Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Handling and Maintenance
2.2. Microinjection
2.3. BiFC Assay
2.4. Live Cell Imaging
2.5. Quantification of Fluorescence Intensity
2.6. Recombinant Protein Expression of Buc-VBM
2.7. Recombinant Protein Expression of zfVasa (227–670) aa
2.8. Coomassie Staining
2.9. GST Pull-Down Assay
2.10. ATPase Assay
2.11. Circular Dichroism (CD) Spectroscopy
2.12. Bioinformatics Methods
2.13. Statistics
3. Results
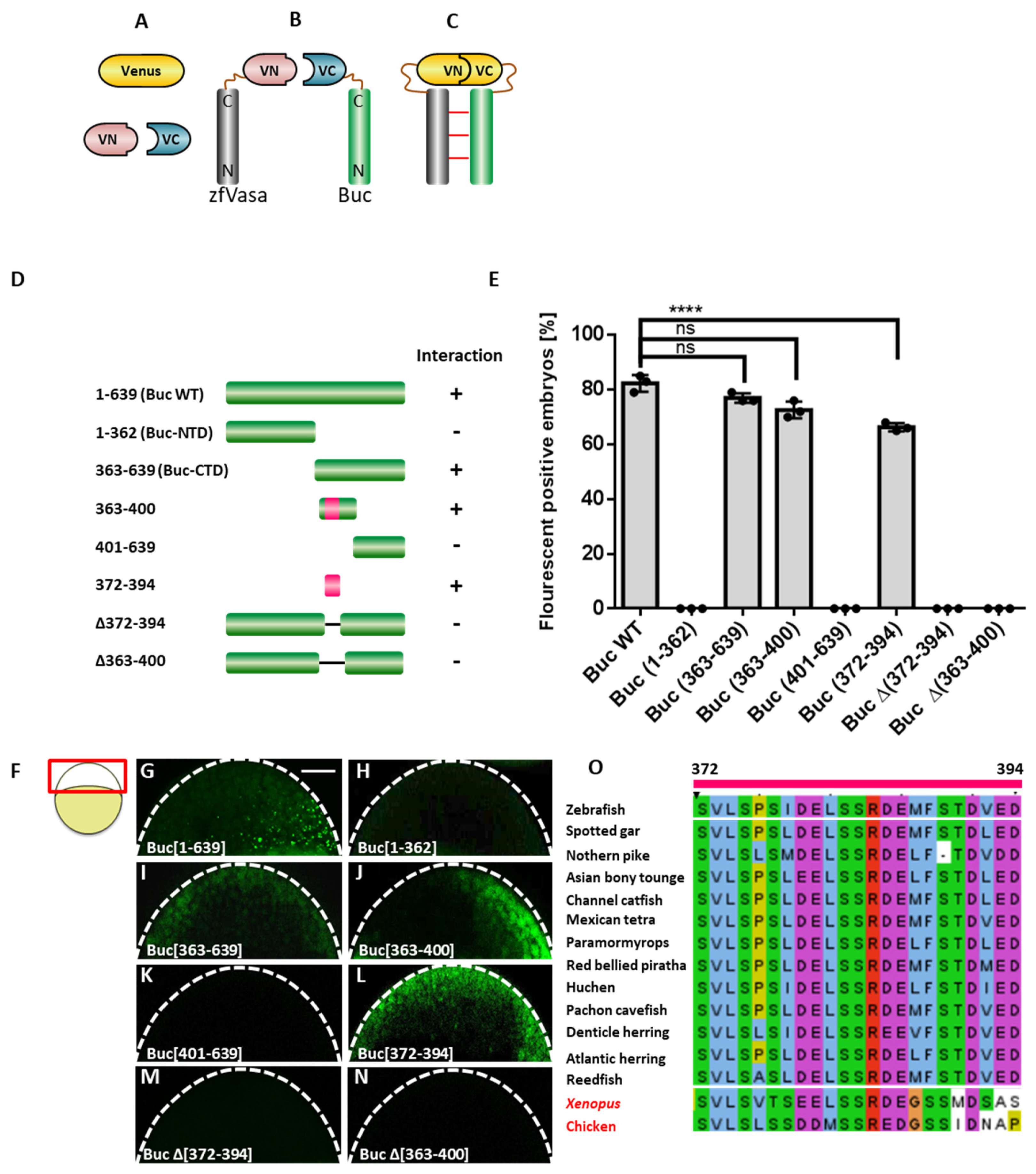
3.1. Identification of the zfVasa-Binding Motif in Buc (Buc-VBM)
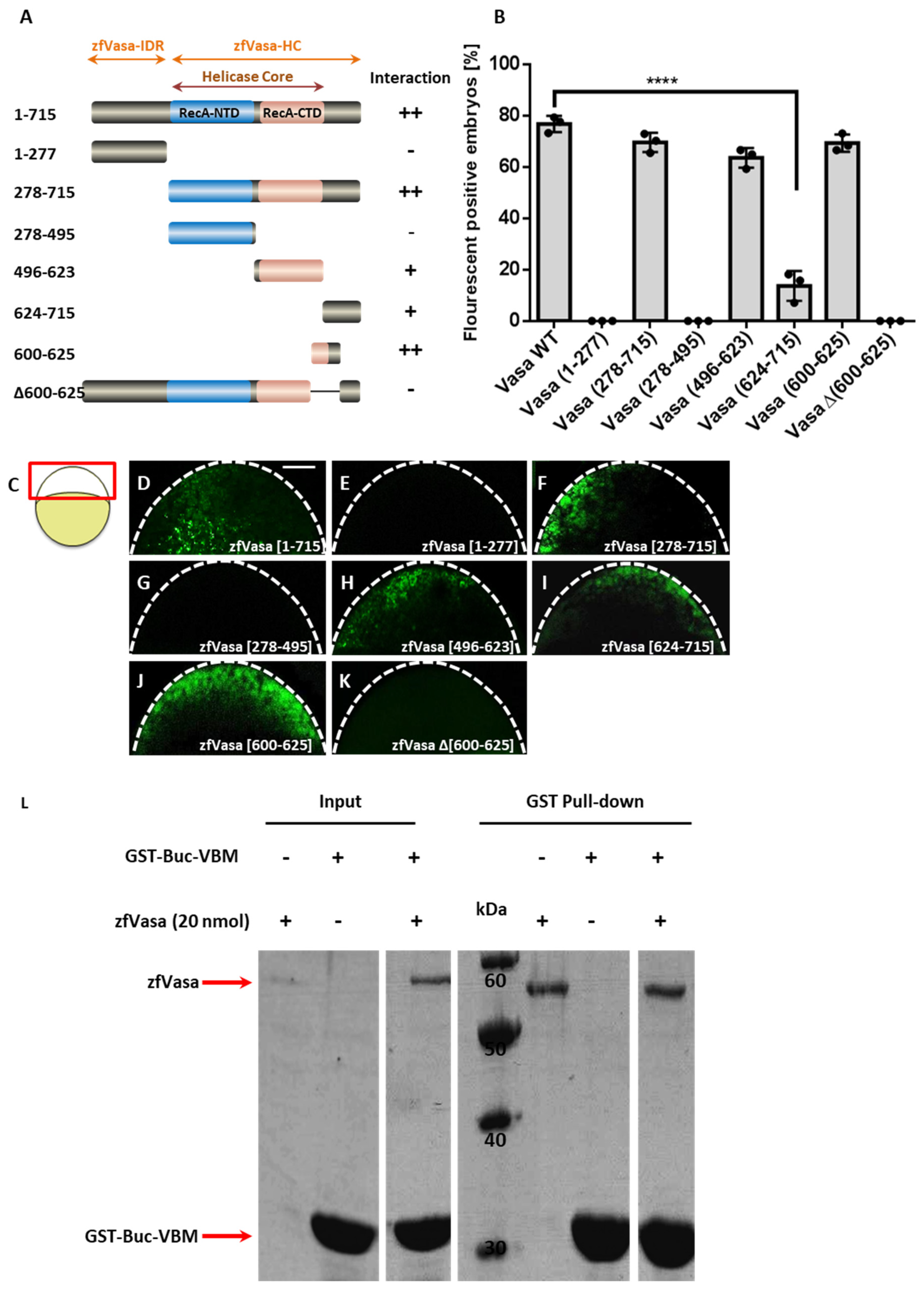
3.2. Identification of the Buc Binding Motif in zfVasa (zfVasa-BBM)
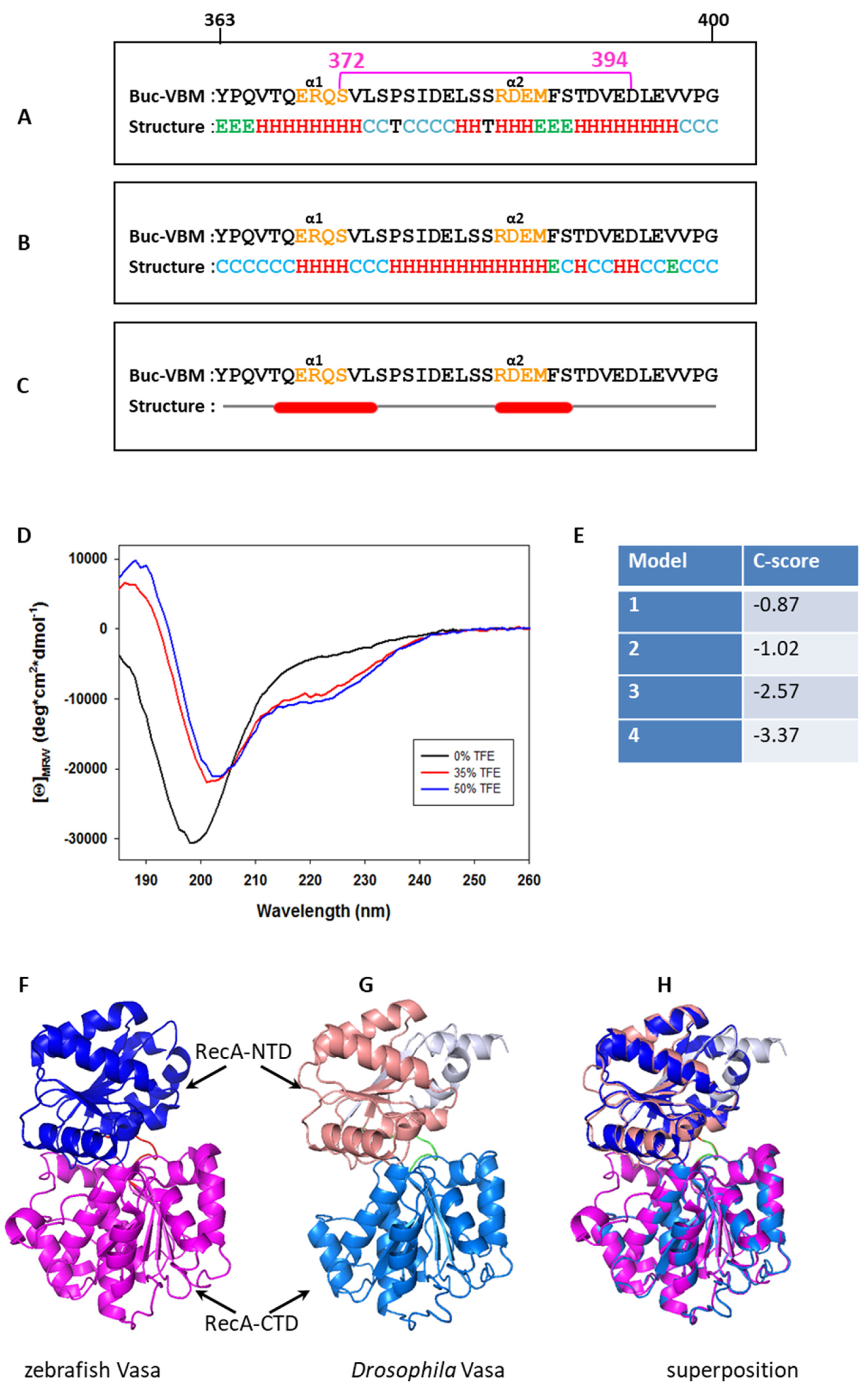
3.3. Buc-VBM Adopts α-Helices from Its Disordered Nature
3.4. Homology Modeling for zfVasa
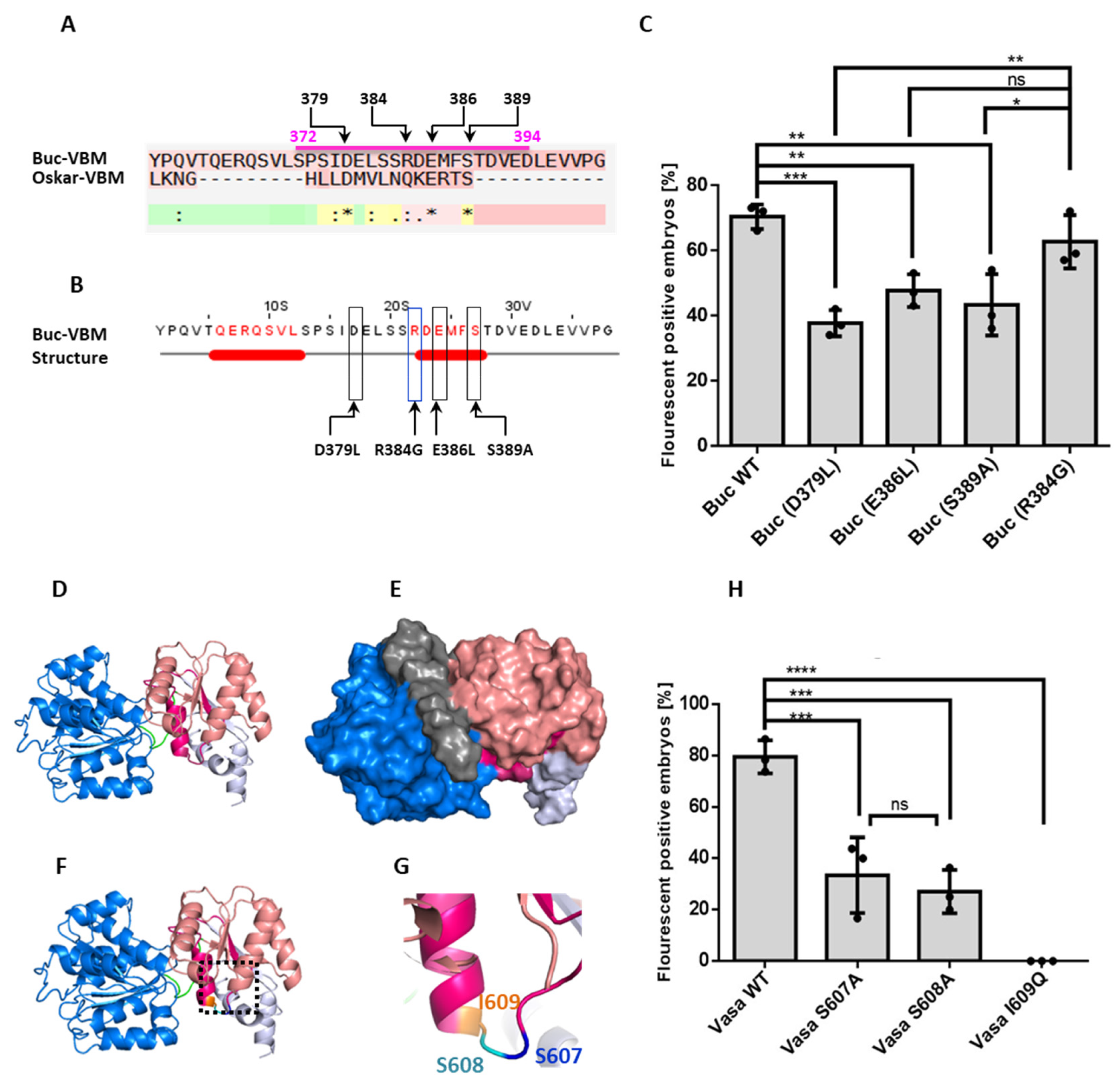
3.5. Identification of Amino Acids in the Buc-VBM, Which Are Required for zfVasa Interaction
3.6. Amino Acids in zfVasa-BBM Required for Buc Interaction
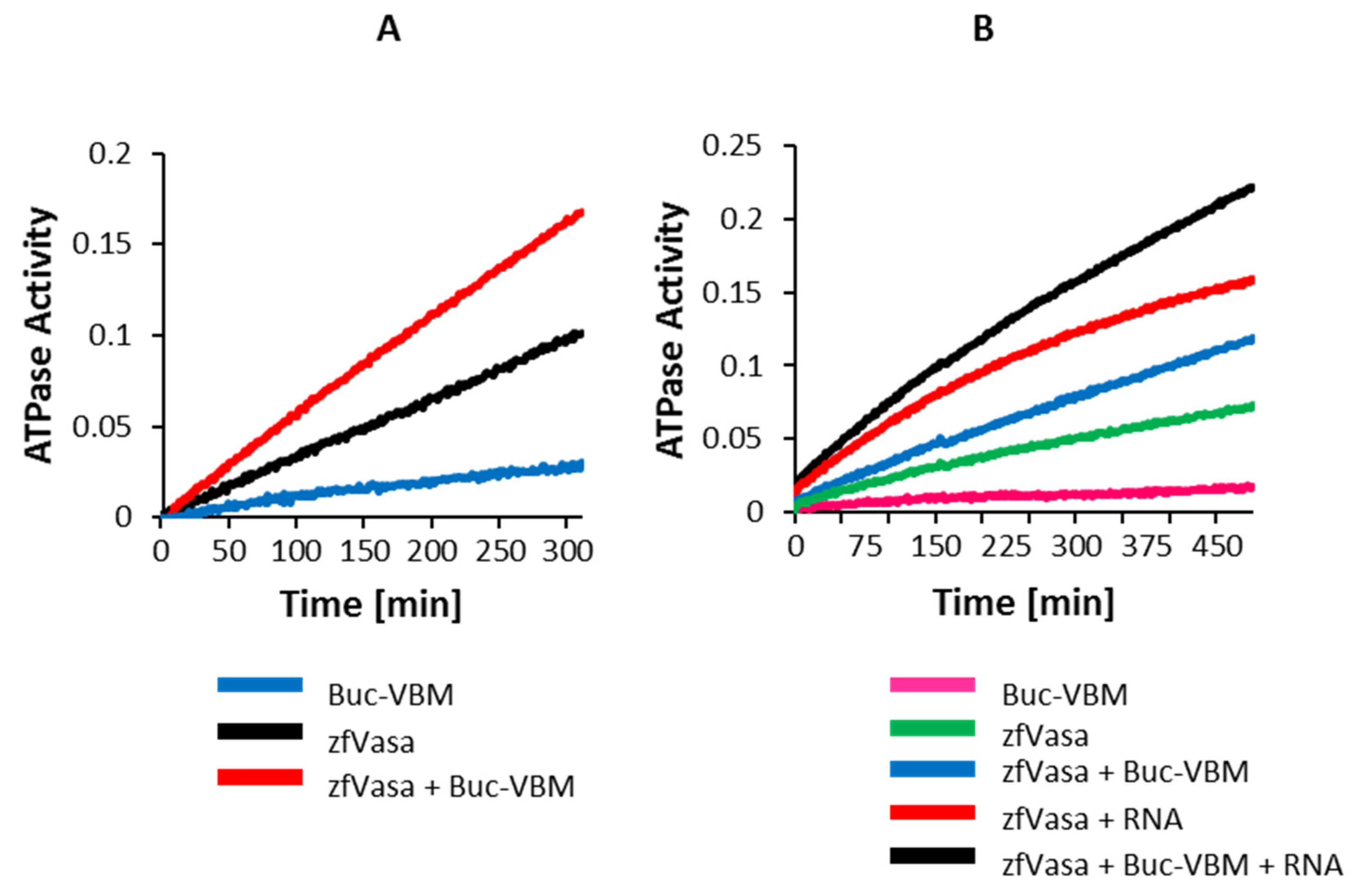
3.7. The Buc-VBM Is a Novel Activator of zfVasa ATPase Activity
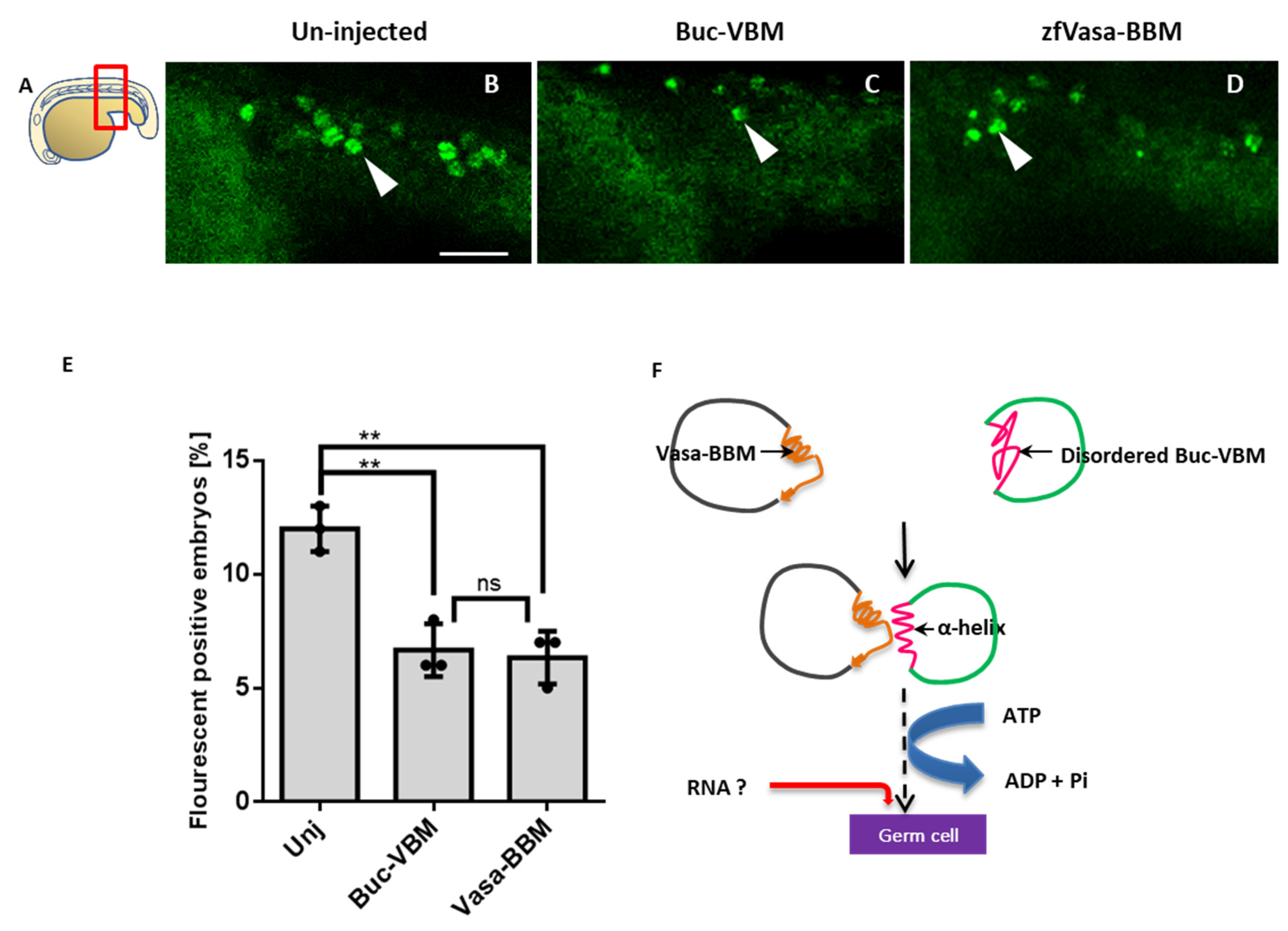
3.8. The Buc-VBM and the zfVasa-BBM Inhibit PGC Formation in Zebrafish
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos, A.C.; Lehmann, R. Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 2004, 14, 578–589. [Google Scholar] [CrossRef]
