Effects of Molecular Iodine/Chemotherapy in the Immune Component of Breast Cancer Tumoral Microenvironment
Abstract
1. Introduction
2. Materials and Methods
2.1. Mammary Tumors
2.2. RNA-Seq and Transcriptomic Analysis
2.3. Gene Set Enrichment Analysis
2.4. Th1 and Th2 Differentiation Genes
2.5. Deconvolution Analysis
2.6. Real Time RT-PCR
2.7. Immunohistochemistry
2.8. Methylation-Specific PCR
2.9. Statistic Analysis
3. Results
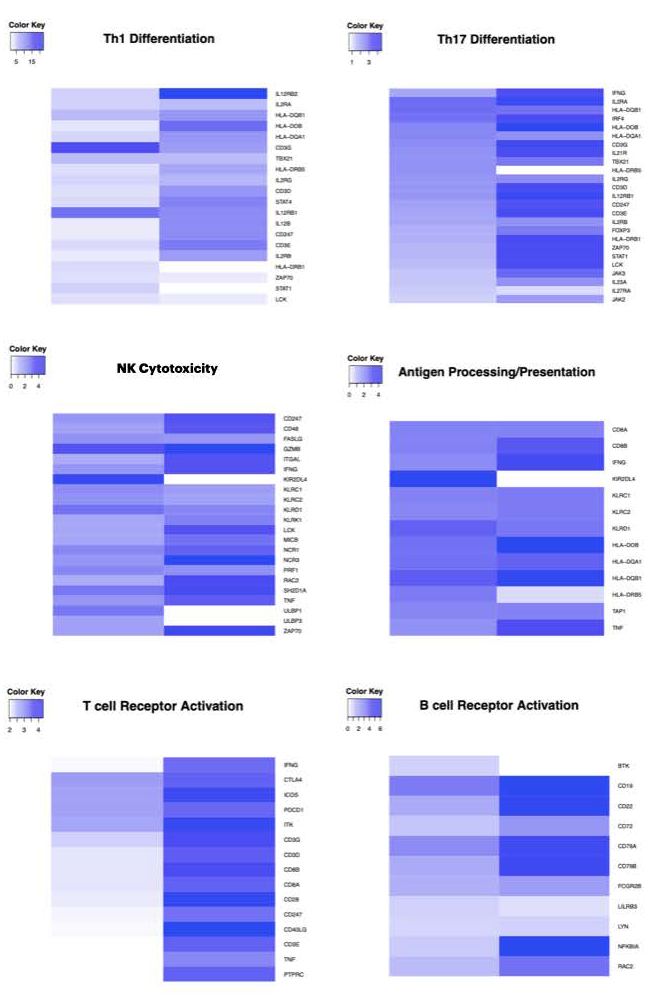
3.1. Supplementation with I2 Increases the Immune Pathways Associated with an Antitumor Response
3.2. I2 Increases the Intratumoral Ratio of Antigen-Presenting Macrophages/Dendritic Cells in Early-Stage Tumors and B Lymphocytes in the Advanced Stage
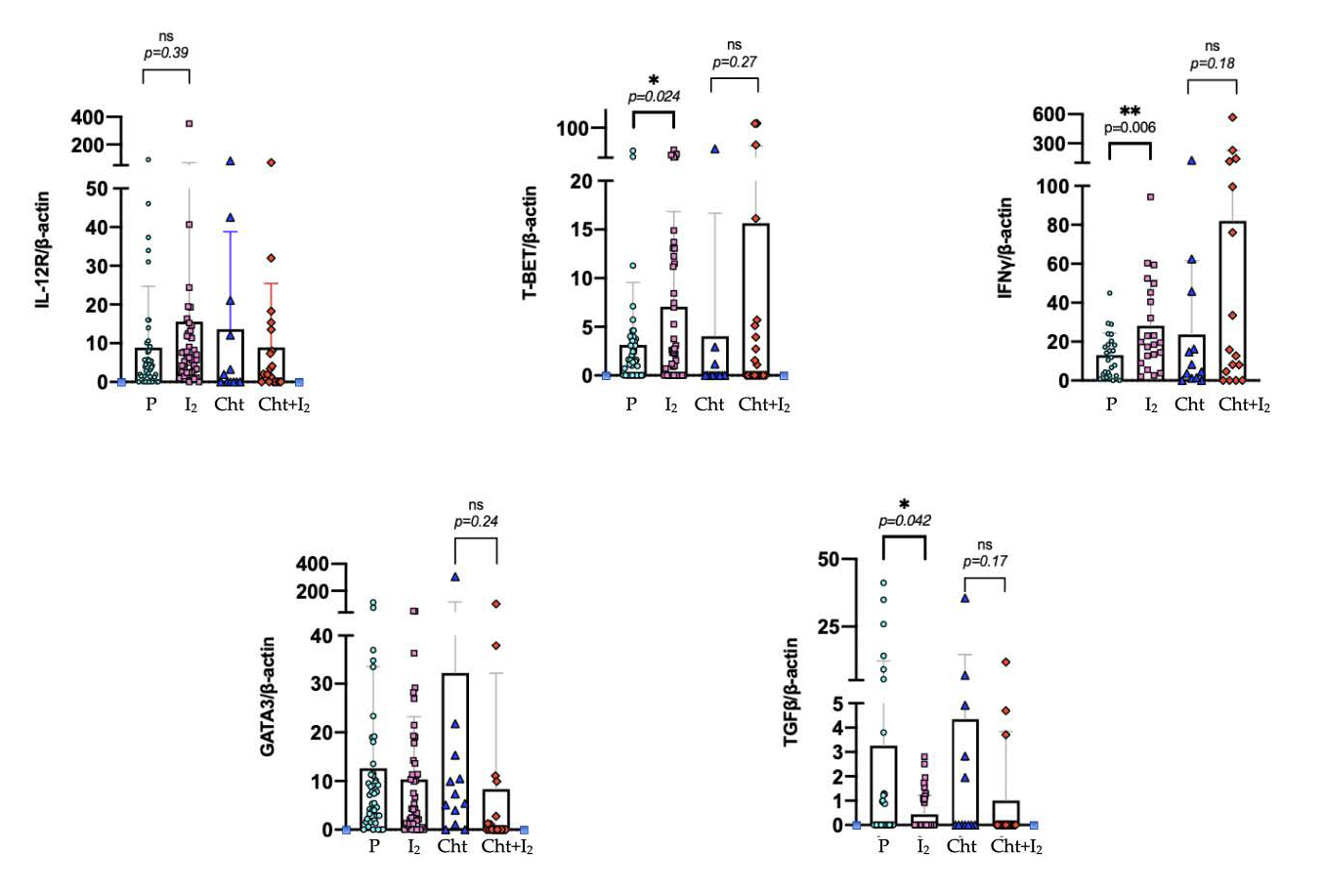
3.3. I2 Activates Th1 Differentiation in the Early Stages of the Disease, While in Advanced Stages It Suppresses Th2 Differentiation
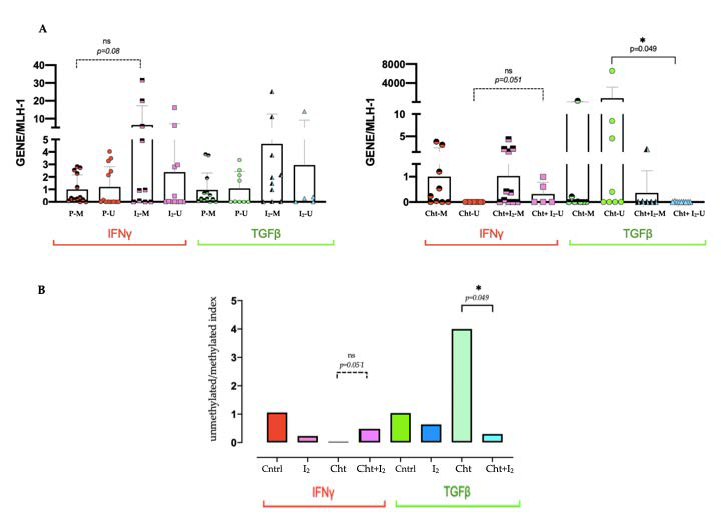
3.4. I2 Modifies the Epigenetic Landscape Activating Antitumor Gene Promoters Expression and Silencing Oncogenic Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taube, J.M.; Galon, J.; Sholl, L.M.; Rodig, S.J.; Cottrell, T.R.; A Giraldo, N.; Baras, A.S.; Patel, S.S.; A Anders, R.; Rimm, D.L.; et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod. Pathol. 2018, 31, 214–234. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lachapelle, J.; Leung, S.; Gao, D.; Foulkes, W.D.; O Nielsen, T. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012, 14, R48. [Google Scholar] [CrossRef] [PubMed]
- Tang, X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013, 332, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Carpi, A.; Rossi, G. Cytokines in breast cancer. Cytokine Growth Factor Rev. 2006, 17, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Paul, W.E. Heterogeneity and plasticity of T helper cells. Cell Res. 2010, 20, 4–12. [Google Scholar] [CrossRef]
