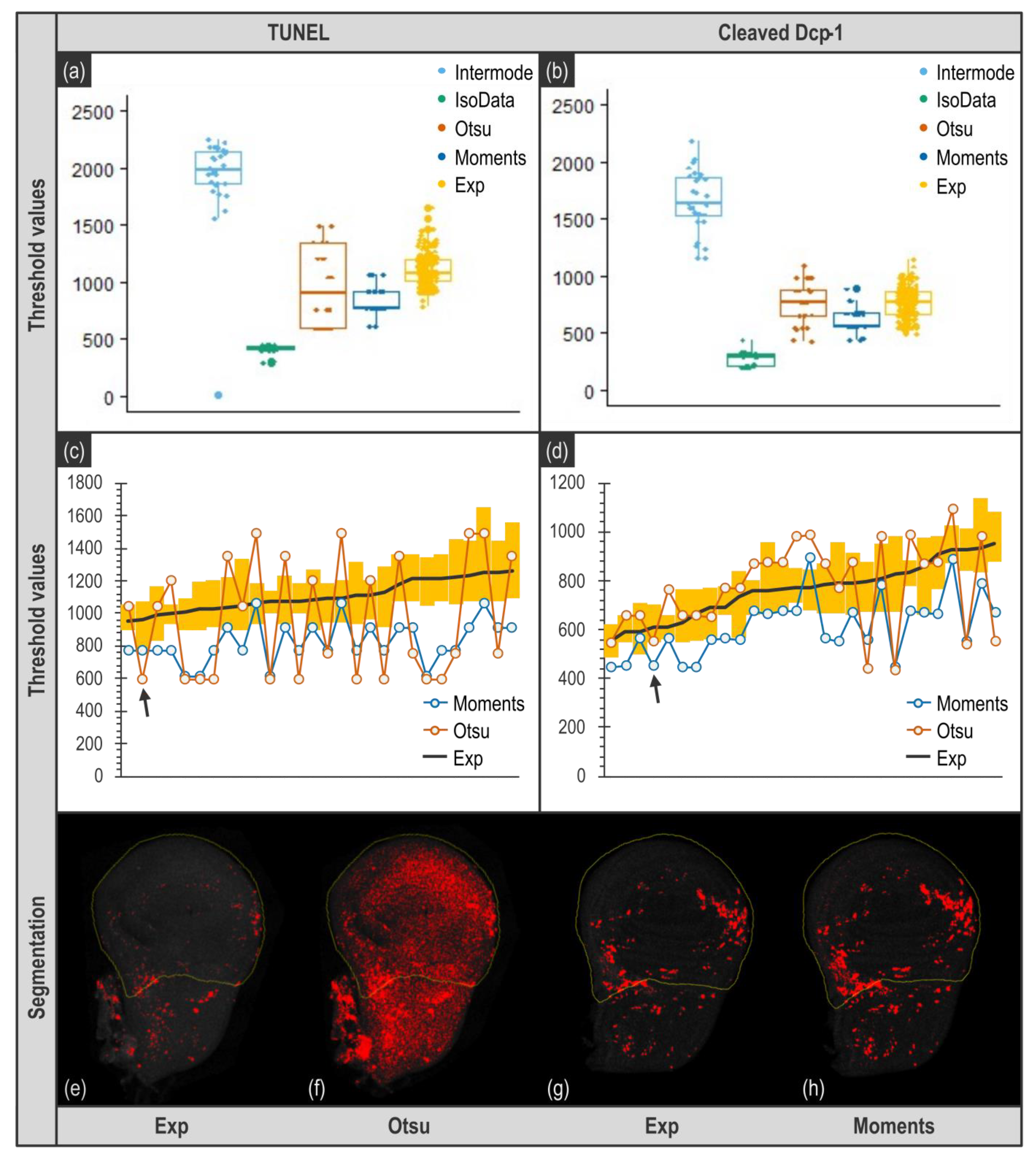
Figure 4a,b; compare IsoData and Intermode with Exp. Data not shown). On the contrary, Moments, Otsu, and experimenter threshold values are in the same range (
Figure 4a,b).