- Marlow, F.L. Maternal Control of Development in Vertebrates; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2010; Volume 1, ISBN 9781615040513. [Google Scholar]
- Jostes, S.; Schorle, H. Signals and transcription factors for specification of human germ cells. Stem Cell Investig. 2018, 5, 1–5. [Google Scholar] [CrossRef][Green Version]
- Krishnakumar, P.; Dosch, R. Germ Cell Specification: The Evolution of a Recipe to Make Germ Cells. Germ Cell 2018, 1–22. [Google Scholar] [CrossRef]
- Carr, R.M.; Oranu, A.; Khungar, V. piRNA Biogenesis in Drosophila Melanogaster. Physiol. Behav. 2016, 176, 139–148. [Google Scholar] [CrossRef]
- Hartung, O.; Forbes, M.M.; Marlow, F.L. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 2015, 81, 946–961. [Google Scholar] [CrossRef]
- Gustafson, E.A.; Wessel, G.M. Vasa genes: Emerging roles in the germ line and in multipotent cells. Bioessays 2010, 32, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Saitou, M.; Yamaji, M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction 2010, 139, 931–942. [Google Scholar] [CrossRef] [PubMed]
- D’Orazio, F.M.; Balwierz, P.J.; González, A.J.; Guo, Y.; Hernández-Rodríguez, B.; Wheatley, L.; Jasiulewicz, A.; Hadzhiev, Y.; Vaquerizas, J.M.; Cairns, B.; et al. Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization. Dev. Cell 2021, 56, 641–656. [Google Scholar] [CrossRef] [PubMed]
- Ikenishi, K.; Kotani, M.; Tanabe, K. Ultrastructural changes associated with UV irradiation in the “germinal plasm” of Xenopus laevis. Dev. Biol. 1974, 36, 155–168. [Google Scholar] [CrossRef]