- Calle-Fabregat, C.; de la Calle-Fabregat, C.; Morante-Palacios, O.; Ballestar, E. Understanding the Relevance of DNA Methylation Changes in Immune Differentiation and Disease. Genes 2020, 11, 110. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Kersh, E.N.; Fitzpatrick, D.R.; Murali-Krishna, K.; Shires, J.; Speck, S.H.; Boss, J.M.; Ahmed, R. Rapid Demethylation of the IFN-γ Gene Occurs in Memory but Not Naive CD8 T Cells. J. Immunol. 2006, 176, 4083–4093. [Google Scholar] [CrossRef]
- Cuenca-Micó, O.; Aceves, C. Micronutrients and Breast Cancer Progression: A Systematic Review. Nutrients 2020, 12, 3613. [Google Scholar] [CrossRef]
- Zambrano-Estrada, X.; Landaverde-Quiroz, B.; Dueñas-Bocanegra, A.A.; De Paz-Campos, M.A.; Hernández-Alberto, G.; Solorio-Perusquia, B.; Trejo-Mandujano, M.; Pérez-Guerrero, L.; Delgado-González, E.; Anguiano, B.; et al. Molecular iodine/doxorubicin neoadjuvant treatment impair invasive capacity and attenuate side effect in canine mammary cancer. BMC Vet. Res. 2018, 14, 87. [Google Scholar] [CrossRef]
- Rösner, H.; Möller, W.; Groebner, S.; Torremante, P. Antiproliferative/cytotoxic effects of molecular iodine, povidone-iodine and Lugol’s solution in different human carcinoma cell lines. Oncol. Lett. 2016, 12, 2159–2162. [Google Scholar] [CrossRef] [PubMed]