From this global analysis, it could seem that algorithms can be as efficient as human eye for threshold determination (compare for instance Otsu and Exp in
Figure 4b). However, visually, Otsu capability to determine a relevant threshold seemed irregular. We thus further deepened our data analysis and compared the threshold values obtained not globally but for each image. As shown in
Figure 4c,d, the values obtained with Otsu, if they tend to be roughly the same on average than the ones obtained by experimenters, are actually most of the time out of the range of experimenters values. This is particularly striking for images obtained from TUNEL (
Figure 4c) as Otsu’s determined values are far higher or lower that the ones obtained by any experimenter. This would not be an issue as long as the threshold values obtained still allow a realistic segmentation of apoptotic signal and subsequent relevant quantification. However, such deviation of the threshold value results in an inappropriate segmentation (compare
Figure 4e,f), that necessarily ends in a biased or most likely totally wrong quantification. When it comes to Moments, it provides threshold values that are usually lower than experimenters’ ones (
Figure 4a,b), which means that using this algorithm tends to include some background noise to the quantification. The question resides then in determining whether this amount of background noise is important enough to alter quantification. In the case of TUNEL staining (
Figure 4c), values are quite low, thus it certainly affects quantification rather significantly. By contrast, when images come from anti-cleaved Dcp-1 staining, the threshold values given by Moments are much closer to the ones obtained by experimenters, they actually seem very similar to the lowest values determined by experimenters (
Figure 4d). Therefore, one could assume that the variability of threshold values between Moments and an experimenter is comparable to the one that exists between experimenters. We tested this by comparing the relationship between the two most distant experimenters’ batch of measurements to the one between Moments and its closest dataset. As shown in
Supplementary Figure S2B, if two experimenters do not determine exactly the same value for the threshold, their estimates remain consistent with each other (
p = 10
−5 and R
2 = 0.53 for the most distant measurements), the difference can be more or less described as a given experimenter tending to set thresholds always lower than the other. This is a systematic error that should affect quantification only moderately. On the contrary, threshold values determined by Moments are not consistent with the values of experimenters (
p = 0.8 for the closest in
Supplementary Figure S2B). This indicates that Moments can set a low threshold value when all experimenters would have chosen a higher one, but it is not always the case, and most of all, the extent of this underestimation (i.e., the range of the difference between experimenters and Moments’ threshold values) is variable. This is more problematic as the extent of background incorporation in the quantification will then vary and might alter quantification relevance.
Contrary to algorithms, manual determination of the threshold values appears quite robust. Indeed, comparison of manually determined threshold values between experimenters for individual images shows a low variability and a good reproducibility both between several determinations of a given experimenter and between experimenters (
Supplementary Figure S1). As all images from an experiment are acquired identically, with the same microscope settings and come from samples treated with the same solutions, at the same time, theoretically, the appropriate threshold value is expected to be the same for every image. Moreover, using the same unique threshold value for all images can be considered as more objective and unbiased.
However, manually determined threshold values display some variability (
Figure 4, Exp). This can justify using a distinct individual, manually determined, threshold value for each image as it might enable a more accurate segmentation and subsequent quantification. In order to assess to which extent these two thresholding methods (unique threshold value for every image or individual, manually determined threshold value per image) can affect quantification and detection of our biological effect, we tried both (
Figure 5).
In the Manual condition, each image was binarized using its own manually determined threshold value (determined by experimenter 1, measure 3). From these individual threshold values, we calculated the median value per genotype and then the median of these medians. This latter value was used as the unique threshold value to binarize all images in the Total condition (1128 for TUNEL and 777 for anti-cleaved Dcp-1). We decided to use the median of the medians per genotype rather than the global median (calculated from the whole of the images independently of their genotype) to avoid giving more weight to a genotype (that might have a larger headcount for instance).