- Smith, L.D. The role of a “germinal plasm” in the formation of primordial germ cells in Rana pipiens. Dev. Biol. 1966, 14, 330–347. [Google Scholar] [CrossRef]
- Tanabe, K.; Kotani, M. Relationship between the amount of the “germinal plasm” and the number of primordial germ cells in Xenopus laevis. J. Embryol. Exp. Morphol. 1974, 31, 89–98. [Google Scholar]
- Wakahara, M. Partial characterization of “primordial germ cell forming activity” localized in vegetal pole cytoplasm in anuran eggs. J. Embryol. Exp. Morphol. 1977, 39, 221–233. [Google Scholar] [PubMed]
- Ikenishi, K.; Sakiko, N.; Okuda, T. Direct Evidence for the Presence of Germ Cell Determinant in Vegetal Pole Cytoplasm of Xenopus laevis and in a Subcellular Fraction of It: (Xenopus laevis/germ cell determinant/germ plasm/PGC induction). Dev. Growth Differ. 1986, 28, 563–568. [Google Scholar] [CrossRef]
- Illmensee, K.; Mahowald, A.P. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc. Natl. Acad. Sci. USA 1974, 71, 1016–1020. [Google Scholar] [CrossRef]
- Lehmann, R.; Nüsslein-Volhard, C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in drosophila. Cell 1986, 47, 141–152. [Google Scholar] [CrossRef]
- Ephrussi, A.; Lehmann, R. Induction of germ cell fate by oskar. Nature 1992, 358, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Wilson, J.E.; Macdonald, P.M. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell 1992, 70, 849–859. [Google Scholar] [CrossRef]