- García-Solís, P.; Alfaro, Y.; Anguiano, B.; Delgado, G.; Guzman, R.C.; Nandi, S.; Díaz-Muñoz, M.; Vázquez-Martínez, O.; Aceves, C. Inhibition of N-methyl-N-nitrosourea-induced mammary carcinogenesis by molecular iodine (I2) but not by iodide (I-) treatment Evidence that I2 prevents cancer promotion. Mol. Cell. Endocrinol. 2005, 236, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Tiwari, M.; Sinha, R.A.; Kumar, A.; Balapure, A.; Bajpai, V.K.; Sharma, R.; Mitra, K.; Tandon, A.; Godbole, M.M. Molecular iodine induces caspase-independent apoptosis in human breast carcinoma cells involving the mitochondria-mediated pathway. J. Biol. Chem. 2006, 281, 19762–19771. [Google Scholar] [CrossRef] [PubMed]
- Aceves, C.; Anguiano, B.; Delgado, G. The extrathyronine actions of iodine as antioxidant, apoptotic, and differentiation factor in various tissues. Thyroid 2013, 23, 938–946. [Google Scholar] [CrossRef]
- Nava-Villalba, M.; Aceves, C. 6-iodolactone, key mediator of antitumor properties of iodine. Prostaglandins Other Lipid Mediat. 2014, 112, 27–33. [Google Scholar] [CrossRef]
- Bilal, M.Y.; Dambaeva, S.; Kwak-Kim, J.; Gilman-Sachs, A.; Beaman, K.D. A Role for Iodide and Thyroglobulin in Modulating the Function of Human Immune Cells. Front. Immunol. 2017, 8, 1573. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Vega, A.; Vega-Riveroll, L.; Ayala, T.; Peralta, G.; Torres-Martel, J.M.; Rojas, J.; Mondragón, P.; Domínguez, A.; De Obaldía, R.; Avecilla-Guerrero, C.; et al. Adjuvant Effect of Molecular Iodine in Conventional Chemotherapy for Breast Cancer. Randomized Pilot Study. Nutrients 2019, 11, 623. [Google Scholar]
- Aceves, C. Immunehistochemistry and Transcriptomic Analysis of Iodine and Breast Cancer. Available online: https://www.protocols.io/view/immunehistochemistry-and-transcriptomic-analysis-o-t7gerjw (accessed on 1 August 2019).
- CIBERSORT. Available online: https://cibersort.stanford.edu (accessed on 1 July 2018).