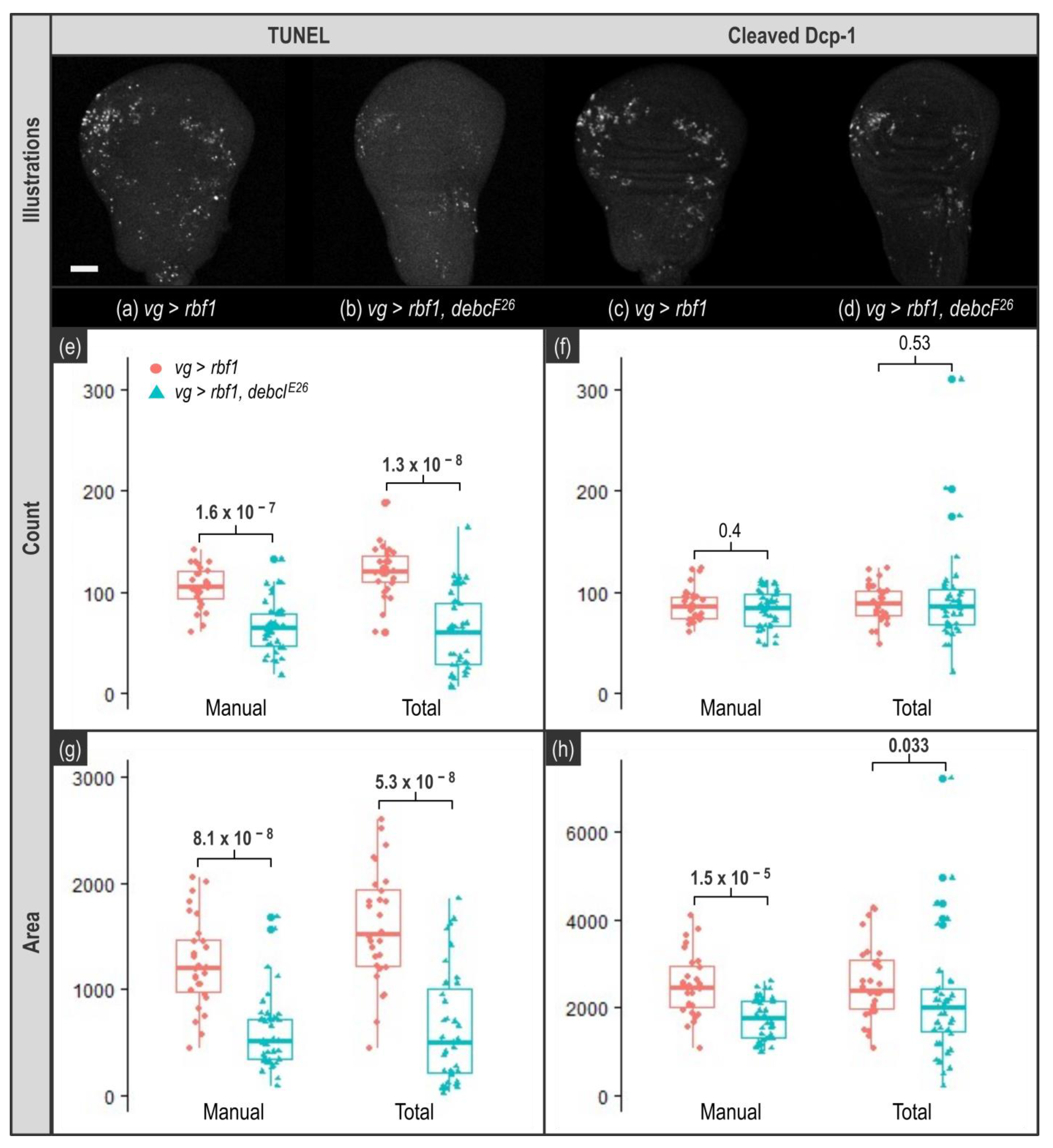
For TUNEL stained images, both thresholding approaches give a workable quantification both for count and area readouts (
Figure 5e,g). Indeed, a significant decrease in apoptosis between the two genotypes is detected in all cases. However, even if Total or Manual thresholding methods enable to detect the biological effect, we noticed that quantification is still somehow altered when a unique threshold is used (
Supplementary Figure S3).