- Ephrussi, A.; Dickinson, L.K.; Lehmann, R. Oskar Organizes the Germ Plasm and Directs Localization of the Posterior Determinant Nanos. Cell 1991, 66, 37–50. [Google Scholar] [CrossRef]
- Dosch, R.; Wagner, D.S.; Mintzer, K.A.; Runke, G.; Wiemelt, A.P.; Mullins, M.C. Maternal control of vertebrate development before the midblastula transition: Mutants from the zebrafish I. Dev. Cell 2004, 6, 771–780. [Google Scholar] [CrossRef]
- Wagner, D.S.; Dosch, R.; Mintzer, K.A.; Wiemelt, A.P.; Mullins, M.C. Maternal control of development at the midblastula transition and beyond: Mutants from the zebrafish II. Dev. Cell 2004, 6, 781–790. [Google Scholar] [CrossRef]
- Bontems, F.; Stein, A.; Marlow, F.; Lyautey, J.; Gupta, T.; Mullins, M.C.; Dosch, R. Bucky Ball Organizes Germ Plasm Assembly in Zebrafish. Curr. Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef]
- Krishnakumar, P.; Riemer, S.; Perera, R.; Lingner, T.; Goloborodko, A.; Khalifa, H.; Bontems, F.; Kaufholz, F.; El-Brolosy, M.A.; Dosch, R. Functional equivalence of germ plasm organizers. PLoS Genet. 2018, 14, e1007696. [Google Scholar] [CrossRef] [PubMed]
- Jeske, M.; Müller, C.W.; Ephrussi, A. The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 2017, 31, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Jeske, M.; Bordi, M.; Glatt, S.; Müller, S.; Rybin, V.; Müller, C.W.; Ephrussi, A. The crystal structure of the Drosophila germline inducer Oskar identifies two domains with distinct Vasa Helicase- and RNA-binding activities. Cell Rep. 2015, 12, 587–598. [Google Scholar] [CrossRef]
