Immunoaffinity Capillary Electrophoresis in the Era of Proteoforms, Liquid Biopsy and Preventive Medicine: A Potential Impact in the Diagnosis and Monitoring of Disease Progression
Abstract
1. Introduction
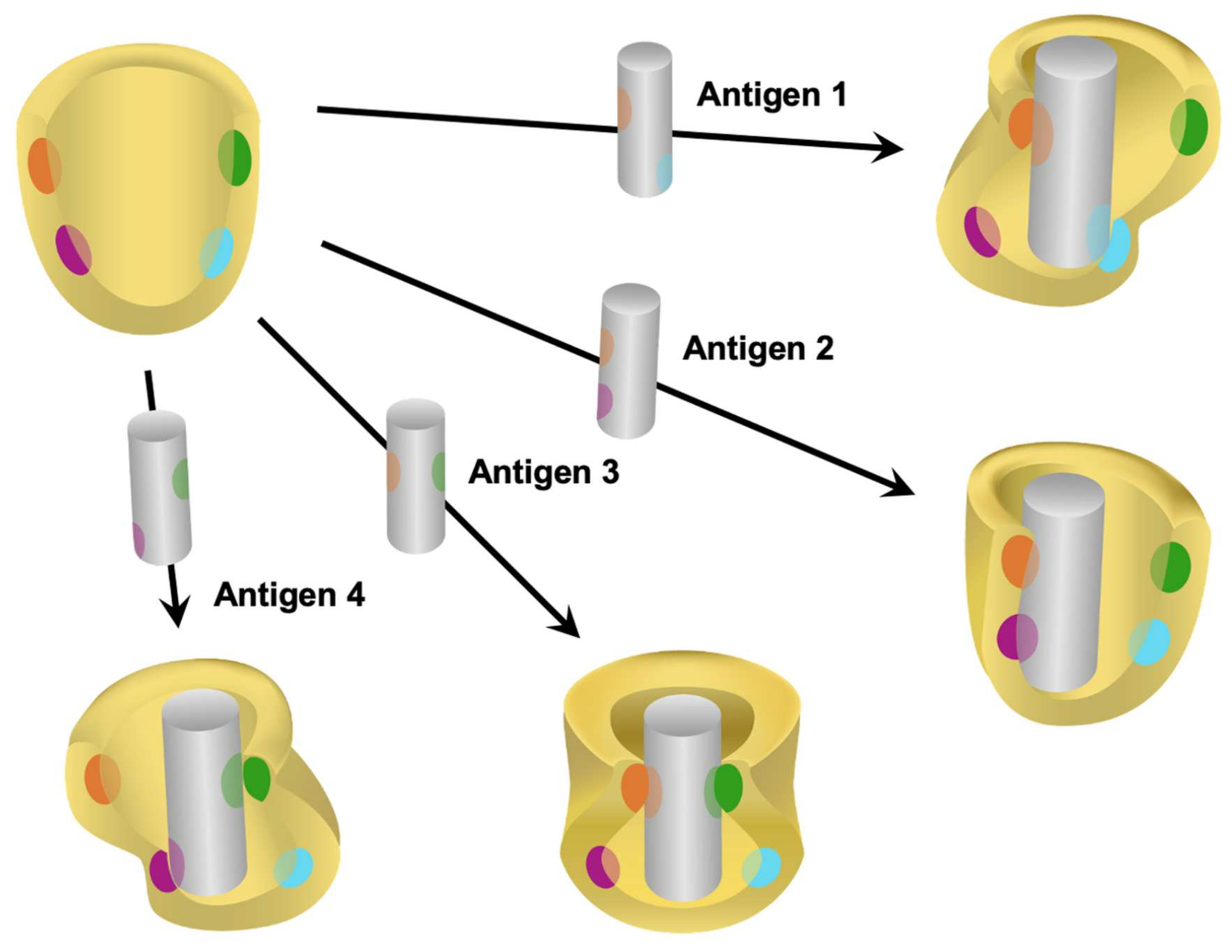
2. Proteoforms as Diagnostic Biomarkers
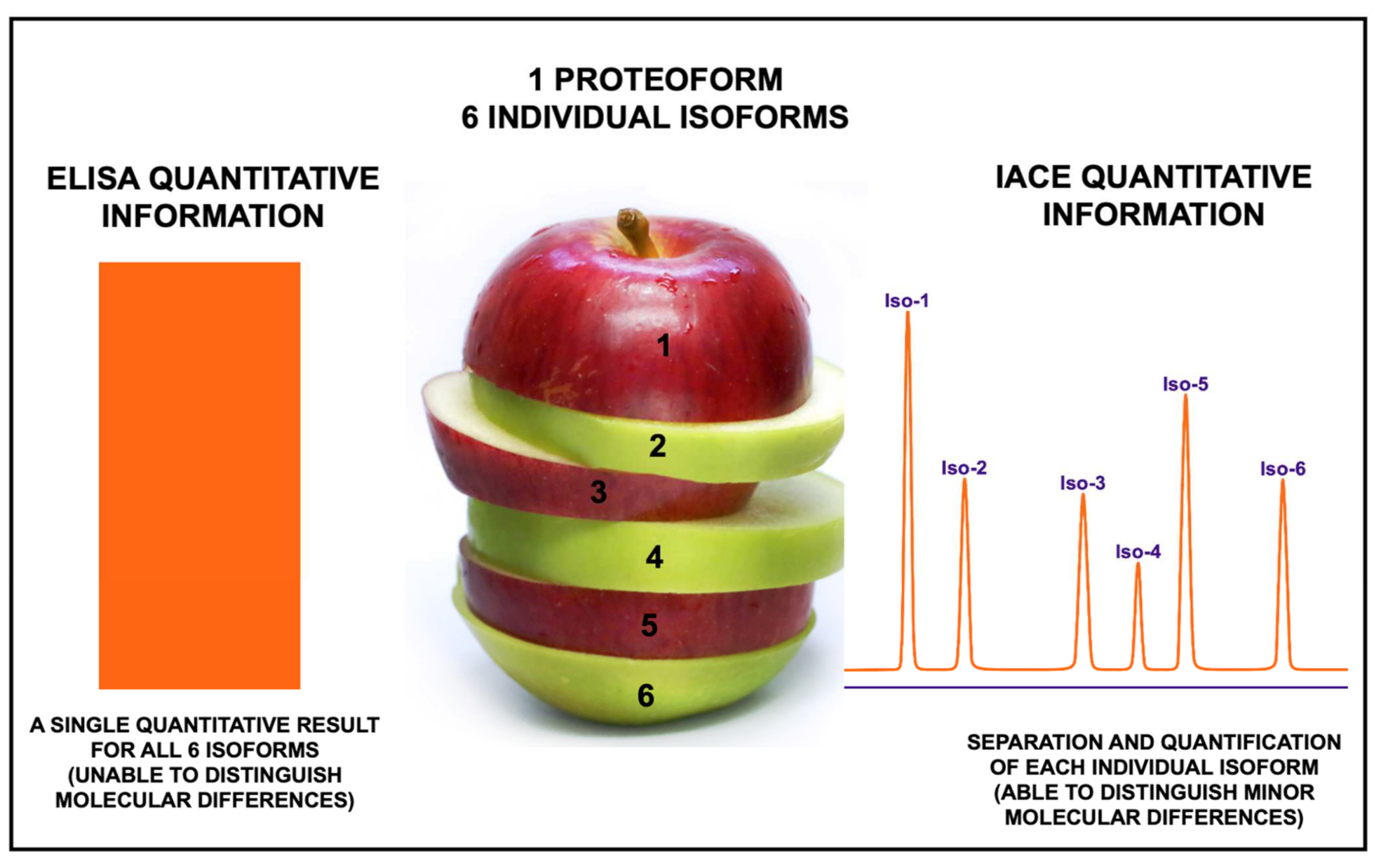
3. Limitations of Existing Immunoassays Used in Clinical Diagnostics
4. Capillary Electrophoresis as a Useful Technology for the Separation of Structurally Related Small Molecular-Weight Substances and Biomolecules
5. Advantages of Immunoaffinity Capillary Electrophoresis for the Detection of Proteoforms
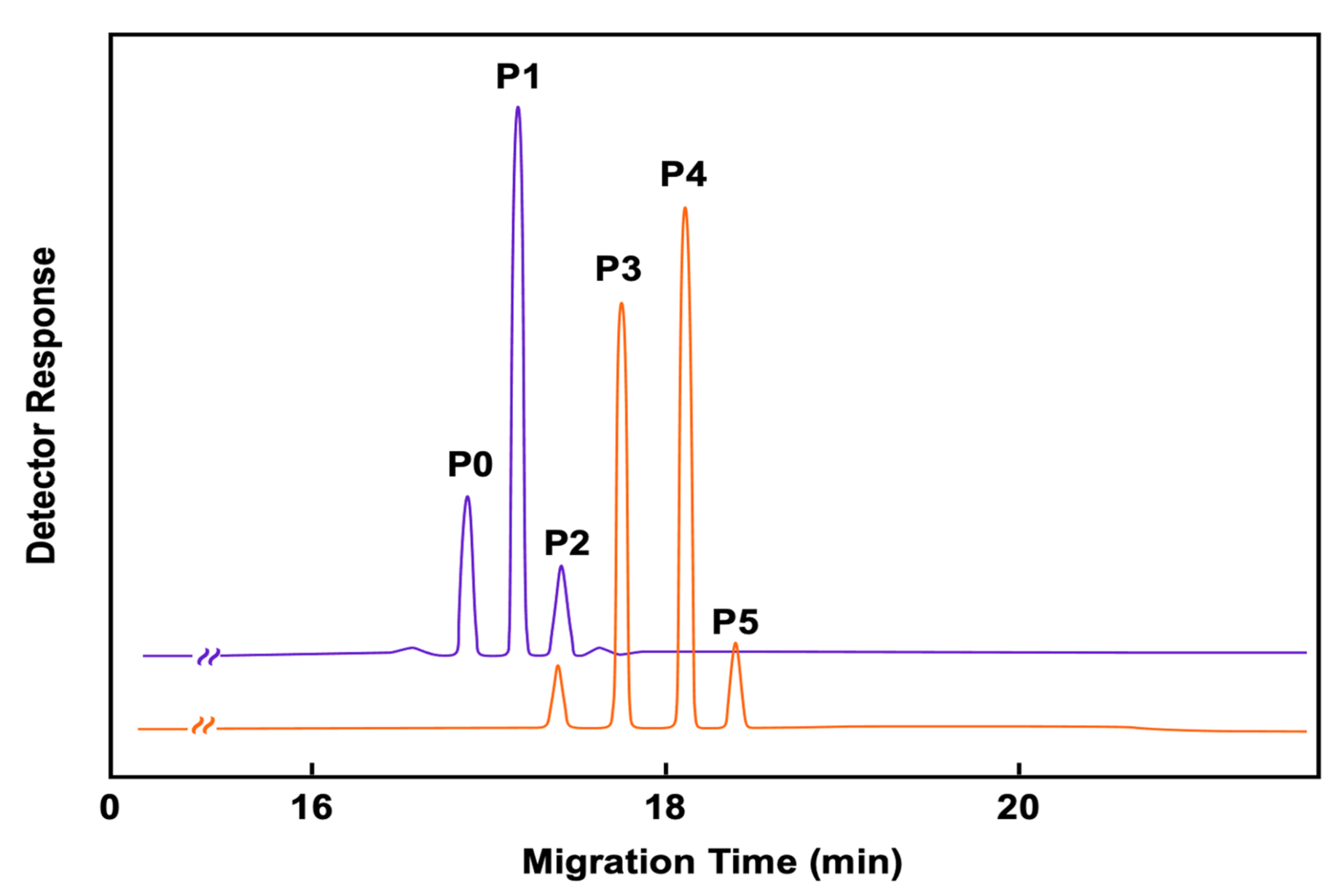
6. Applications of Immunoaffinity Capillary Electrophoresis. The Importance of Detecting Molecular Modifications of Proteins during the Progression of a Disease
7. Future of Immunoaffinity Capillary Electrophoresis for the Analysis of Proteoforms
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stone, J.; Carson, A.; Sharpe, M. Functional symptoms and signs in neurology: Assessment and diagnosis. J. Neurol. Neurosurg. Psychiatry 2005, 76 (Suppl. S1), i2–i12. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Votey, S. Sign and Symptoms in Emergency Medicine 2006, 2nd ed.; Mosby-Elsevier: Amsterdam, The Netherlands, 2006. [Google Scholar]
- King, L.S. Signs and symptoms. J. Am. Med. Assoc. 1968, 206, 1063–1065. [Google Scholar] [CrossRef]
- Llewelyn, H.; Ang, H.A.; Lewis, K.; Al-Abdullah, A. Oxford Handbook of Clinical Diagnosis, 3rd ed.; Oxford University Press: Oxford, UK, 2014; ISBN 13 9780199679867. [Google Scholar] [CrossRef]
- National Center for Chronic Disease Prevention and Health Promotion. Health and Economic Costs of Chronic Diseases; Centers for Disease Control and Prevention: Atlanta, GA, USA; US Department of Health and Human Services: Washington, DC, USA, 2018. Available online: https://www.cdc.gov/chronicdisease/about/costs/index.htm (accessed on 1 January 2021).