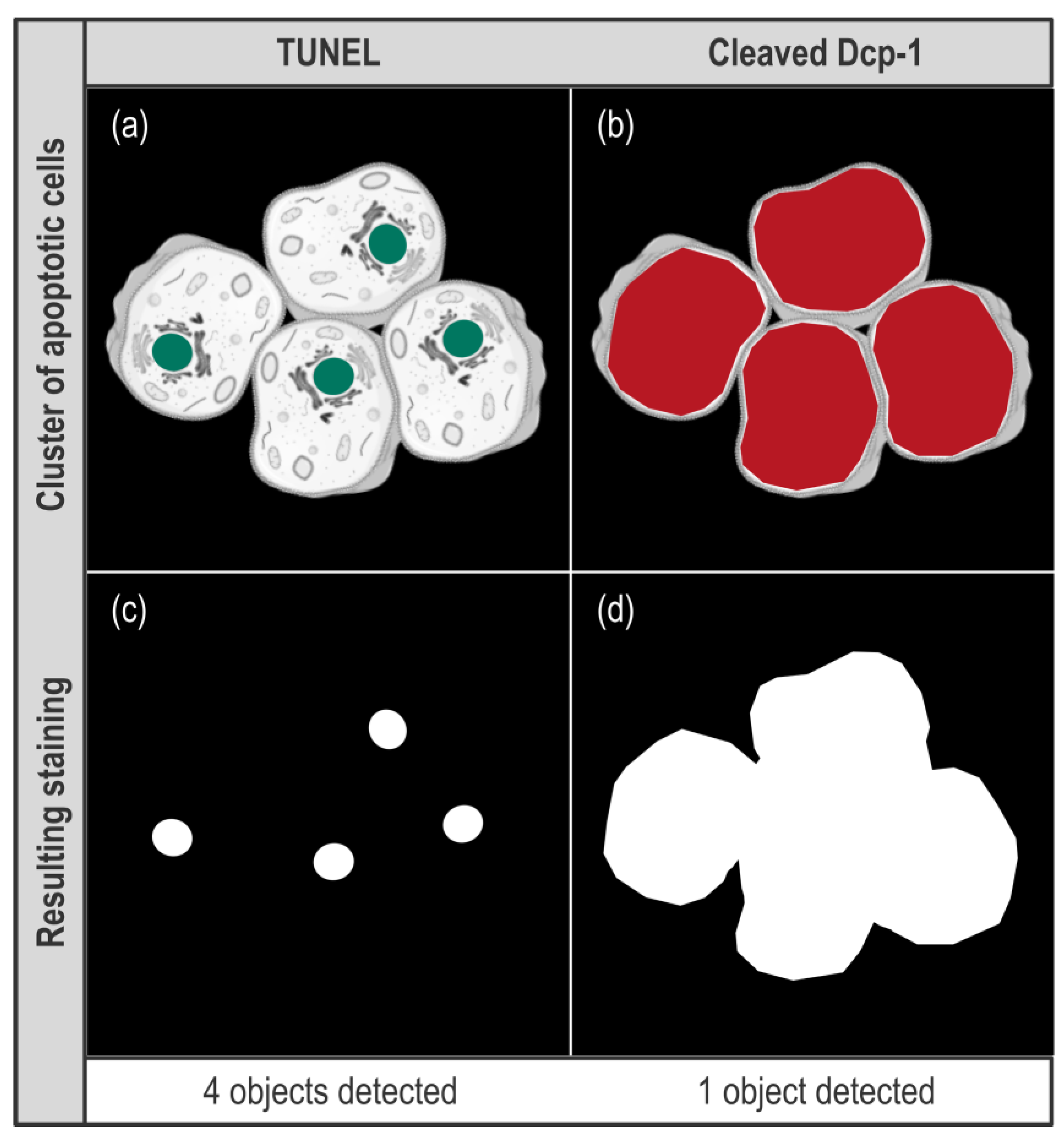
As for anti-cleaved Dcp-1-stained images, the first observation is that the count readout was not usable. Indeed, we knew that apoptotic cells clusters might alter quantification as these clusters might be considered as a single object. Moreover, such underestimation is enhanced when the apoptosis rate rises, eventually leading to the flattening of the difference between two samples. However, we chose to keep this readout, as it was not possible to anticipate the extent of this phenomenon in our samples. As shown in
Figure 5f, with this readout, the difference in the apoptosis rate between the two genotypes becomes undetectable. This indicates that the level of apoptosis induced by
rbf1 generates apoptotic cells clusters frequently enough to significantly alter quantification, and this, whatever the thresholding method, thus prohibiting the use of the count readout. By contrast, when anti-cleaved Dcp-1 staining is quantified using the area readout, the difference between the two thresholding methods (unique versus individual thresholds) becomes obvious. As shown in
Figure 5h, when a unique threshold value is used for all images (Total), the difference of apoptosis rate between the two genotypes is barely detectable (
p = 0.033). Moreover, even with an individual, manually determined threshold value per image, all the quantification results show a noticeable variability (
Figure 5, Manual) due to differences between specimen, variation of the UAS/gal4 system intensity and small asynchrony in the development of the larvae. However, use of a unique threshold value to process every image tends to increase this variability and induces the appearance of extreme values compatible with an overestimation due to inadequate segmentation (see highest values for
vg >
rbf1,
debclE26 genotype in
Figure 5h and also
Supplementary Figure S3). On the contrary, the use of individual specific threshold values (Manual) enables to readily detect the difference of apoptosis rate between the two genotypes (
p = 1.5 × 10
−5).
In the end, this analysis shows that, in our case, using an individual threshold per image is more appropriate and turns out to be the safest option for accurate segmentation, and thus relevant quantification.