- Heim, A.E.; Hartung, O.; Rothhämel, S.; Ferreira, E.; Jenny, A.; Marlow, F.L. Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development 2014, 141, 842–854. [Google Scholar] [CrossRef]
- Roovers, E.F.; Kaaij, L.J.T.; Redl, S.; Bronkhorst, A.W.; Wiebrands, K.; de Jesus Domingues, A.M.; Huang, H.Y.; Han, C.T.; Riemer, S.; Dosch, R.; et al. Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification. Dev. Cell 2018, 46, 285–301.e9. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Miao, R.; Xiao, R.; Mei, J. m6A reader Igf2bp3 enables germ plasm assembly by m6A-dependent regulation of gene expression in zebrafish. Sci. Bull. 2021, 66, 1119–1128. [Google Scholar] [CrossRef]
- Perera, R.P.; Dosch, R. Chapter 24 with Bimolecular Fluorescent Complementation; Springer: New York, NY, USA; Humana: New York, NY, USA, 2021; Volume 2218, pp. 303–317. [Google Scholar] [CrossRef]
- Riemer, S.; Bontems, F.; Krishnakumar, P.; Gömann, J.; Dosch, R. A functional Bucky ball-GFP transgene visualizes germ plasm in living zebrafish. Gene Expr. Patterns 2015, 18, 44–52. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–41. [Google Scholar] [CrossRef]