- Partnership to Fight Chronic Disease. Available online: https://www.fightchronicdisease.org/sites/default/files/docs/PFCD_ChronDisease_FactSheet3Final.pdf (accessed on 1 January 2020).
- Golics, C.J.; Basra, M.K.; Salek, M.S.; Finlay, A.Y. The impact of patients’ chronic disease on family quality of life: An experience from 26 specialties. Int. J. Gen. Med. 2013, 6, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Raghupathi, W.; Raghupathi, V. An empirical study of chronic diseases in the United States: A visual analytics approach to public health. Int. J. Environ. Res. Pub. Health 2018, 15, 431. [Google Scholar] [CrossRef] [PubMed]
- Prince, A.E.R.; Berkman, B.E. When does an illness begin: Genetic discrimination and disease manifestation. J. Law Med. Ethics 2012, 40, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P. The inflammation theory of disease. The growing realization that chronic inflammation is crucial in many diseases opens new avenues for treatment. EMBO Rep. 2012, 13, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Menozzi, E.; Mcnaughtan, J.; Schapira, A.H.V. The gut-brain axis and Parkinson disease: Clinical pathogenetic relevance. Ann. Med. 2021, 53, 611–624. [Google Scholar] [CrossRef]
- Cinque, A.; Vago, R.; Trevisani, F. Circulating RNA in kidney cancer: What we know and what we still suppose. Genes 2021, 12, 835. [Google Scholar] [CrossRef]
- Özgur, Y. Relationship between vitamin D deficiency, albuminuria, peripheral artery disease and 5-year mortality in chronic kidney disease. J. Coll. Physicians Surg. Pak. 2021, 30, 644–650. [Google Scholar] [CrossRef]
- Carmichael, J.; Fadavi, H.; Ishibashi, F.; Shore, A.C.; Tavakoli, M. Advances in screening, early diagnosis and accurate staging of diabetic neuropathy. Front. Endocrinol. 2021, 12, 671257. [Google Scholar] [CrossRef]
- Shaw, G. The silent disease. Nature 2016, 537, S98–S99. [Google Scholar] [CrossRef] [PubMed]
- Hanmer, J.; Yu, L.; Li, J.; Kavalieratos, D.; Peterson, L.; Hess, R. The diagnosis of asymptomatic disease is associated with fewer healthy days: A cross sectional analysis from the national health and nutrition examination survey. Br. J. Health Psychol. 2019, 24, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Saquib, N.; Saquib, J.; Ioannidis, J.P.A. Does screening for disease save lives in asymptomatic adults? Systemic review of meta-analyses and randomized trials. Int. J. Epidemiol. 2015, 44, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Cebey-López, M.; Salas, A. Recognising the asymptomatic enemy. Lancet Infect. Dis. 2021, 21, 305–306. [Google Scholar] [CrossRef]
- King, P.; Peacock, I.; Donnelly, R. The UK Prospective Diabetes Study (UKPDS): Clinical and therapeutic implications for type 2 diabetes. Br. J. Clin. Pharmacol. 1999, 48, 643–648. [Google Scholar] [CrossRef]
- Russo, G.T.; Giorda, C.B.; Cercone, S.; Nicolucci, A.; Cucinotta, D.; on behalf of BetaDecline Group. Factors associated with beta-cell dysfunction in type 2 diabetes: The BetaDecline study. PLoS ONE 2014, 9, e109702. [Google Scholar] [CrossRef]
- Kahn, S.E. Clinical review 135: The importance of beta-cell failure in the development and progression of type 2 diabetes. J. Clin. Endocr. Metabol. 2001, 86, 4047–4058. [Google Scholar] [CrossRef]
- White, M.G.; Shaw, J.A.M.; Taylor, R. Type 2 diabetes: The pathologic basis of reversible β-cell dysfunction. Diabetes Care 2016, 39, 2080–2088. [Google Scholar] [CrossRef]
- Holman, R.R. Assesing the potential for alpha-glucosidase inhibitors in prediabetic states. Diab. Res. Clin. Pract. 1998, 40 (Suppl. S1), S21–S25. [Google Scholar] [CrossRef]
- Gerstung, M.; Jolly, C.; Leshchiner, I.; Dentro, S.C.; Gonzalez, S.; Rosebrock, D.; Mitchell, T.J.; Rubanova, Y.; Anur, P.; Yu, K.; et al. The evolutionary history of 2,658 cancers. Nature 2020, 578, 122–128. [Google Scholar] [CrossRef]
- Tang, Y.; Qiao, G.; Xu, E.; Xuan, Y.; Liao, M.; Ying, G. Biomarkers for early diagnosis, prognosis, prediction, and recurrence monitoring of non-small cell cancer. OncoTargets Ther. 2017, 10, 4527–4534. [Google Scholar] [CrossRef]
- Hassaneim, M.; Callison, J.C.; Callaway-Lane, C.; Aldrich, M.C.; Grogan, E.L.; Massion, P.P. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev. Res. 2012, 5, 992–1006. [Google Scholar] [CrossRef]
- Vashit, S.K.; Luong, J.H.T. (Eds.) Handbook of Immunoassay Technologies: Approaches, Performances, and Applications; Academic Press-Elsevier: San Diego, CA, USA, 2018; ISBN 13 9780128117620. [Google Scholar]
- Notkins, A.L. Polyreactivity of antibody molecules. Trends Immunol. 2004, 25, 174–179. [Google Scholar] [CrossRef]
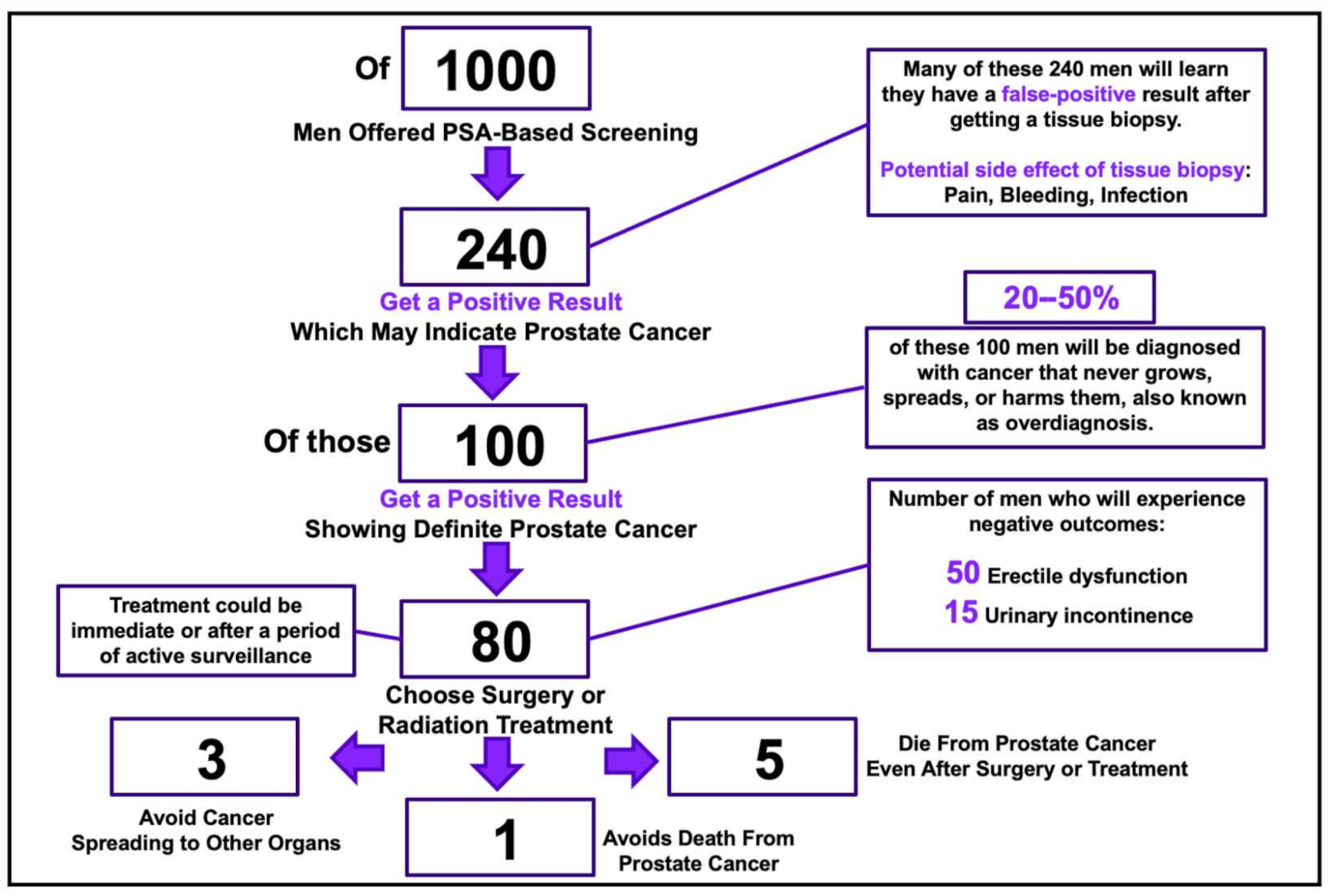
- U.S. Preventive Services Task Force; Grossman, D.C.; Curry, S.J.; Owens, D.K.; Bibbins-Domingo, K.; Caughey, A.B.; Davidson, K.W.; Doubeni, C.A.; Ebell, M.; Epling, J.W.J.; et al. Screening for prostate cancer: US Preventive Task Force recommendation statement. J. Am. Med. Assoc. 2018, 319, 1901–1913. [Google Scholar] [CrossRef]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False negative test for SARS-CoV-2 infection–Challenges and implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef]
- Lin, J.; Dai, W.; Li, W.; Xiao, L.; Luo, T.; Guo, Y.; Yang, Y.; Han, Y.; Zhu, P.; Wu, Q.; et al. Potential false-positive and false-negative results for COVID-19 IgG/IgM antibody testing after heat-inactivation. Front. Med. 2021, 7, 589080. [Google Scholar] [CrossRef] [PubMed]
- Kassirer, J.P. Our stubborn quest for diagnostic certainty. A cause of excessive testing. N. Engl. J. Med. 1989, 320, 1489–1491. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 1–25. [Google Scholar] [CrossRef]
- Katathiya, U.; Padariya, M.; Faktor, J.; Coyaud, E.; Alfaro, J.A.; Fahraeus, R.; Hupp, T.R.; Goodlett, D.R. Interfaces with structure dynamics of the workhorses from cells revealed through cross-linking mass spectrometry (CLMS). Biomolecules 2021, 11, 382. [Google Scholar] [CrossRef] [PubMed]
- Roehrl, M.H.; Roehrl, V.B.; Wang, J.Y. Proteome-based pathology: The next frontier in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2021, 6, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L.; Consortium for Top Down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next proteomics currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef]
- Smith, L.; Agar, J.; Chamot-Rooke, J.; Danis, P.; Ge, Y.; Loo, J.; Pasa-Tolic, L.; Tsybin, Y.; Kelleher, N. The human proteoform project: A plan to define the Human Proteome. Preprints 2020, 2020100368. [Google Scholar] [CrossRef]
- Aebersold, R.; Agar, J.N.; Amster, I.J.; Bager, M.S.; Bertozzi, C.R.; Boja, E.S.; Costello, C.E.; Cravatt, B.J.; Fenselau, B.F.; Garcia, B.A.; et al. How many human proteoforms are there? Nat. Chem. Bio. 2018, 14, 206–214. [Google Scholar] [CrossRef]
- Hoff, F.W.; van Dijk, A.D.; Kornblau, S.M. Proteoforms in acute leukemia: Evaluation of age- and disease-specific proteoform patterns. Intechopen 2019, 5, 62–79. [Google Scholar] [CrossRef]
- Bogaert, A.; Fernandez, E.; Gevaert, K. N-terminal proteoforms in human disease. Trends Biochem. Sci. 2020, 45, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The size of the Human Proteome: The width and the depth. Intern. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef] [PubMed]
- Da Silva-Araújo, J.; Tavares-Machado, O.L. Proteoforms: General concepts and methodological process for identification. Intechopen 2019, 2, 523–555. [Google Scholar] [CrossRef]
- Gregorich, Z.R.; Ge, Y. Top-down proteomics in health and disease: Challenges and opportunities. Proteomics 2014, 14, 1195–1210. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H. Cardiac markers: From enzymes to proteins, diagnosis, laboratory to bedside. Ann. Clin. Lab. Sci. 1999, 29, 18–23. [Google Scholar] [PubMed]
- Mayeux, R. Biomarkers: Potential uses and limitations. NeuroRx 2004, 1, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.L.; Chan, D.W. Enzymes and related proteins as cancer biomarkers: A proteomic approach. Clin. Chim. Acta 2007, 381, 93–97. [Google Scholar] [CrossRef]
- Yassine, H.N.; Trenchevska, O.; Dong, Z.; Bashawri, Y.; Koska, J.; Reaven, P.D.; Nelson, R.W.; Nedelkov, D. The association of plasma cystatin C proteoforms with diabetic chronic kidney disease. Proteome Sci. 2016, 14, 7. [Google Scholar] [CrossRef]
- Trenchevska, O.; Sherma, N.D.; Oran, P.E.; Reaven, P.D.; Nelson, R.W.; Nedelkov, D. Quantitative mass spectrometric immunoassay for the chemokine RANTES and its variants. J. Proteom. 2015, 116, 15–23. [Google Scholar] [CrossRef]
- Noor, A.; Zafar, S.; Zerr, I. Neurodegenerative proteinopathies in the proteoform spectrum–Tools and challenges. Int. J. Mol. Sci. 2021, 22, 1085. [Google Scholar] [CrossRef]
- Muchtar, E.; Blauwet, L.A.; Gert, M.A. Restrictive cardiomyopathy. Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circul. Res. 2017, 121, 819–837. [Google Scholar] [CrossRef]
- Wildburger, N.C.; Esparza, T.J.; LeDuc, R.D.; Fellers, R.T.; Thomas, P.M.; Cairns, N.J.; Kelleher, N.L.; Bateman, R.J.; Brody, D.L. Diversity of amyloid-beta proteoforms in the Alzheimer’s disease brain. Sci. Rep. 2017, 7, 9520. [Google Scholar] [CrossRef]
- Georgiev, G.A.; Gh, M.S.; Romano, J.; Teixeira, K.L.D.; Struble, C.; Ryan, D.S.; Sia, R.K.; Kitt, J.P.; Harris, J.M.; Hsu, K.-L.; et al. Lacritin proteoforms prevent tear film collapse and maintain epithelial homeostasis. J. Biol. Chem. 2021, 296, 100070. [Google Scholar] [CrossRef]
- Justis, B.M.; Coburn, C.E.; Tyler, E.M.; Showalter, R.S.; Dissler, B.J.; Li, M.; McNamara, N.A.; Laurie, G.M.; McKown, R.L. Development of a quantitative immunoassay for tear lacritin proteoforms. Trans. Vis. Sci. Tech. 2020, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Malaney, P.; Uversky, V.; Davé, V. PTEN proteoforms in biology and disease. Cell Mol. Life Sci. 2017, 74, 2783–2794. [Google Scholar] [CrossRef] [PubMed]
- Tiambeng, T.N.; Tucholski, T.; Wu, Z.; Zhu, Y.; Mitchell, S.D.; Roberts, D.S.; Jin, Y.; Ge, Y. Analysis of cardiac troponin proteoforms by top-down mass spectrometry. Methods Enzymol. 2019, 626, 347–374. [Google Scholar] [CrossRef]
- Soetkamp, D.; Raedschelders, K.; Mastali, M.; Sobhani, K.; Bairey Merz, C.N.; Van Eyk, J. The continuing evolution of cardiac troponin I biomarker analysis: From protein to proteoform. Expert. Rev. Proteom. 2017, 14, 973–986. [Google Scholar] [CrossRef]