- Chang, W.; Wan, C.; Lu, X.; Tu, S.W.; Sun, Y.; Zhang, X.; Zang, Y.; Zhang, A.; Huang, K.; Liu, Y.; et al. ICTD: A semi-supervised cell type identification and deconvolution method for multi-omics data. bioRxiv 2020. [CrossRef]
- Nava-Villalba, M.; Nuñez-Anita, R.E.; Bontempo, A.; Aceves, C. Activation of peroxisome proliferator-activated receptor gamma is crucial for antitumor effects of 6-iodolactone. Mol. Cancer 2015, 14, 168. [Google Scholar] [CrossRef]
- Tower, H.; Ruppert, M.; Britt, K. The Immune Microenvironment of Breast Cancer Progression. Cancers 2019, 11, 1375. [Google Scholar] [CrossRef]
- Campo, A.B.; del Campo, A.B.; Carretero, J.; Aptsiauri, N.; Garrido, F. Targeting HLA class I expression to increase tumor immunogenicity. Tissue Antigens 2012, 79, 147–154. [Google Scholar] [CrossRef]
- Arroyo-Helguera, O.; Anguiano, B.; Delgado, G.; Aceves, C. Uptake and antiproliferative effect of molecular iodine in the MCF-7 breast cancer cell line. Endocr.-Relat. Cancer 2006, 13, 1147–1158. [Google Scholar] [CrossRef][Green Version]
- Alfaro, Y.; Delgado, G.; Cárabez, A.; Anguiano, B.; Aceves, C. Iodine and doxorubicin, a good combination for mammary cancer treatment: Antineoplastic adjuvancy, chemoresistance inhibition, and cardioprotection. Mol. Cancer 2013, 12, 45. [Google Scholar] [CrossRef]
- Aceves, C.; García-Solís, P.; Arroyo-Helguera, O.; Vega-Riveroll, L.; Delgado, G.; Anguiano, B. Antineoplastic effect of iodine in mammary cancer: Participation of 6-iodolactone (6-IL) and peroxisome proliferator-activated receptors (PPAR). Mol. Cancer 2009, 8, 33. [Google Scholar] [CrossRef]
- Chaudhari, M.; Jayaraj, R.; Bhaskar, A.S.B.; Lakshmana Rao, P.V. Oxidative stress induction by T-2 toxin causes DNA damage and triggers apoptosis via caspase pathway in human cervical cancer cells. Toxicology 2009, 262, 153–161. [Google Scholar] [CrossRef]
- Nunez-Anita, R.E.; Cajero-Juarez, M.; Aceves, C. Peroxisome proliferator-activated receptors: Role of isoform gamma in the antineoplastic effect of iodine in mammary cancer. Curr. Cancer Drug Targets 2011, 11, 775–786. [Google Scholar] [CrossRef]
- Wu, X.Z.; Shi, X.Y.; Zhai, K.; Yi, F.S.; Wang, Z.; Wang, W.; Pei, X.B.; Xu, L.L.; Wang, Z.; Shi, H.Z. Activated naïve B cells promote development of malignant pleural effusion by differential regulation of T1 and T17 response. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 315, L443–L455. [Google Scholar] [CrossRef] [PubMed]
- Largeot, A.; Pagano, G.; Gonder, S.; Moussay, E.; Paggetti, J. The B-side of Cancer Immunity: The Underrated Tune. Cells 2019, 8, 449. [Google Scholar] [CrossRef] [PubMed]
- Stout, R.D.; Jiang, C.; Matta, B.; Tietzel, I.; Watkins, S.K.; Suttles, J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J. Immunol. 2005, 175, 342–349. [Google Scholar] [CrossRef] [PubMed]
- van den Ham, H.-J.; de Boer, R.J. From the two-dimensional Th1 and Th2 phenotypes to high-dimensional models for gene regulation. Int. Immunol. 2008, 20, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Oriss, T.B.; McCarthy, S.A.; Morel, B.F.; Campana, M.A.; Morel, P.A. Crossregulation between T helper cell (Th)1 and Th2: Inhibition of Th2 proliferation by IFN-gamma involves interference with IL-1. J. Immunol. 1997, 158, 3666–3672. [Google Scholar]
- Beatty, G.; Paterson, Y. IFN-gamma-dependent inhibition of tumor angiogenesis by tumor-infiltrating CD4+ T cells requires tumor responsiveness to IFN-gamma. J. Immunol. 2001, 166, 2276–2282. [Google Scholar] [CrossRef]