- Kiianitsa, K.; Solinger, J.A.; Heyer, W.D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003, 321, 266–271. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M.; Taly, J.F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006, 7, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev. Biophys. 2008, 37, 465–487. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. Available online: http://academic.oup.com/nar/article/35/suppl_1/D61/1099759?login=true (accessed on 20 July 2019). [CrossRef]
- Karimi, K.; Fortriede, J.D.; Lotay, V.S.; Burns, K.A.; Wang, D.Z.; Fisher, M.E.; Pells, T.J.; James-Zorn, C.; Wang, Y.; Ponferrada, V.G.; et al. Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018, 46, D861–D868. Available online: http://academic.oup.com/nar/article/46/D1/D861/4559118 (accessed on 15 August 2019). [CrossRef]
- Ashok Kumar, T. CFSSP: Chou and Fasman Secondary Structure Prediction server. Wide Spectr. 2013, 1, 15–19. [Google Scholar] [CrossRef]
- Singh, H.; Singh, S.; Singh Raghava, G.P. Peptide Secondary Structure Prediction using Evolutionary Information. bioRxiv 2019, 558791. [Google Scholar] [CrossRef]
- Cole, C.; Barber, J.D.; Barton, G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008, 36, 197–201. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural Basis for RNA Unwinding by the DEAD-Box Protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef]
- Norma, J. Greenfield Using circular dichroism spectra to estimate protein secondary structure. ProQuest Diss. Theses 2012, 1, 218. [Google Scholar] [CrossRef]