- Nuti, E.; Rossello, A.; Cuffaro, D.; Camodeca, C.; Van Bael, J.; van der Matt, D.; Martens, E.; Fiten, P.; Pereira, R.V.S.; Ugarte-Berzal, E.; et al. Bivalent inhibitor with selectivity for trimeric MMP-9 amplifies neutrophil chemotaxis and enables functional studies on MMP-9 proteoforms. Cells 2020, 9, 1634. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Cobbaert, C.M. Quantifying apolipoprotein(a) in the era of proteoforms and precision medicine. Clin. Chim. Acta 2020, 511, 260–268. [Google Scholar] [CrossRef]
- Fania, C.; Arosio, B.; Capitanio, D.; Torretta, E.; Gussago, C.; Ferri, E.; Mari, D.; Gelfi, C. Protein signature in cerebrospinal fluid and serum of Alzheimer’s disease patients: The case of apolipoprotein A-1 proteoforms. PLoS ONE 2017, 12, e0179280. [Google Scholar] [CrossRef]
- Scumaci, D.; Olivo, E.; Fiumara, V.; La Chimia, M.; De Angelis, M.T.; Mauro, S.; Costa, G.; Ambrosio, F.A.; Alcaro, S.; Agosti, V.; et al. DJ-1 proteoforms in breast cancer cells: The escape of metabolic epigenetic misregulation. Cells 2020, 9, 1968. [Google Scholar] [CrossRef] [PubMed]
- Moran, A.B.; Domínguez-Vega, E.; Nouta, J.; Pongracz, T.; de Reijke, T.M.; Wuhrer, M.; Lageveen-Kammeijer, G.S.M. Profiling the proteoforms of urinary prostate-specific antigen by capillary electrophoresis-mass spectrometry. J. Proteom. 2021, 238, 104148. [Google Scholar] [CrossRef] [PubMed]
- Tamara, S.; Franc, V.; Heck, A.J.R. A wealth of genotype-specific proteoforms fine-tunes hemoglobin scavenging by haptoglobin. Proc. Natl. Acad. Sci. USA 2020, 117, 15554–15564. [Google Scholar] [CrossRef] [PubMed]
- Falck, D.; Haberger, M.; Plomp, R.; Hook, M.; Bulau, P.; Wuhrer, M.; Reusch, D. Affinity purification of erythropoietin from cell culture supernatant combined with MALDI-TOF-MS analysis of erythropoietin N-glycosylation. Sci. Rep. 2017, 7, 5324. [Google Scholar] [CrossRef]
- Ng, T.; Marx, G.; Littlewood, T.; Macdougall, I. Recombinant erythropoietin in clinical practice. Postgrad. Med. J. 2003, 79, 367–376. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Čaval, T.; Tian, W.; Yang, Z.; Clausen, H.; Heck, A.J.R. Direct quality control of glycoengineered erythropoietin variants. Nat. Commun. 2018, 9, 3342. [Google Scholar] [CrossRef] [PubMed]
- Savaryn, J.P.; Toby, T.K.; Catherman, A.D.; Fellers, R.T.; LeDuc, R.D.; Thomas, P.M.; Friedewald, J.J.; Salomon, D.R.; Abecassis, M.M.; Kelleher, N.L. Comparative top down proteomics of peripheral blood mononuclear cells from kidney transplant recipients with normal kidney biopsies or acute rejection. Proteomics 2016, 16, 2048–2058. [Google Scholar] [CrossRef] [PubMed]
- Ahn, C.-S.; Han, X.; Bae, Y.-A.; Ma, X.; Kim, J.-T.; Cai, H.; Yang, H.-J.; Kang, I.; Wang, H.; Kong, Y. Alteration of immunoproteome profile of Echinococcus granulosus hydatid fluid with progression of cystic echinococcosis. Parasites Vectors 2015, 8, 10. [Google Scholar] [CrossRef]
- Li, B.; Wang, X.; Yang, C.; Wen, S.; Li, J.; Li, N.; Long, Y.; Mu, Y.; Liu, Q.; Li, X.; et al. Human growth hormone proteoform pattern changes in pituitary adenomas: Potential biomarkers for 3P medical approaches. EPMA J. 2021, 12, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Garranzo-Asensio, M.; Guzmán-Aránguez, A.; Povés, C.; Fernández-Aceñero, M.J.; Montero-Calle, A.; Ceron, M.A.; Fernandez-Diez, S.; Rodríguez, N.; de Cedrón, M.; Ramírez de Molina, A.; et al. The specific seroreactivity to ∆Np73 isoforms shows higher diagnostic ability in colorectal cancer patients than the canonical p73 protein. Sci. Rep. 2019, 9, 13547. [Google Scholar] [CrossRef] [PubMed]
- Banfi, C.; Brioschi, M.; Mapelli, M.; Gianazza, E.; Mallia, A.; Zoanni, B.; Salvioni, E.; Gugliando, P.; Capra, N.; Veglia, F.; et al. Immature circulating SP-B, bound to HDL, represents an early sign of smoke-induced pathophysiological alterations. Biomolecules 2021, 11, 551. [Google Scholar] [CrossRef]
- Tucholski, T.; Cai, W.; Gregorich, Z.R.; Bayne, E.F.; Mitchell, S.D.; Mcllwain, S.J.; de Lange, W.J.; Wrobbel, M.; Karp, H.; Hite, Z.; et al. Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics. Proc. Natl. Acad. Sci. USA 2020, 117, 24691–24700. [Google Scholar] [CrossRef]
- Turner, A.; Schiel, J.E. Qualification of NISTmAb charge heterogeneity control assays. Anal. Bioanal. Chem. 2018, 410, 2079–2093. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Nishikimi, T.; Kuwahara, K. Atrial and brain natriuretic peptides: Hormones secreted from the heart. Peptides 2019, 111, 18–25. [Google Scholar] [CrossRef]
- Hammerer-Lercher, A.; Halfinger, B.; Sarg, B.; Mair, J.; Puschendorf, B.; Griesmacher, A.; Guzman, N.A.; Lindner, H.H. Analysis of circulating forms of proBNP and NT-proBNP in patients with severe heart failure. Clin. Chem. 2008, 54, 858–865. [Google Scholar] [CrossRef]
- Bianchi, L.; Sframeli, M.; Vantaggiato, L.; Vita, G.L.; Ciranni, A.; Polito, F.; Oteri, R.; Gitto, E.; Di Giuseppe, F.; Angelucci, S.; et al. Nursinersen modulates proteomics profiles of cerebrospinal fluid in spinal muscular atrophy type 1 patients. Int. J. Mol. Sci. 2021, 22, 4329. [Google Scholar] [CrossRef] [PubMed]
- Sproston, N.R.; Ashworth, J.J. Role of C-reactive protein at sites of inflammation and infection. Front. Immunol. 2018, 9, 754. [Google Scholar] [CrossRef]
- Mir, H.A.; Ali, R.; Mushtaq, U.; Khanday, F.A. Structure-functional implications of longevity protein p66Shc in health and disease. Ageing Res. Rev. 2020, 63, 101139. [Google Scholar] [CrossRef]
- Jelinek, H.F.; Helf, C.; Khalaf, K. Human SHC-transforming protein 1 and its isoform p66shc: A novel marker for prediabetes. J. Diabetes Investig. 2021. [Google Scholar] [CrossRef]
- Cutroneo, K.R.; Guzman, N.A.; Liebelt, A.G. Elevation of peptidylproline hydroxylase activity and collagen synthesis in spontaneous primary mammary cancers of inbred mice. Cancer Res. 1972, 32, 2828–2833. [Google Scholar]
- Gorres, K.L.; Raines, R.T. Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 106–124. [Google Scholar] [CrossRef]
- Guzman, N.A.; Oronsky, A.L.; Desnick, R.J.; Cutroneo, K.R.; Prockop, D.J. A new isoenzyme of prolyl 4-hydroxylase from human placenta. Purification and partial characterization. In Federation Proceedings; Federation of American Societies for Experimental Biology: St. Louis, MO, USA, 1981; Volume 40, p. 1706. [Google Scholar]
- Guzman, N.A.; Ascari, W.Q.; Cutroneo, K.R.; Desnick, R.J. Comparison between avian and human prolyl 4-hydroxylases: Studies on the holomeric enzymes and their constituent subunits. J. Cell. Biochem. 1992, 48, 172–189. [Google Scholar] [CrossRef]
- Guzman, N.A. Prolyl hydroxylase: An overview. In Prolyl Hydroxylase, Protein Disulfide Isomerase, and Other Structurally Related Proteins; Guzman, N.A., Ed.; Marcel Dekker Inc: New York, NY, USA, 1998; Chapter 1; pp. 1–64. [Google Scholar]
- Gilkes, D.M.; Chaturvedi, P.; Bajpai, S.; Wong, C.C.-L.; Wei, H.; Pitcairn, S.; Hubbi, M.E.; Wirtz, D.; Semenza, G.L. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013, 73, 3285–3296. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K. HIF-a prolyl hydroxylase inhibitors and their implications for biomedicine: A comprehensive review. Biomedicines 2021, 9, 468. [Google Scholar] [CrossRef]
- Li, M.; Wang, Q.; Zheng, Q.; Wu, L.; Zhao, B.; Wu, Y. Prognostic and diagnostic roles of prolyl 4-hydroxylase subunit alpha members in breast cancer. Biomark. Med. 2021, 15, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Gao, S.; Zhang, J.; Xu, J.; Graham, L.M.; Yang, X.; Li, C. Collagen prolyl-4-hydroxylases modify tumor progression. Acta Biochim. Biophys. Sin. 2021, 53, 805–814. [Google Scholar] [CrossRef]
- Mahan, A.E.; Tedesco, J.; Dionne, K.; Baruah, K.; Cheng, H.D.; De Jager, P.L.; Barouch, D.H.; Suscovich, T.; Ackerman, M.; Crispin, M.; et al. A method for high-throughput, sensitive analysis of IgG Fc and Fab glycosylation by capillary electrophoresis. J. Immunol. Meth. 2015, 417, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Gudelj, I.; Lauc, G.; Pezer, M. Immunoglobulin G glycosylation in aging and diseases. Cell. Immunol. 2018, 333, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Markina, Y.V.; Gerasimova, E.V.; Markin, A.M.; Glanz, V.Y.; Wu, W.-K.; Sobenin, I.A.; Orekhov, A.N. Sialylated immunoglobulins for the treatment of immune-inflammatory diseases. Int. J. Mol. Sci. 2020, 21, 5472. [Google Scholar] [CrossRef]
- Anderson, L.C.; Karch, K.R.; Ugrin, S.A.; Coradin, M.; Engish, A.M.; Sidoli, S.; Shabanowitz, J.; Garcia, B.A.; Hunt, D.F. Analyses of histone proteoforms using front-end electron transfer dissociation-enabled orbitrap instruments. Mol. Cell. Proteom. 2016, 15, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Coradin, M.; Porter, E.G.; Garcia, B.A. Accelerating the field of epigenetic histone modification through mass spectrometry-based approaches. Mole. Cell. Proteom. 2021, 20, 10006. [Google Scholar] [CrossRef]
- Martire, S.; Banaszynski, L.A. The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell. Biol. 2020, 21, 522–541. [Google Scholar] [CrossRef]
- Lindner, H.H. Analysis of histones, histones variants, and their post-translational modified forms. Electrophoresis 2008, 29, 2516–2532. [Google Scholar] [CrossRef]
- Ntai, I.; Fornelli, L.; DeHart, C.J.; Hutton, J.E.; Doubleday, P.F.; LeDuc, R.D.; van Nispen, A.J.; Fellers Whiteley, G.; Boja, E.S.; Rodriguez, H.; et al. Precise characterization of KRAS4b proteoforms in human colorectal cells and tumor reveals mutation/modification cross-talk. Proc. Natl. Acad. Sci. USA 2018, 115, 4140–4145. [Google Scholar] [CrossRef]
- Seeley, C.; Kegel-Gleason, K.B. Taming the Huntington’s disease proteome: What have we learned? J. Huntingt. Dis. 2021, 10, 239–257. [Google Scholar] [CrossRef]
- Wildburger, N.C.; Hartle, A.-S.; Schidlitzki, A.; Richter, F. Current evidence for a bidirectional loop between the lysosome and alpha-synuclein proteoforms. Front. Cell. Dev. Biol. 2020, 8, 598446. [Google Scholar] [CrossRef]
- Nocentini, A.; Cuffaro, D.; Ciccone, L.; Orlandini, E.; Nencetti, S.; Nuti, E.; Rosello, A.; Supuran, C.T. Activation of carbonic anhydrases from human brain by alcohol oxime ethers: Towards human carbonic anhydrase VII selective activators. J. Enzyme Inhib. Med. Chem. 2021, 36, 48–57. [Google Scholar] [CrossRef]
- Nedelkov, D. Mass spectrometric studies of apolipoprotein proteoforms and their role in lipid mebabolism and type 2 diabetes. Proteomes 2017, 5, 27. [Google Scholar] [CrossRef]