- Das, A.; Sinha, M.; Datta, S.; Abas, M.; Chaffee, S.; Sen, C.K.; Roy, S. Monocyte and Macrophage Plasticity in Tissue Repair and Regeneration. Am. J. Pathol. 2015, 185, 2596–2606. [Google Scholar] [CrossRef]
- Mori, H.; Kubo, M.; Kai, M.; Yamada, M.; Kurata, K.; Kawaji, H.; Kaneshiro, K.; Osako, T.; Nishimura, R.; Arima, N.; et al. T-bet lymphocytes infiltration as an independent better prognostic indicator for triple-negative breast cancer. Breast Cancer Res. Treat. 2019, 176, 569–577. [Google Scholar] [CrossRef]
- Yamashita, M.; Ukai-Tadenuma, M.; Miyamoto, T.; Sugaya, K.; Hosokawa, H.; Hasegawa, A.; Kimura, M.; Taniguchi, M.; DeGregori, J.; Nakayama, T. Essential Role of GATA3 for the Maintenance of Type 2 Helper T (Th2) Cytokine Production and Chromatin Remodeling at the Th2 Cytokine Gene Loci. J. Biol. Chem. 2004, 279, 26983–26990. [Google Scholar] [CrossRef]
- Mirza, S.; Shah, K.; Patel, S.; Jain, N.; Rawal, R. Natural Compounds as Epigenetic Regulators of Human Dendritic Cell-mediated Immune Function. J. Immunother. 2018, 41, 169–180. [Google Scholar] [CrossRef]
- Gerhauser, C. Cancer chemoprevention and nutriepigenetics: State of the art and future challenges. Top. Curr. Chem. 2013, 329, 73–132. [Google Scholar] [PubMed]
- Shenoy, N.; Bhagat, T.; Nieves, E.; Stenson, M.; Lawson, J.; Choudhary, G.S.; Habermann, T.; Nowakowski, G.; Singh, R.; Wu, X.; et al. Upregulation of TET activity with ascorbic acid induces epigenetic modulation of lymphoma cells. Blood Cancer J. 2017, 7, e587. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Mao, S.Q.; Zhao, B.; Chong, Z.; Yang, Y.; Zhao, C.; Zhang, D.; Huang, H.; Gao, J.; Li, Z.; et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013, 135, 10396–10403. [Google Scholar] [CrossRef] [PubMed]
- Luchtel, R.A.; Bhagat, T.; Pradhan, K.; Jacobs, W.R., Jr.; Levine, M.; Verma, A.; Shenoy, N. High-dose ascorbic acid synergizes with anti-PD1 in a lymphoma mouse model. Proc. Natl. Acad. Sci. USA 2020, 117, 1666–1677. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuenca-Micó, O.; Delgado-González, E.; Anguiano, B.; Vaca-Paniagua, F.; Medina-Rivera, A.; Rodríguez-Dorantes, M.; Aceves, C. Effects of Molecular Iodine/Chemotherapy in the Immune Component of Breast Cancer Tumoral Microenvironment. Biomolecules 2021, 11, 1501. https://doi.org/10.3390/biom11101501
Cuenca-Micó O, Delgado-González E, Anguiano B, Vaca-Paniagua F, Medina-Rivera A, Rodríguez-Dorantes M, Aceves C. Effects of Molecular Iodine/Chemotherapy in the Immune Component of Breast Cancer Tumoral Microenvironment. Biomolecules. 2021; 11(10):1501. https://doi.org/10.3390/biom11101501
Chicago/Turabian StyleCuenca-Micó, Olga, Evangelina Delgado-González, Brenda Anguiano, Felipe Vaca-Paniagua, Alejandra Medina-Rivera, Mauricio Rodríguez-Dorantes, and Carmen Aceves. 2021. "Effects of Molecular Iodine/Chemotherapy in the Immune Component of Breast Cancer Tumoral Microenvironment" Biomolecules 11, no. 10: 1501. https://doi.org/10.3390/biom11101501
APA StyleCuenca-Micó, O., Delgado-González, E., Anguiano, B., Vaca-Paniagua, F., Medina-Rivera, A., Rodríguez-Dorantes, M., & Aceves, C. (2021). Effects of Molecular Iodine/Chemotherapy in the Immune Component of Breast Cancer Tumoral Microenvironment. Biomolecules, 11(10), 1501. https://doi.org/10.3390/biom11101501