- Laureto, P.P.; De Donadi, M.; Scaramella, E.; Frare, E.; Fontana, A. Trifluoroethanol-assisted protein folding: Fragment 53-103 of bovine α-lactalbumin. Biosystems 2001, 1548, 29–37. [Google Scholar]
- Walgers, R.; Lee, T.C.; Cammers-Goodwin, A. An indirect chaotropic mechanism for the stabilization of helix conformation of peptides in aqueous trifluoroethanol and hexafluoro-2- propanol. J. Am. Chem. Soc. 1998, 120, 5073–5079. [Google Scholar] [CrossRef]
- Kaczka, P.; Winiewska, M.; Zhukov, I.; Rempoła, B.; Bolewska, K.; Łozinski, T.; Ejchart, A.; Poznańska, A.; Wierzchowski, K.L.; Poznański, J. The TFE-induced transient native-like structure of the intrinsically disordered σ_4^704 domain of Escherichia coli RNA polymerase. Eur. Biophys. J. 2014, 43, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Kohji Ikenishi, T.S.T. Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev. Growth Differ. 1997, 39, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, K.; Mukumoto, Y.; Tanigawa, Y.; Komiya, T. Xenopus Vasa Homolog XVLG1 is Essential for Migration and Survival of Primordial Germ Cells. Zoolog. Sci. 2017, 34, 93–104. [Google Scholar] [CrossRef]
- Cipriani, P.G.; Bay, O.; Zinno, J.; Gutwein, M.; Gan, H.H.; Mayya, K.; Chung, G.; Chen, J.; Fahs, H.; Guan, Y.; et al. Novel LOTUS-domain proteins are organizational hubs that recruit C. elegans Vasa to germ granules. eLife 2021, 10, 1–41. [Google Scholar] [CrossRef]
- Perera R and RD. Unpublished work. 2021.