- Lorentzian, A.; Uzozie, A.; Lange, P.F. Origins and clinical relevance of proteoforms in pediatric malignancies. Expert Rev. Proteom. 2019, 16, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Mnatsakanyan, R.; Shema, G.; Basik, M.; Batist, G.; Borchers, C.H.; Sickmann, A.; Zahedi, R.P. Detecting post-translational modification signatures as potential biomarkers in clinical mass spectrometry. Expert Rev. Proteom. 2018, 15, 515–535. [Google Scholar] [CrossRef] [PubMed]
- Toby, T.K.; Fornelly, L.; Kelleher, N.L. Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, Y.; Lin, S.; Deng, W.; Peng, D.; Cui, Q.; Xue, Y. PTMD: A database of human disease-associated post-translational modifications. Genom. Proteom. Bioinf. 2018, 16, 244–251. [Google Scholar] [CrossRef]
- Weng, S.S.H.; Demir, F.; Ergin, E.K.; Dirnberger, S.; Uzozie, A.; Tuscher, D.; Nierves, L.; Tsui, J.; Huesgen, P.F.; Lange, P.F. Sensitive determination of proteolytic proteoforms in limited microscale proteome samples. Mol. Cell. Proteom. 2019, 18, 2335–2347. [Google Scholar] [CrossRef]
- Eckhard, U.; Marino, G.; Butler, G.S.; Overall, C.M. Positional proteomics in the era of the human proteome project on the doorstep of precision medicine. Biochimie 2016, 122, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, R.E.; Libert, C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat. Rev. Drug Discov. 2014, 13, 904–927. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [PubMed]
- Gioia, M.; Ciaccio, C.; Calligari, P.; De Simone, G.; Sbardella, D.; Tundo, G.; Fasciglione, G.F.; Di Masi, A.; Di Pierro, D.; Bocedi, A.; et al. Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem. Pharmacol. 2020, 182, 114225. [Google Scholar] [CrossRef] [PubMed]
- Seth, S.; Batra, J.; Srinivasan, S. COVID-19: Targeting proteases in viral invasion and host immune response. Front. Mol. Biosci. 2020, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Nair, S. Proteases in cardiometabolic diseases: Pathophysiology, molecular mechanisms and clinical applications. Biochem. Biophys. Acta 2015, 185, 195–208. [Google Scholar] [CrossRef]
- Peterson, J.T.; Hallak, H.; Johnson, L.; Li, H.; O’Brien, P.M.; Sliskovic, D.R.; Bocan, T.M.; Coker, M.L.; Etoh, T.; Spinale, F.G. Matrix metalloproteinase inhibition attenuates left ventricular remodeling and dysfunction in a rat model of progressive heart failure. Circulation 2001, 103, 2303–2309. [Google Scholar] [CrossRef]
- Miksztowicz, V.; Siseles, N.; Fernandez-Machulsky, N.; Schreier, L.; Berg, G. Increase in MMP-2 activity in overweight and obese women is associated with menopausal status. Climateric 2012, 15, 602–606. [Google Scholar] [CrossRef]
- Glowinska-Olszewska, B.; Urban, M. Elevated matrix metalloproteinase 9 and tissue inhibitor of metalloproteinase 1 in obese children and adolescents. Metabolism 2007, 56, 799–805. [Google Scholar] [CrossRef]
- Wild, D.G. (Ed.) The Immunoassay Handbook. In Theory and Applications of Ligand Binding, ELISA and Related Techniques, 4th ed.; Elsevier Science: Amsterdam, The Netherlands, 2013; ISBN 9780080970370. [Google Scholar]
- Saah, A.J.; Hover, D.R. “Sensitivity” and “specificity” reconsidered: The meaning of these terms in analytical and diagnostic settings. Ann. Intern. Med. 1997, 126, 91–94. [Google Scholar] [CrossRef]
- Li, Y.; Cassone, V.M. A simple, specific high-throughput enzyme-linked immunosorbent assay (ELISA) for quantitative determination of melatonin in cell culture medium. Int. Immunopharmacol. 2015, 28, 230–234. [Google Scholar] [CrossRef]
- Rao, T.N. Validation of Analytical Methods. In Calibration and Validation of Analytical Methods—A Sampling of Current Approaches, Chapter 7; IntechOpen Limited: London, UK, 2018. [Google Scholar] [CrossRef]
- Kwok, J.; Yang, M.; Wu, J.K.; Carter, C.J.; Jackson, S. High false positive rate of an ELISA screen for the detection of an anti-factor VIII antibodies in congenital hemophilia A. Blood 2013, 122, 3586. [Google Scholar] [CrossRef]
- Wai, C.T.; Tambyah, P.A. False-positive HIV-1 ELISA in patients with hepatitis B. Am. J. Med. 2002, 112, 737. [Google Scholar] [CrossRef]
- Kharlamova, N.; Dunn, N.; Bedri, S.K.; Jerling, S.; Almgren, M.; Faustini, F.; Gunnarsson, I.; Rönnelid, J.; Pullerits, R.; Gjertsson, I.; et al. False positive results in SARS-CoV-2 serological tests for samples from patients with chronic inflammatory diseases. Front. Immunol. 2021, 12, 666114. [Google Scholar] [CrossRef] [PubMed]
- Guzman, N.A.; Guzman, D.E. An emerging micro-scale immune-analytical diagnostic tool to see the unseen. Holding promise for precision medicine and P4 medicine. J. Chromatogr. B 2016, 1021, 14–29. [Google Scholar] [CrossRef]
- Jarossay, A.; Hadzhieva, M.; Kaveri, S.V.; Lacroix-Desmazes, S.; Dimitrov, J.D. Natural and induced antibody polyreactivity. Anti-Cancer Agents Med. Chem. 2015, 15, 1230–1241. [Google Scholar] [CrossRef]
- Hadzhieva, M.; Vassilev, T.; Bayry, J.; Kaveri, S.; Lacroix-Desmazes, S.; Dimitrov, J.D. Relationship between natural and heme-mediated antibody polyreactivity. Biochem. Biophys. Res. Commun. 2016, 472, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Ruiz, A.; Dimitrov, J.D. How can polyreactive antibodies conquer rapidly evolving viruses? Trends Immunol. 2021, 42, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Augustyniak, D.; Majkowska-Skrobek, G.; Roszkowiak, J.; Dorotkiewics-Jach, A. Defensive and offensive cross-reactive antibodies elicited by pathogens: The good, the bad and the ugly. Curr. Med. Chem. 2017, 24, 4002–4437. [Google Scholar] [CrossRef]
- Jain, D.; Salunke, D.M. Antibody specificity and promiscuity. Biochem. J. 2019, 476, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Thaper, D.; Prabha, V. Molecular mimicry: An explanation for autoimmune diseases and infertility. Scand. J. Immunol. 2018, 88, e12697. [Google Scholar] [CrossRef]
- Madhavaram, H.; Patel, T.; Kyle, C. Kavain interference with amphetamine immunoassay. J. Anal. Toxicol. 2020, bkaa178. [Google Scholar] [CrossRef]
- Tapryal, S.; Gaur, V.; Kaur, K.J.; Salunke, D.M. Structural evaluation of a mimicry-recognizing paratope: Plasticity in antigen-antibody interactions manifests in molecular mimicry. J. Immunol. 2013, 191, 456–463. [Google Scholar] [CrossRef]
- Obando-Pereda. Can molecular mimicry expain the cytokine storm of SARS-CoV-2?: An in silico approach. J. Med. Virol. 2021, 93, 5350–5357. [Google Scholar] [CrossRef]
- Bergman, D.; Larsson, A.; Hansson-Hamlin, H.; Ahlén, E.; Holst, B.S. Characterization of canine anti-mouse antibodies highlights that multiple strategies are need to combat immunoassay interference. Sci. Rep. 2019, 9, 14521. [Google Scholar] [CrossRef]
- Jara-Aguirre, J.C.; Baumann, N.A.; Block, D.R.; Algeciras-Schimnich, A. Human chorionic gonadotrophin suspected heterophile interference investigations in immunoassays: A recommended approach. Clin. Chem. Lab. Med. 2019, 57, 1192–1196. [Google Scholar] [CrossRef]
- Emerson, J.F.; Ngo, G.; Emerson, S.S. Screening for interference in immunoassays. Clin. Chem. 2003, 49, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Selby, C. Interference in immunoassay. Ann. Clin. Biochem. 1999, 36, 704–721. [Google Scholar] [CrossRef]
- Tate, J.; Ward, G. Interferences in immunoassays. Clin. Biochem. Rev. 2004, 25, 105–120. [Google Scholar] [PubMed]
- Sturgeon, C.M.; Viljoen, A. Analytical error and interference in immunoassay: Minimizing risk. Ann. Clin. Biochem. 2011, 48, 418–432. [Google Scholar] [CrossRef] [PubMed]
- Rotmensch, S.; Cole, L.A. False diagnosis and needless therapy of presumed malignant disease in women with false-positive human chorionic gonadotropin concentrations. Lancet 2000, 355, 712–715. [Google Scholar] [CrossRef]
- Emerson, J.F.; Lai, K.K.Y. Endogenous antibody interferences in immunoassays. Lab. Med. 2013, 44, 69–73. [Google Scholar] [CrossRef]
- Halvorsen, T.G.; Reubsaet, L. Determination of very low abundance diagnostic proteins in serum using immunocapture LC-MS/MS. Spectroscopy 2017, 15, 16–22. [Google Scholar]
- Vehus, T. Performing quantitative determination of low-abundant proteins by targeted mass spectrometry liquid chromatography. Intechopen 2017, 3, 79–94. [Google Scholar] [CrossRef]
- Harney, D.J.; Hutchison, A.T.; Su, Z.; Hatchwell, L.; Heilbronn, L.K.; Hocking, S.; James, D.E.; Larance, M. Small-protein enrichment assay enables the rapid, unbiased analysis of over 100 low abundance factors from human plasma. Mol. Cell. Proteom. 2019, 18, 1899–1915. [Google Scholar] [CrossRef]
- Voeten, R.L.C.; Ventouri, I.K.; Haselberg, R.; Somsen, G.W. Capillary electrophoresis: Trends and recent advances. Anal. Chem. 2018, 90, 1464–1481. [Google Scholar] [CrossRef]
- Sastre-Toraño, J.; Ramautar, R.; de Jong, G. Advances in capillary electrophoresis for life sciences. J. Chromatogr. B 2019, 1118, 116–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Adeoye, A.; Ogunkunle, E.O.; Wei, I.-A.; Filla, R.T.; Roper, M.G. Affinity capillary electrophoresis: A critical review of the literature from 2018 to 2020. Anal. Chem. 2021, 93, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Dortez, S.; Sierra, T.; Crevillén, A.G.; Escarpa, A. CE/microchip electrophoresis of carbohydrates and glycoconjugates with electrochemical detection. In Carbohydrate Analysis by Modern Liquid Phase Separation, Techniques, 2nd ed.; El Rassi, Z., Ed.; Elsevier: Amsterdam, The Netherlands, 2021; Chapter 12; pp. 563–594. ISBN 9780128214473. [Google Scholar]
- Vitorino, R.; Guedes, S.; Pinto da Costa, J.; Kasicka, V. Microfluidics for peptidomics, proteomics, and cell analysis. Nanomaterials 2021, 11, 1118. [Google Scholar] [CrossRef] [PubMed]
- El Deeb, S.; Fonseca-Silva, C.; Nascimento-Junior, C.; Hanafi, R.S.; Borges, K.B. Chiral capillary electrokinetic chromatography: Principle and applications, detection and identification, design of experiment, and exploration of chiral recognition using molecular modeling. Molecules 2021, 26, 2841. [Google Scholar] [CrossRef]
- Peluso, P.; Chankvetadze, B. Native and substituted cyclodextrins as chiral selectors for capillary electrophoresis enantioseparations: Structures, features, application, and molecular modeling. Electrophoresis 2021. [Google Scholar] [CrossRef]
- Guzman, N.A. (Ed.) Capillary Electrophoresis Technology; Marcel Dekker Inc.: New York, NY, USA, 1993. [Google Scholar]
- Pan, J.-Z.; Fang, P.; Fang, X.-X.; Hu, T.-T.; Fang, J.; Fang, Q. A low-cost palmtop high-speed capillary electrophoresis bioanalyzer with laser induced fluorescence detection. Sci. Rep. 2018, 8, 1791. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Dorsey, G.; Hubbard, A.E.; Rosenthal, P.J.; Greenhouse, B. Gel versus capillary electrophoresis genotyping for categorizing treatment outcomes in two anti-malarial trials in Uganda. Malar. J. 2010, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lu, J.J.; Liu, S. Protein separation by capillary electrophoresis: A review. Anal. Chim. Acta 2012, 709, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.B.; Quirino, J.P. Chiral selectors in capillary electrophoresis: Trends during 2017. Molecules 2019, 24, 1135. [Google Scholar] [CrossRef] [PubMed]
- Kalíková, K.; Riesová, M.; Tesařová, E. Recent chiral selectors for separation in HPLC and CE. Open Chem. 2012, 10, 450–471. [Google Scholar] [CrossRef]
- Francotte, E.R. Polysaccharide derivatives as unique chiral selectors for enantioselective chromatography. Chimia 2017, 71, 430–450. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Du, Y.; Hu, X.; Luo, L.; Guo, X.; Guo, X.; Yu, J. Carboxymethyl β-cyclodextrin as chiral selector in capillary electrophoresis: Enantioseparation of 16 basic chiral drugs and its chiral recognition mechanism associated with drugs’ structural features. Biomed. Chromatogr. 2017, 31, e3991. [Google Scholar] [CrossRef] [PubMed]
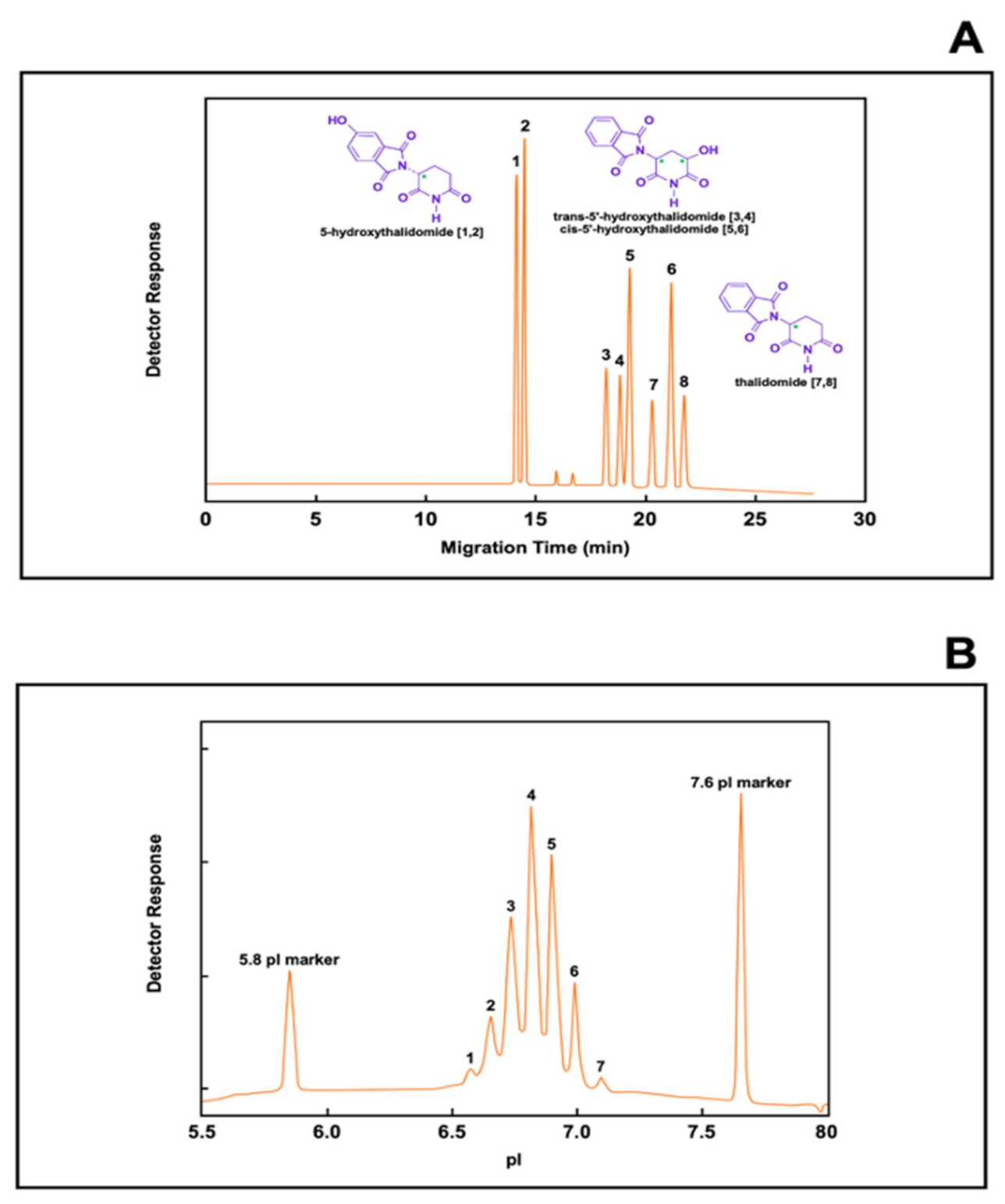
- Blaschke, G.; Meryring, M.; Muhlenbrock, C.; Chankvetadze, B. Recent results of biotransformation of drugs: Investigation of the in vitro biotransformation of thalidomide using a dual cyclodextrin system in capillary electrophoresis. Il Farmaco 2020, 57, 551–554. [Google Scholar] [CrossRef]
- Suba, D.; Urbányl, Z.; Salgó, A. Capillary isoelectric focusing method development and validation for investigation of recombinant therapeutic monoclonal antibody. J. Pharm. Biomed. Anal. 2015, 114, 53–61. [Google Scholar] [CrossRef]
- Xu, T.; Sun, L. A mini review on capillary isoelectric focusing-mass spectrometry for top-down proteomics. Front. Chem. 2021, 9, 651757. [Google Scholar] [CrossRef]
- Costello, M.A.; Woititz, C.; DeFeo, J.; Stremlo, D.; Wen, L.-F.L.; Palling, D.J.; Iqbal, K.; Guzman, N.A. Characterization of humanized anti-Tac monoclonal antibody by traditional separation techniques and capillary electrophoresis. J. Liq. Chromatogr. Rel. Technol. 1992, 15, 1081–1097. [Google Scholar] [CrossRef]
- Dadouch, M.; Ladner, Y.; Perrin, C. Analysis of monoclonal antibodies by capillary electrophoresis: Sample preparation, separation, and detection. Separations 2021, 8, 4. [Google Scholar] [CrossRef]
- Salas-Solano, O.; Kennel, B.; Park, S.S.; Roby, K.; Sosic, Z.; Boumajny, B.; Free, S.; Reed-Bogan, A.; Michels, D.; McElroy, W.; et al. Robustness of iCIEF methodology for the analysis of monoclonal antibodies: An interlaboratory study. J. Sep. Sci. 2012, 35, 3124–3129. [Google Scholar] [CrossRef]
- Osbourn, D.M.; Weiss, D.J.; Lunte, C.E. On-line preconcentration methods for capillary electrophoresis. Electrophoresis 2000, 21, 2768–2779. [Google Scholar] [CrossRef]
- Glatz, Z. On-capillary derivatization as an approach to enhancing sensitivity in capillary electrophoresis. Electrophoresis 2015, 36, 744–763. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, F.; Liu, Y. Recent advances in enhancing the sensitivity and resolution of capillary electrophoresis. J. Chromatogr. Sci. 2013, 51, 666–683. [Google Scholar] [CrossRef]
- Kawai, T. Recent advances in trace bioanalysis by capillary electrophoresis. Anal. Sci. 2021, 37, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.H.; Rakestraw, D.J.; Schoeniger, J.S.; Lopez-Avila, V.; Van Emon, J. Selective trace enrichment by immunoaffinity capillary electrochromatography on-line with capillary electrophoresis–laser induced fluorescence. Electrophoresis 1999, 20, 57–66. [Google Scholar] [CrossRef]
- Figeys, D.; Zhang, Y.; Aebersold, R. Optimization of solid phase microextraction-capillary zone electrophoresis-mass spectrometry for high sensitivity protein identification. Electrophoresis 1998, 19, 2338–2347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, L.; Chen, X. Improving sensitivity by large-volume sample stacking combined with sweeping without polarity switching by capillary electrophoresis coupled to photodiode array ultraviolet detection. Electrophoresis 2008, 29, 1556–1564. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Su, P.; Zhang, X.-X.; Chang, W.B. Enhancement of the sensitivity of a capillary electrophoresis immunoassay for estradiol with laser-induced fluorescence based on a fluorescein-labeled secondary antibody. Anal. Chem. 2001, 73, 5616–5619. [Google Scholar] [CrossRef]
- Marie, A.L.; Ray, S.; Lu, S.; Jones, J.; Ghiran, I.; Ivanov, A.R. High-sensitivity glycan profiling of blood-derived immunoglobulin G, plasma, and extracellular vesicles isolates with capillary zone electrophoresis-mass spectrometry. Anal. Chem. 2021, 93, 1991–2002. [Google Scholar] [CrossRef]
- Guzman, N.A.; Guzman, D.E. A two-dimensional affinity capture and separation mini-platform for the isolation, enrichment, and quantification of biomarkers and its potential use for liquid biopsy. Biomedicines 2020, 8, 255. [Google Scholar] [CrossRef]
- Guzman, N.A.; Guzman, D.E. From a central laboratory to the beside: A point-of-care instrument to monitoring wellness and disease using two-dimensional immunoaffinity capillary electrophoresis technology. Archiv. Biomed. Res. 2018, 1. [Google Scholar]
- Gao, Z.; Zhong, W. Recent (2018–2020) development in capillary electrophoresis. Anal. Bioanal. Chem. 2021, 1–16. [Google Scholar] [CrossRef]
- Ta, H.Y.; Collin, F.; Perquis, L.; Poinsot, V.; Ong-Meang, V.; Couderc, F. Twenty years of amino acid determination using capillary electrophoresis: A review. Anal. Chim. Acta 2021, 1174, 338233. [Google Scholar] [CrossRef] [PubMed]
- John, A.S.; Sidek, M.M.; Thang, L.Y.; Sami, S.; Tey, H.Y.; See, H.H. Online sample preconcentration techniques in nonaqueous capillary and microchip electrophoresis. J. Chromatogr. A 2021, 1638, 461868. [Google Scholar] [CrossRef] [PubMed]
- Kristoff, C.J.; Bwanali, L.; Veltri, L.M.; Gautam, G.P.; Rutto, P.K.; Newton, E.O.; Holland, L.A. Challenging bioanalyses with capillary electrophoresis. Anal. Chem. 2020, 92, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Ouahabi, O.E.; Salim, H.; Pero-Gascon, R.; Benavente, F. A simple method for the analysis of extracellular vesicles enriched for exosomes from human serum by capillary electrophoresis with ultraviolet diode array detection. J. Chromatogr. A 2021, 1635, 461752. [Google Scholar] [CrossRef] [PubMed]
- Oedit, A.; Hankemeier, T.; Lindenburg, P.W. On-line coupling of two-phase microelectroextraction to capillary electrophoresis-mass spectrometry for metabolomics analyses. Microchem. J. 2021, 162, 105741. [Google Scholar] [CrossRef]
- Lim, R.R.X.; Fung, F.M.; Feng, H.-T.; Li, S.F.Y. Analysis of lipopolysaccharides by coupling microscale solid-phase extraction with capillary electrophoresis-laser induced fluorescence. Microchem. J. 2021, 161, 105771. [Google Scholar] [CrossRef]
- Zhang, Z.; Park, J.; Barrett, H.; Dooley, S.; Davies, C.; Verhagen, M.C. Capillary electrophoresis-sodium dodecyl sulfate with laser-induced fluorescence detection as a highly sensitive and quality control-friendly method for monitoring adeno-associated virus capsid protein purity. Hum. Gene Ther. 2021, 32, 628–637. [Google Scholar] [CrossRef]
- Claude, B.; Nehmé, R.; Morin, P. Analysis of urinary neurotransmitters by capillary electrophoresis: Sensitivity enhancement using field-amplified sample injection and molecular imprinted polymer solid phase extraction. Anal. Chim. Acta 2011, 699, 242–248. [Google Scholar] [CrossRef]
- Simonet, B.M.; Ríos Valcárcel, M. Enhancing sensitivity in capillary electrophoresis. Trends Anal. Chem. 2003, 22, 605–614. [Google Scholar] [CrossRef]
- Almeda, S.; Arce, L.; Valcárcel, M. The more and less common approaches to enhancing sensitivity in capillary electrophoresis. Curr. Anal. Chem. 2010, 6, 126–143. [Google Scholar] [CrossRef]
- Drevinskas, T.; Telksnys, L.; Maruska, A.; Gorbatsova, J.; Kaljurand, M. Capillary electrophoresis sensitivity enhancement based on adaptive moving average method. Anal. Chem. 2018, 90, 6773–6780. [Google Scholar] [CrossRef] [PubMed]
- Galievsky, V.A.; Stasheushi, A.S.; Krylov, S.K. “Getting the best sensitivity from on-capillary fluorescence detection in capillary electrophoresis”–A tutorial. Anal. Chim. Acta 2016, 935, 58–81. [Google Scholar] [CrossRef]
- Wang, Y.; Fonslow, B.R.; Wong, C.C.; Nakorchevsky, A.; Yates, J.R. III. Improving the comprehensiveness and sensitivity of sheathless CE-MS/MS for proteomic analysis. Anal. Chem. 2012, 84, 8505–8513. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; McCool, E.N.; Yang, Z.; Shen, X.; Lubeckyj, R.A.; Xu, T.; Wang, Q.; Sun, L. Recent advances (2019–2021) of capillary electrophoresis-mass spectrometry for multilevel proteomics. Mass Spectrom. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Heegaard, N.H.H.; Nilsson, S.; Guzman, N.A. Affinity capillary electrophoresis: Important applications areas and some recent developments. J. Chromatogr. B 1998, 715, 29–54. [Google Scholar] [CrossRef]
- Olabi, M.; Stein, M.; Wätzig, H. Affinity capillary electrophoresis for studying interactions in life sciences. Methods 2018, 146, 76–92. [Google Scholar] [CrossRef]
- Nevídalová, H.; Michalcová, L.; Glatz, Z. Capillary electrophoresis-based approaches for the study of affinity interactions combined with various sensitive and nontraditional detection techniques. Electrophoresis 2019, 40, 625–642. [Google Scholar] [CrossRef]
- Nevídalová, H.; Michalcová, L.; Glatz, Z. Capillary electrophoresis-based immunoassay and aptamer assay: A review. Electrophoresis 2020, 41, 414–433. [Google Scholar] [CrossRef]
- Guzman, N.A. Disease Detection System and Method. U.S. Patent 10,408,789, 10 September 2019. [Google Scholar]
- Guzman, N.A.; Guzman, D.E. A home-based portable instrument to monitor wellness and disease. Atlas of Science–Another View on Science. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015. Available online: https://atlasofscience.org/a-home-based-portable-instrument-to-monitor-wellness-and-disease/ (accessed on 21 March 2016).