- Marnik, E.A.; Fuqua, J.H.; Sharp, C.S.; Rochester, J.D.; Xu, E.L.; Holbrook, S.E.; Updike, D.L. Germline maintenance through the multifaceted activities of GLH/Vasa in caenorhabditis elegans P Granules. Genetics 2019, 213, 923–939. [Google Scholar] [CrossRef] [PubMed]
- Boke, E.; Ruer, M.; Wühr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Hyman, A.A.; Mitchison, T.J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166, 637–650. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perera, R.P.; Shaikhqasem, A.; Rostam, N.; Dickmanns, A.; Ficner, R.; Tittmann, K.; Dosch, R. Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator. Biomolecules 2021, 11, 1507. https://doi.org/10.3390/biom11101507
Perera RP, Shaikhqasem A, Rostam N, Dickmanns A, Ficner R, Tittmann K, Dosch R. Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator. Biomolecules. 2021; 11(10):1507. https://doi.org/10.3390/biom11101507
Chicago/Turabian StylePerera, Roshan Priyarangana, Alaa Shaikhqasem, Nadia Rostam, Achim Dickmanns, Ralf Ficner, Kai Tittmann, and Roland Dosch. 2021. "Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator" Biomolecules 11, no. 10: 1507. https://doi.org/10.3390/biom11101507
APA StylePerera, R. P., Shaikhqasem, A., Rostam, N., Dickmanns, A., Ficner, R., Tittmann, K., & Dosch, R. (2021). Bucky Ball Is a Novel Zebrafish Vasa ATPase Activator. Biomolecules, 11(10), 1507. https://doi.org/10.3390/biom11101507