- Pero-Gascon, R.; Pont, L.; Sanz-Nebot, V.; Benavente, F. On-line immunoaffinity solid-phase extraction capillary electrophoresis-mass spectrometry for the analysis of transthyretin. Methods Mol. Biol. 2019, 1972, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Puerta, A.; Martin-Alvarez, P.J.; Ongay, S.; Diez-Masa, J.C.; de Frutos, M. Immunoaffinity, capillary electrophoresis, and statistics for studying intact alpha 1-acid glycoprotein isoforms as an atherothrombosis biomarkers. Methods Mol. Biol. 2013, 919, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.-H.; Zhang, L.W.; Zhang, X.-X. Simultaneous determination of nandrolone, testosterone, and methyltestosterone by multi-immunoaffinity column and capillary electrophoresis. Electrophoresis 2008, 29, 3398–3405. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Medina, R.; Farina-Gomez, N.; Diez-Masa, J.C.; de Frutos, M. Immunoaffinity chromatographic isolation of prostate-specific antigen from seminal plasma for capillary electrophoresis analysis of its isoforms. Anal. Chim. Acta 2014, 820, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Phillips, T.M.; Wellner, E.F. Analysis of inflammatory mediators in newborn dried blood spot samples by chip-based immunoaffinity capillary electrophoresis. Methods Mol. Biol. 2019, 1972, 185–198. [Google Scholar] [CrossRef]
- Phillips, T.M.; Wellner, E.F. Detection of cerebral spinal fluid associated chemokines in birth traumatized premature babies by chip-based immunoaffinity CE. Electrophoresis 2013, 34, 1530–1538. [Google Scholar] [CrossRef]
- Guzman, N.A. Method and System for Simultaneous Determination of Multiple Measurable Biomarkers during the Development of Communicable Disease. U.S. Patent Application No. 2020/16/894,316, 5 June 2020. [Google Scholar]
- Vermassen, T.; Van Praet, C.; Vanderschaeghe, D.; Maenhout, T.; Lumen, N.; Callewaert, N.; Hoebeke, P.; Van Belle, S.; Rottey, S.; Delanghe, J. Capillary electrophoresis of urinary prostate glycoproteins assists in the diagnosis of prostate cancer. Electrophoresis 2014, 35, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Kammeijer, G.S.M.; Nouta, J.; de la Rosette, J.J.M.C.H.; de Reijke, T.M.; Wuher, M. An in-depth glycosylation assay for urinary prostate-specific antigen. Anal. Chem. 2018, 90, 4414–4421. [Google Scholar] [CrossRef] [PubMed]
- Reider, B.; Gacsi, E.; Jankovics, H.; Vonderviszt, F.; Szarvas, T.; Guttman, A.; Jarvas, G. Integrated workflow for urinary prostate specific antigen N-glycosylation anlysis using sdAb partitioning and downstream capillary electrophoresis separation. Anal. Chim. Acta 2021. [Google Scholar] [CrossRef]
- Dimitrov, D.S. Therapeutic proteins. Methods Mol. Biol. 2012, 899, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Leader, B.; Baca, Q.J.; Golan, D.E. Protein therapeutics: A summary and pharmacological classification. Nat. Rev. Drug Discov. 2008, 7, 21–39. [Google Scholar] [CrossRef]
- Chennamsetty, N.; Voynov, V.; Kayser, V.; Helk, B.; Trout, B.L. Design of therapeutic proteins with enhanced stability. Proc. Natl. Acad. Sci. USA 2009, 106, 11937–11942. [Google Scholar] [CrossRef]
- Kariolis, M.S.; Wells, R.C.; Getz, J.A.; Kwan, W.; Mahon, C.S.; Tong, R. Brain delivery of therapeutic proteins using Fc fragment blood-brain barrier transport vehicle in mice and monkeys. Sci. Transl. Med. 2020, 12, eaay1359. [Google Scholar] [CrossRef]
- Schadt, S.; Hauri, S.; Lopes, F.; Edelmann, M.R.; Staack, R.F.; Villaseñor, R.; Kettenberger, H.; Roth, A.B.; Schuler, F.; Richter, W.F.; et al. Are biotransformation studies of therapeutic proteins needed? Scientific considerations and technical challenges. Drug Metabol. Dispos. 2019, 47, 1443–1456. [Google Scholar] [CrossRef]
- Scientific Advice from European Medicines Agency (EMA). Guideline on Immunogenicity Assessment of Therapeutic Proteins. 2017. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-immunogenicity-assessment-therapeutic-proteins-revision-1_en.pdf (accessed on 18 May 2017).
- Rosenberg, A.S. Immunogenicity Risk Assessment for Therapeutic Proteins: A Guide to Risk Assessment for Therapeutic Peptides and Generic Peptides. 2014. Available online: https://www.fda.gov/media/146112/download (accessed on 31 August 2014).
- Ratanji, K.D.; Derrick, J.P.; Dearman, R.J.; Kimber, I. Immunogenicity of therapeutic proteins: Influence of aggregation. J. Immunotoxicol. 2014, 11, 99–109. [Google Scholar] [CrossRef]
- Solá, R.J.; Griebenow, K. Glycosylation of therapeutic proteins: An effective strategy to optimize efficacy. BioDrugs 2010, 24, 9–21. [Google Scholar] [CrossRef]
- Roseman, D.S.; Weinberger, R. Quantitative capillary zone electrophoresis method for the precise determination of charge differences arising from the manufacture of heparan-N-sulfatase. J. Pharm. Biomed. Anal. 2013, 85, 67–73. [Google Scholar] [CrossRef]
- Papathanasiou, M.M.; Kontoravdi, C. Engineering challenges in therapeutic protein product and process design. Curr. Opin. Chem. Engin. 2020, 27, 81–88. [Google Scholar] [CrossRef]
- Morrow, T.; Felcone, L.H. Defining the difference: What makes biologics unique. Biotechnol. Health 2004, 1, 24–29. [Google Scholar]
- Suresh, S.; Rajvanshi, P.K.; Noguchi, C.T. The many facets of erythropoietin physiologic and metabolic response. Front. Physiol. 2020, 10, 1534. [Google Scholar] [CrossRef]
- Peng, B.; Kong, G.; Yang, C.; Ming, Y. Erythropoietin and its derivatives: From tissue protection to immune regulation. Cell Death Dis. 2020, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Debeljak, N.; Solár, P.; Sytkowski, A.J. Erythropoietin and cancer: The unintended consequences of anemia correction. Front. Immunol. 2014, 5, 563. [Google Scholar] [CrossRef] [PubMed]
- Nagano, M.; Stübiger, G.; Marchetti, M.; Gmeiner, G.; Allmaier, G.; Reichel, C. Detection of isoforms of recombinant human erythropoietin by various plant lectins after isoelectric focusing. Electrophoresis 2005, 26, 1633–1645. [Google Scholar] [CrossRef] [PubMed]
- Bristow, A.; Charton, E. Assessment of the suitability of a capillary electrophoresis method for determining isoform distribution of erythropoietin. Pharmeuropa 1999, 11, 290–300. [Google Scholar]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Alocci, D.; Ghraichy, M.; Barletta, E.; Gastaldello, A.; Mariethoz, J.; Lisacek, F. Understanding the glycome: An interactive view of glycosylation from glycocompositions to glycoepitopes. Glycobiology 2018, 28, 349–362. [Google Scholar] [CrossRef]
- Lauc, G.; Pezer, M.; Rudan, I.; Campbell, H. Mechanisms of disease: The human N-glycome. Biochim. Biophys. Acta 2016, 1860, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.H.; Johnson, A.G.; George, B.M.; Majzoub, K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021, 184, 3109–3124. [Google Scholar] [CrossRef] [PubMed]
- Franc, V.; Zhu, J.; Heck, A.J.R. Comprehensive proteoform characterization of plasma complement component C8abj by hybrid mass spectrometry approaches. J. Am. Soc. Mass Spectrom. 2018, 29, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Li, H.; Guan, F.; Li, X. Multiple roles of glycans in hematological malignancies. Front. Oncol. 2018, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- Meany, D.L.; Chan, D.W. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin. Proteom. 2011, 8, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Poiroux, G.; Barre, A.; Rougé, P.; Benoist, H. Targeting glycosylation aberrations to improve the efficiency of cancer phototherapy. Curr. Cancer Drug Targ. 2019, 19, 349–359. [Google Scholar] [CrossRef]
- Tuccillo, F.M.; de Laurentiis, A.; Palmieri, C.; Fiume, G.; Bonelli, P.; Borrelli, A.; Tassone, P.; Scala, I.; Buonaguro, F.M.; Quinto, I.; et al. Aberrant glycosylation as biomarker for cancer: Focus on CD43. BioMed Res. Int. 2014, 2014, 742831. [Google Scholar] [CrossRef]
- Ip, C.K.M.; Yin, J.; Ng, P.K.S.; Lin, S.-Y.; Mills, G. Genomic-glycosylation aberrations in tumor initiation, progression and management. AIMS Med. Sci. 2016, 3, 386–416. [Google Scholar] [CrossRef]
- Venkitachalam, S.; Revoredo, L.; Varadan, V.; Fecteau, R.E.; Ravi, L.; Lutterbaugh, J.; Markowitz, S.D.; Willis, J.E.; Gerken, T.A.; Guda, K. Biochemical and functional characterization of glycosylation-associated mutational landscapes in colon cancer. Sci. Rep. 2016, 6, 23642. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.W.; Qiu, Y.; Ligeralde, A.; Raybon, A.Y.; Yoo, S.Y.; Coombes, K.R.; Qutub, A.A.; Kornblau, S.M. A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia. Nat. Biomed. Enginer. 2019, 3, 889–901. [Google Scholar] [CrossRef]
- Gratacós-Mulleras, A.; Duran, A.; Shehni, A.A.; Ferrer-Batallé, M.; Ramírez, M.; Comet, J.; de Llorens, R.; Saldova, R.; Llop, E.; Peracaula, R. Characterisation of the main PSA glycoforms in aggressive prostate cancer. Sci. Rep. 2020, 10, 18974. [Google Scholar] [CrossRef]
- Saldova, R.; Fan, Y.; Fitzpatrick, J.M.; Watson, R.W.G.; Rudd, P.M. Core fucosylation and α2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology 2011, 21, 195–205. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, W.; Hu, Y.; Höti, N.; Liu, Y.; Shah, P.; Sun, S.; Clark, D.; Thomas, S.; Zhang, H. Site-specific fucosylation analysis identifying glycoproteins associated with aggressive prostate cancer cell lines using tandem affinity enrichments of intact glycopeptides followed by mass spectrometry. Anal. Chem. 2017, 89, 7623–7630. [Google Scholar] [CrossRef]
- Besarab, A.; Drueke, T.B. The problem with transferrin saturation as a key indicator of iron ‘sufficient’ in chronic kidney disease. Nephrol. Dial. Transplant. 2021, 36, 1377–1383. [Google Scholar] [CrossRef]
- Formanowicz, D.; Formanowicz, P. Transferrin changes in haemodialised patients. Int. Urol. Nephrol. 2012, 44, 907–919. [Google Scholar] [CrossRef]
- Vogt, A.-C.; Arsiwala, T.; Mohsen, M.; Manolova, V.; Bachmann, M.F. On iron metabolism and its regulation. Int. J. Mol. Sci. 2021, 22, 4591. [Google Scholar] [CrossRef]
- Bartnikas, T.B. Known and potential roles of transferrin in iron biology. Biometals 2012, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Arndt, T. Carbohydrate-deficient transferrin as a marker of chronic alcohol abuse: A critical review of preanalysis, analysis, and interpretation. Clin. Chem. 2001, 47, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Zühlsdorf, A.; Said, M.; Seger, C.; Park, J.H.; Reunert, J.; Rust, S.; Wada, Y.; Grüneber, M.; DuChesne, I.; Marquardt, T. It is not always alcohol abuse-A transferrin variant impairing the CTC test. Alcohol Alcohol. 2016, 51, 148–153. [Google Scholar] [CrossRef][Green Version]
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Medina-Casanellas, S.; Benavente, F.; Giménez, E.; Barbosa, J.; Sanz-Nebot, V. On-line immunoaffinity solid phase extraction capillary spectrometry for the analysis of large biomolecules: A preliminary report. Electrophoresis 2014, 35, 2130–2136. [Google Scholar] [CrossRef]
- Barrabés, S.; Farina-Gomez, N.; Llop, E.; Puerta, A.; Diez-Masa, J.C.; Perry, A.; de Llorens, R.; de Frutos, M.; Peracaula, R. Comparative analysis of prostate-specific antigen by two-dimensional gel electrophoresis and capillary electrophoresis. Electrophoresis 2017, 38, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Horiba Scientific-Capillary Electrophoresis Application Note CEAN02-Protein Analysis-Analysis of Transferrin Glycoforms Using EVA and GST Algorithms. Available online: https://www.horiba.com/fileadmin/uploads/Scientific/Documents/Capillary_Electrophoresis/ce02-app-note.pdf (accessed on 1 January 2021).
- Nuevo-Ordonez, Y.; Anton, R.F.; Davis, W.C. Quantification of total serum transferrin and transferrin sialoforms in human serum; an alternative method for the determination of carbohydrate-deficient transferrin in clinical samples. Anal. Methods 2014, 6, 3967. [Google Scholar] [CrossRef]
- Zühlsdorf, A.; Park, J.H.; Wada, Y.; Rust, S.; Reunert, J.; DuChesne, I.; Grüneberg, M.; Marquardt, T. Transferrin variants: Pitfalls in the diagnosis of congenital disorders of glycosylation. Clin. Biochem. 2015, 48, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Gruszewska, E.; Cylwik, B.; Gudowska, M.; Kedra, B.; Szmitkowski, M.; Chrostek, L. Changes in transferrin isoforms in pancreatic cancer. Ann. Clin. Lab. Sci. 2016, 46, 286–290. [Google Scholar]
- Guzman, N.A. Consecutive protein digestion and peptide derivatization employing an on-line analyte concentrator to map proteins using capillary electrophoresis. In CRC Series in Analytical Biotechnology; Righetti, P.G., Ed.; CRC Press: Boca Raton, FL, USA, 1996; Chapter 4; pp. 101–121. [Google Scholar]
- Guzman, N.A.; Blanc, T.; Phillips, T.M. Immunoaffinity capillary electrophoresis as a powerful strategy for the quantification of low-abundance biomarkers, drugs, and metabolites in biological matrices. Electrophoresis 2008, 29, 3259–3278. [Google Scholar] [CrossRef]
- Chang, W.W.P.; Hobson, C.; Bomberger, D.C.; Schneider, L.V. Rapid separation of protein isoforms by capillary zone electrophoresis with dynamic coatings. Electrophoresis 2005, 26, 2179–2186. [Google Scholar] [CrossRef] [PubMed]
- Carillo, S.; Jakes, C.; Bones, J. In-depth analysis of monoclonal antibodies using microfluidic capillary electrophoresis and native mass spectrometry. J. Pharm. Biomed. Anal. 2020, 185, 113218. [Google Scholar] [CrossRef]
- Stolz, A.; Hedeland, Y.; Salzer, L.; Römer, J.; Heiene, R.; Leclercq, L.; Cottet, H.; Bergquist, J.; Neusüß, C. Capillary zone electrophoresis-top-down tandem mass spectrometry for in-depth characterization of hemoglobin proteoforms in clinical veterinary samples. Anal. Chem. 2020, 92, 10531–10539. [Google Scholar] [CrossRef]
- Shen, X.; Yang, Z.; McCool, E.N.; Lubeckyj, R.A.; Chen, D.; Sun, L. Capillary electrophoresis-mass spectrometry for top-down proteomics. Trends Analyt. Chem. 2019, 120, 115644. [Google Scholar] [CrossRef]
- Chen, C.-H.; Feng, H.; Guo, R.; Li, P.; Laserna, A.K.C.; Ji, B.; Ng, B.H.; Li, S.F.Y.; Khan, S.H.; Paulus, A.; et al. Intact NIST monoclonal antibody characterization-Proteoforms, glycoforms-using CE-MS and CE-LIF. Cogent Chem. 2018, 4, 1480455. [Google Scholar] [CrossRef]
- Thomas, S.L.; Thacker, J.B.; Schug, K.A.; Maráková, K. Sample preparation and fractionation techniques for intact proteins for mass spectrometric analysis. J. Sep. Sci. 2021, 44, 211–246. [Google Scholar] [CrossRef]
- Kaur, H.; Beckman, J.; Zhang, Y.; Li, Z.J.; Szigeti, M.; Guttman, A. Capillary electrophoresis and the biopharmaceutical industry: Therapeutic protein analysis and characterization. Trends Anal. Chem. 2021, 144, 116407. [Google Scholar] [CrossRef]
- Giorgetti, J.; Beck, A.; Leize-Wagner, E.; François, Y.-N. Combination of intact, middle-up and bottom-up levels to characterize 7 therapeutic monoclonal antibodies by capillary electrophoresis – Mass spectrometry. J. Pharm. Biomed. Anal. 2020, 182, 113107. [Google Scholar] [CrossRef] [PubMed]
- McCool, E.N.; Lubeckyj, R.A.; Shen, X.; Chen, D.; Kou, Q.; Liu, X.; Sun, L. Deep top-down proteomics using capillary zone electrophoresis-tandem mass spectrometry: Identification of 5700 proteoforms from the Escherichia coli proteome. Anal. Chem. 2018, 90, 5529–5533. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Tang, K. Highly sensitive and robust capillary electrophoresis-electrospray ionization-mass spectrometry: Interfaces, preconcentration techniques and applications. Rev. Anal. Chem. 2020, 39, 45–55. [Google Scholar] [CrossRef]
- Sarg, B.; Faserl, K.; Kremser, L.; Halfinger, B.; Sebastiano, R.; Lindner, H.H. Comparing and combining capillary electrophoresis electrospray ionization mass spectrometry and nano-liquid chromatography electrospray ionization mass spectrometry for the characterization of post-translationally modified histones. Mol. Cell. Proteom. 2013, 12, 2640–2656. [Google Scholar] [CrossRef]
- Ren, T.-J.; Zhang, X.-X.; Li, X.; Chen, H.-X. Isoforms of recombinant human erythropoietin by polarity-reversed capillary isoelectric focusing. Electrophoresis 2020, 41, 2055–2061. [Google Scholar] [CrossRef]
- Andrasi, M.; Pajaziti, B.; Sipos, B.; Nagy, C.; Hamidli, N.; Gaspar, A. Determination of deamidated isoforms of human insulin using capillary electrophoresis. J. Chromatogr. A 2020, 1626, 461344. [Google Scholar] [CrossRef]
- Crihfield, C.L.; Kristoff, C.J.; Veltri, L.M.; Penny, W.M.; Holland, L.A. Semi-permanent cationic coating for protein separations. J. Chromatogr. A 2019, 1607, 460397. [Google Scholar] [CrossRef]



| Proteoform | Function | Clinical Significance | Reference |
|---|---|---|---|
| Cystatin C | Endogenous cysteine protease inhibitor. | Assessment of progression of kidney function. Increased levels of Cystatin C can be found in acute and chronic kidney disease. | [48] |
| Chemokine RANTES | Key role in inflammation, cell recruitment, and T cell activation. | Associated with autoimmune diseases, arthritis, diabetes, obesity, cardiovascular diseases, breast and cervical cancers, and other diseases. | [49] |
| Amyloid proteoforms | Complication of normally soluble proteins that can join together to form an insoluble protein. | Associated with infiltrative proteinopathies, including Alzheimer’s disease, restrictive cardiomyopathy, infiltrative kidney disease, and other diseases. | [50,51] |
|
Amyloid-beta (Aβ) proteoforms | Amyloid-beta (Aβ) plays a key role in the pathogenesis of Alzheimer’s disease. Post-mortem identification of 26 proteoforms in the brain of Alzheimer’s disease patients has provided significant information in patients suffering from Alzheimer’s disease (AD) and dementia. | The heterogeneity of Aβ proteoforms deepens our understanding of Alzheimer’s disease and offers new avenues for investigation into pathological mechanisms of the disease, with implications for therapeutic development. | [52] |
| Lacritin proteoforms | Prevent tear film collapse and maintain epithelial homeostasis.Proteoforms include an active monomer, inactive polymers, and a splice variant termed lacritrin-c. | Associated with dry eye disease. Important in visual acuity and promotes basal tearing.Quantitation of the different proteoforms of tear lacritin may provide a diagnostic tool for ocular diseases. | [53,54] |
| Tumor suppressor PTEN | Control various aspects of cellular function, grouping them into three categories: intrinsic, function-induced, and inducible proteoforms. | PTEN proteoforms offer novel therapeutic opportunities in the treatment of various cancers and other diseases. | [55] |
| Cardiac troponin | Critical regulator of cardiac muscle contraction and relaxation. | Assessment of cardiac troponin proteoforms in serum of patients with acute myocardial infarction and other forms of myocardial injury. | [56,57] |
| MM-9 proteoforms | Matrix metalloproteinases (MMPs) are a class of secreted or cell bound endopeptidases, implicated in every step of the process of inflammatory cell migration. | Specific inhibition of MMPs has been suggested to be an interesting approach to control inflammation. | [58] |
| Apolipoprotein(a) | Lipoprotein(a) (Lp(a)) is an LDL-like particle, that contains a single copy of the apolipoprotein(a) covalently linked by a disulfide bridge to apolipoprotein(b). Its function is in wound healing, where it promotes tissue repair and vascular remodeling. | Lipoprotein(a) plays a role as an independent risk factor in the development of atherosclerotic cardiovascular diseases and calcified aortic valve disease. New research indicates that Lp(a) should be evaluated in terms of its apo(a) component and no longer in terms of Lp(a) mass. | [59] |
| Apolipoprotein A-1 | Apolipoprotein A1 (Apo A-1) is the major constituent of human high-density lipoproteins, which plays a key role in reverse cholesterol transport and lipid homeostasis. | Apo A-1 exhibits antioxidant and anti-inflammatory properties and inhibits the aggregation and neurotoxicity of amyloid-beta peptide in Alzheimer’s disease. Apo A-1 may possibly provide protection against neurological disorders. | [60] |
| DJ-1 proteoforms | DJ-1 is a cancer associated protein that protects cells from oxidative stress, functioning as a deglycase enzyme. DJ-1 acts as a redox-sensitive chaperone and as an oxidative stress sensor. | Associated with breast cancer. The modulation of specific DJ-1 function might produce substantial anticancer effects. | [61] |
| Prostate specific antigen proteoforms | Prostate specific antigen (PSA) is a glycoprotein with protease activity. This androgen-regulated serine-protease participates in the dissolution of the seminal fluid coagulum and plays an important role in fertility. PSA is produced by both prostate epithelial cells and prostate cancer cells. | Commonly used as serum biomarker for prostate cancer. Analysis of PSA proteoforms in urine and assessment of intact protein and glycopeptide analysis might be useful in improving prostate cancer screening. | [62] |
| Haptoglobin proteoforms | Serum haptoglobin (Hp) is a glycoprotein that scavenges freely circulating hemoglobin leaked into the blood stream when erythrocytes are damaged or die. Hp possesses four N-glycosylation sites on the beta-chain. | Studies have shown differences in the glycosylation pattern among patients with liver cirrhosis and hepatocellular carcinoma. | [63] |
| Erythropoietin | Erythropoietin (EPO) is a glycoprotein hormone of significant importance in the formation of red blood cells, as well as in other physiological functions. | EPO is used as a therapeutic protein for the treatment of anemia in chronic kidney disease and cancer. | [64,65,66] |
| Kidney allograft proteoforms | Proteins identified in peripheral blood mononuclear cells (PBMCs) as molecular signatures of kidney allograft pathology. | Non-invasive differential diagnostics of dysfunction of a transplanted kidney, or biomarkers of the kidney graft rejection. | [67] |
| EgAg B proteoforms | EgAg B proteoforms are parasite antigen proteins present in cystic echinococcocis disease caused by the Echininococcus granulosus metacestode parasite. These antigens are immunopotent. | Biomarkers for early detection and monitoring of the progression of the cystic echinococcosis disease. Specific immunodominant epitopes change as the disease progresses. | [68] |
| Human growth hormone proteoforms | Human growth hormone (hGH) is synthesized by, stored in, and secreted by the pituitary gland. It promotes human growth and metabolism. | Monitoring of the proteoform pattern changes in a growth hormone-secreting pituitary adenoma, when compared to control pituitary tissues, can be of significant value for the predictive diagnosis, targeted prevention, and treatment of pituitary adenoma. | [69] |
| ∆Np73 proteoforms | The p53 family of proteins, including p53, p63 and p73, have a role in tumor suppression. The ∆Np63 and ∆Np73 proteoforms are frequently overexpressed in a wide range of tumors, where they are associated with poorer prognosis. Furthermore, it has been demonstrated that the presence of autoantibodies to p53, p63 and p73 proteins exists in the serum of cancer patients. | ∆Np73 proteoforms shows a specific seroreactivity different from that of p73 with a higher diagnostic ability to discriminate between colorectal cancer patients and controls, and especially premalignant individuals and controls which may have an important impact on cancer prevention to predict premalignant tumors. | [70] |
| Surfactant protein B immature proteoform (proSP-B) | Surfactant protein B (SP-B) is a protein vital for normal lung function. Higher levels of circulating SP-B have been found in heavy smokers. Immature SP-B (proSP-B) flow into the bloodstream, where it binds high-density lipoprotein (HDL), modifying its function. Impairing the alveolar cell SP-B metabolism is likely the trigger of the smoke-induced pro-atherosclerotic cascade. | Circulating immature proteoform of surfactant protein B (proSP-B) has been proposed as the most reliable lung-specific marker for alveolar-capillary membrane dysfunction and overall clinical status of heart failure. | [71] |
| Sarcomeric proteoforms | The post-translational modifications (PTMs) of sarcomeric proteins are known to be important mediators of cardiac signaling and exert various effects on contractile function. Hypertrophic cardiomyopathy (HCM) is the most common inherited disease and a leading cause of sudden death in young adults. It is characterized by abnormal thickening of the myocardium, which imposes a mechanical burden on the heart. HCM is highly heterogeneous and has been linked to mutations in the genes that encode proteins of the sarcomere. | Obtaining a comprehensive view of the changes in the sarcomeric proteome is an important first step toward understanding the molecular underpinnings of HCM. Future proteomics studies covering a wide range of HCM phenotypes will hold promise to help define disease progression and prognosis based on the proteoform landscape. | [72] |
| NISTmAB proteoform | The NISTmAb is a recombinant humanized monoclonal antibody Reference Material from the National Institute of Standards and Technology. It is a class representative IgG1κ intended to serve as a pre-competitive platform for harmonization and technology development in the biopharmaceutical industry. | Complex biotherapeutics, in particular monoclonal antibodies, increasingly dominate the arena of new drugs submitted for regulatory approval. Quality control mechanisms and their accompanying analytics are still evolving to meet increasingly sophisticated needs of the biotherapeutics market. Deviations in quality control may be linked to pathological conditions, such as immunogenicity or toxicity. | [73] |
| Natriuretic peptides proteoforms | The natriuretic peptide family consists of three biologically active peptides: atrial natriuretic peptide (ANP), brain (or B-type) natriuretic peptide (BNP), and C-type natriuretic peptide (CNP). Among these, ANP and BNP are secreted by the heart and act as cardiac hormones. ProANP and B-ANP are minor forms in the circulation but increases in patients with heart failure. | The human BNP precursor proBNP is proteolytically processed to BNP1–32 and N-terminal proBNP (NT-proBNP) within ventricular myocytes. Uncleaved proBNP as well as mature BNP1–32 and NT-proBNP are secreted from the heart, and its secretion is increased in patients with heart failure. Furthermore, NT-proBNP is O-glycosylated in the plasma of patients with severe heart failure. | [74,75] |
| Transthyretin proteoforms | Transthyrein (TTR) is a serum and cerebrospinal fluid (CSF) protein with metalloprotease activity. It is one of the most abundant proteins in CSF. Despite being best known for transporting thyroxine (T4) and retinol through the blood–brain barrier (BBB), TTR has been suggested to play a role in a broad range of functions in the central nervous system. | Dysregulations of TTR levels characterize several neurological disorders. In patients with stroke, the detection of TTR has been described as a positive prognostic indicator of clinical outcomes. TTR proteoforms are changed in the CSF of patients affected with spinal muscular atrophy (SMA) type, after treatment with the antisense oligonucleotide nusinersen. | [76] |
| C-reactive protein proteoforms | C-reactive protein (CRP) is an acute inflammatory protein that increases up to 1,000-fold at sites of infection or inflammation. CRP is produced as a homopentameric protein, termed native CRP (nCRP), which can irreversibly dissociate at sites of inflammation and infection into five monomers, termed monomeric CRP (mCRP). CRP is synthesized primarily in liver hepatocytes but also by smooth muscle cells, macrophages, endothelial cells, lymphocytes, and adipocytes. Having been traditionally utilized as a marker of infection and cardiovascular events, there is now growing evidence that CRP plays important roles in inflammatory processes and host responses to infection including the complement pathway, apoptosis, phagocytosis, niric oxide (NO) release, and the production of cytokines, particularly interleukin-6 and tumor necrosis factor-α. | CRP isoforms have distinct biological properties, with nCRP often having more anti-inflammatory activities compared to mCRP. The nCRP isoform activates the classical complement pathway, induces phagocytosis, and promotes apoptosis. On the other hand, mCRP promotes the chemotaxis and recruitment of circulating leukocytes to areas of inflammation and can delay apoptosis. mCRP increases interleuin-8 and monocyte chemoattractant protein-1 production. | [77] |
| SHC-Transforming protein 1 proteoforms | Human SHC-Transforming protein 1 (Shc1) has been found to be important in the regulation of apoptosis and drug resistance in mammalian cells. Shc1 is an intracellular scaffold protein, which is involved in downstream pathways of cell surface signaling receptors, such as the insulin signaling pathway. One of the isoforms of Shc1 is p66shc, a mitochondrial associated oxidative stress biomarker, that may be involved in regulating the life span and the effects of reactive oxygen species (ROS). P66shc has been implicated in several metabolic pathways, being able to act as an adaptor protein as well. Furthermore, p66shc is significantly altered in patients with elevated blood glucose levels. | Evaluation of p66shc/Shc1 has been reported to be a useful addition to the regularly used biomarkers, such as HbA1c, to increase diagnostic sensitivity for the identification of prediabetes (T2DM). Knowledge of the presence of oxidative stress and inflammatory processes, forming a complex pattern of disease progression, is therefore an important step towards early, effective treatment. | [78,79] |
| Prolyl hydroxylase alpha subunit proteoforms | Mammalian prolyl 4-hydroxylase (P4H) is a tetramer composed of two unique subunits, alpha and beta. One gene makes the beta subunit that functions independently as a protein disulfide isomerase, and the other genes make three alpha subunit isoforms. | P4H catalyzes selective proline-containing peptides to hydroxy-proline-containing peptides. The best-known role of hydroxyproline is in stabilizing the collagen triple helix. A prolyl hydroxylase domain protein acts on the hypoxia inducible factor alpha subunits, which plays a key role in sensing molecular oxygen. Prolyl hydroxylases are essential for breast cancer metastasis, and a prolyl hydroxylase inhibitor decreases tumorogenesis. | [80,81,82,83,84,85,86,87,88] |
| Immunoglobulin G (IgG) proteoforms | Beyond their ability to neutralize pathogens, antibodies can mediate an array of effector functions through their interactions with Fc-receptors, complement molecules, and mammalian lectin-like molecules. Immunoglobulin G (IgG) fragment antigen binding (Fab) region binds a specific antigen, while its fragment crystallizable (Fc) region binds different receptors on the surface of various immune cells, thereby dictating the type of immune response elicited by the antigen binding. | Analysis of Fc-specific IgG glycosylation is critical for population-level studies of how antibodies may vary in response to vaccination or infection, and across disease states ranging from autoimmunity to cancer in both clinical and animal studies. IgG glycans are an excellent biomarker of biological age. Sialylation of Fc glycan has been reported to have anti-inflammatory activity. | [89,90,91] |
| Histone proteoforms | Chromatin is the structural framework that packages DNA into chromosomes within the nucleus of a cell. Histones comprises the principal protein component of chromatin and are involved in the regulation of gene expression. This epigenetic configuration is achieved through complex patterns of post-translational modifications, the incorporation of histones variants, and through controlled histone proteolysis. | Histone post-translational modifications (PTMs) are one of the main mechanisms of epigenetic regulation. Dysregulation of histones PTMs leads to many diseases, such as cancer. | [92,93,94,95] |
| KRAS proteoforms | Mutations of the KRAS gene are found in human cancers with high frequency and result in the constitutive activation of its protein products. | Mutations affecting post-translational modifications leads to aberrant regulation of downstream pathways, promoting cell survival, proliferation, and tumorigenesis that drive cancer progression and negatively affect treatment outcomes. | [96] |
| Huntingtin proteoforms | Huntington’s disease (HD) is a rare neurodegenerative disorder caused by the aberrant expression of mutant Huntingtin (HTT) protein containing an expanded polyglutamine tract. The expression of this protein is highly enriched in the brain relative to other tissues with highest expression in neurons. | HTT is now known to have a wide variety of post-translational modifications, including phosphorylation, sumoylation, acetylation, ubiquitination, and protease cleavage. Correlating these HTT proteoforms with functions should be fruitful in identifying mechanism of pathology that can be targeted for intervention. | [97] |
| Alpha-synuclein proteoforms | Cumulative evidence suggests that lysosomal dysfunction contributes to neurodegenerative diseases, especially if amyloid proteins are involved. Among these, alpha-synuclein that progressively accumulates and aggregates in Lewis bodies is undisputedly a main culprit in Parkinson disease. | Alpha-synuclein can possess diverse post-translational modifications, aggregate formations, and truncations, all of which contribute to a growing set of proteoforms. These interfere directly or indirectly with lysosome function, reducing their own degradation, and thereby accelerating the protein aggregation and disease process. | [98] |
| Carbonic anhydrases proteoforms | Metalloenzymes carbonic anhydrases intervene in the second and rate limiting step of the catalytic mechanism of carbon dioxide reversible hydration. Genetic deficiencies of several carbonic anhydrases have been reported to be associated with diseases such as osteopetrosis, cerebral calcifications, hyperammonemia, retinal problems, and hyperchlorhydrolysis. | The loss of function of carbonic anhydrases would be in principle be treatable with selective activators of these enzymes. Carbonic anhydrase proteoforms represent a crucial family of new targets for improving cognition, but also in therapeutic, such as phobias, obsessive compulsive disorder, generalized anxiety, and post-traumatic disorders, for which few therapies are available. Studies on carbonic anhydrase proteoforms are important to elucidate the role of these modified proteins and their potential activators in brain processes. | [99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzman, N.A.; Guzman, D.E. Immunoaffinity Capillary Electrophoresis in the Era of Proteoforms, Liquid Biopsy and Preventive Medicine: A Potential Impact in the Diagnosis and Monitoring of Disease Progression. Biomolecules 2021, 11, 1443. https://doi.org/10.3390/biom11101443
Guzman NA, Guzman DE. Immunoaffinity Capillary Electrophoresis in the Era of Proteoforms, Liquid Biopsy and Preventive Medicine: A Potential Impact in the Diagnosis and Monitoring of Disease Progression. Biomolecules. 2021; 11(10):1443. https://doi.org/10.3390/biom11101443
Chicago/Turabian StyleGuzman, Norberto A., and Daniel E. Guzman. 2021. "Immunoaffinity Capillary Electrophoresis in the Era of Proteoforms, Liquid Biopsy and Preventive Medicine: A Potential Impact in the Diagnosis and Monitoring of Disease Progression" Biomolecules 11, no. 10: 1443. https://doi.org/10.3390/biom11101443
APA StyleGuzman, N. A., & Guzman, D. E. (2021). Immunoaffinity Capillary Electrophoresis in the Era of Proteoforms, Liquid Biopsy and Preventive Medicine: A Potential Impact in the Diagnosis and Monitoring of Disease Progression. Biomolecules, 11(10), 1443. https://doi.org/10.3390/biom11101443







