Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Plasmid
2.2. Preparation of HIV-1 Virions
2.3. Construction of pNL4-3 MIRCAI201V Plasmid and Preparation of HIV-1 Maturation Inhibitor Resistant (MIR) Virus
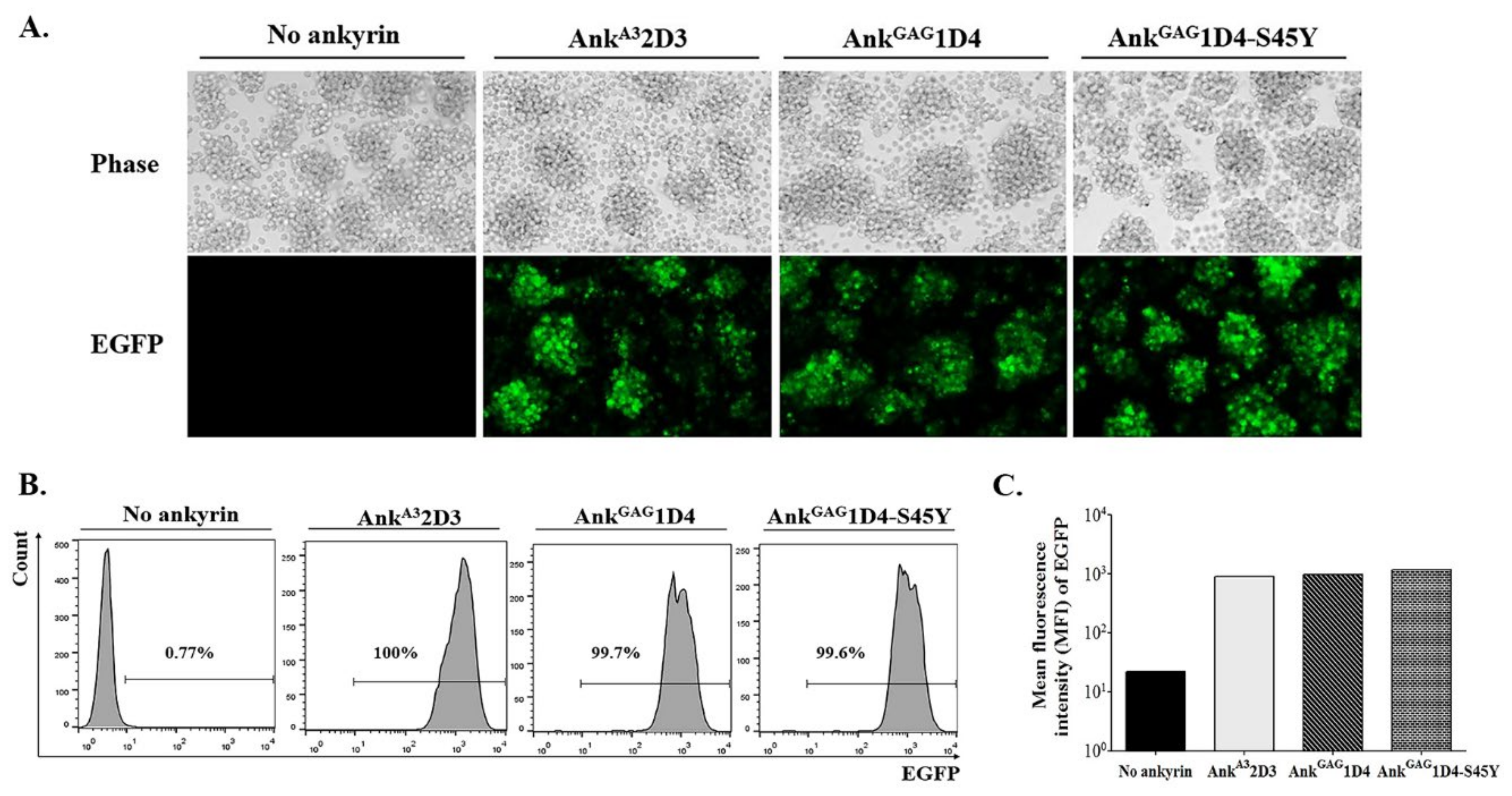
2.4. Generation of SupT1 Cells Stably Expressing Ankyrin Protein, AnkGAG1D4-EGFP, AnkGAG1D4-S45Y-EGFP, and AnkA32D3-EGFP by Lentiviral Gene Transferring Method
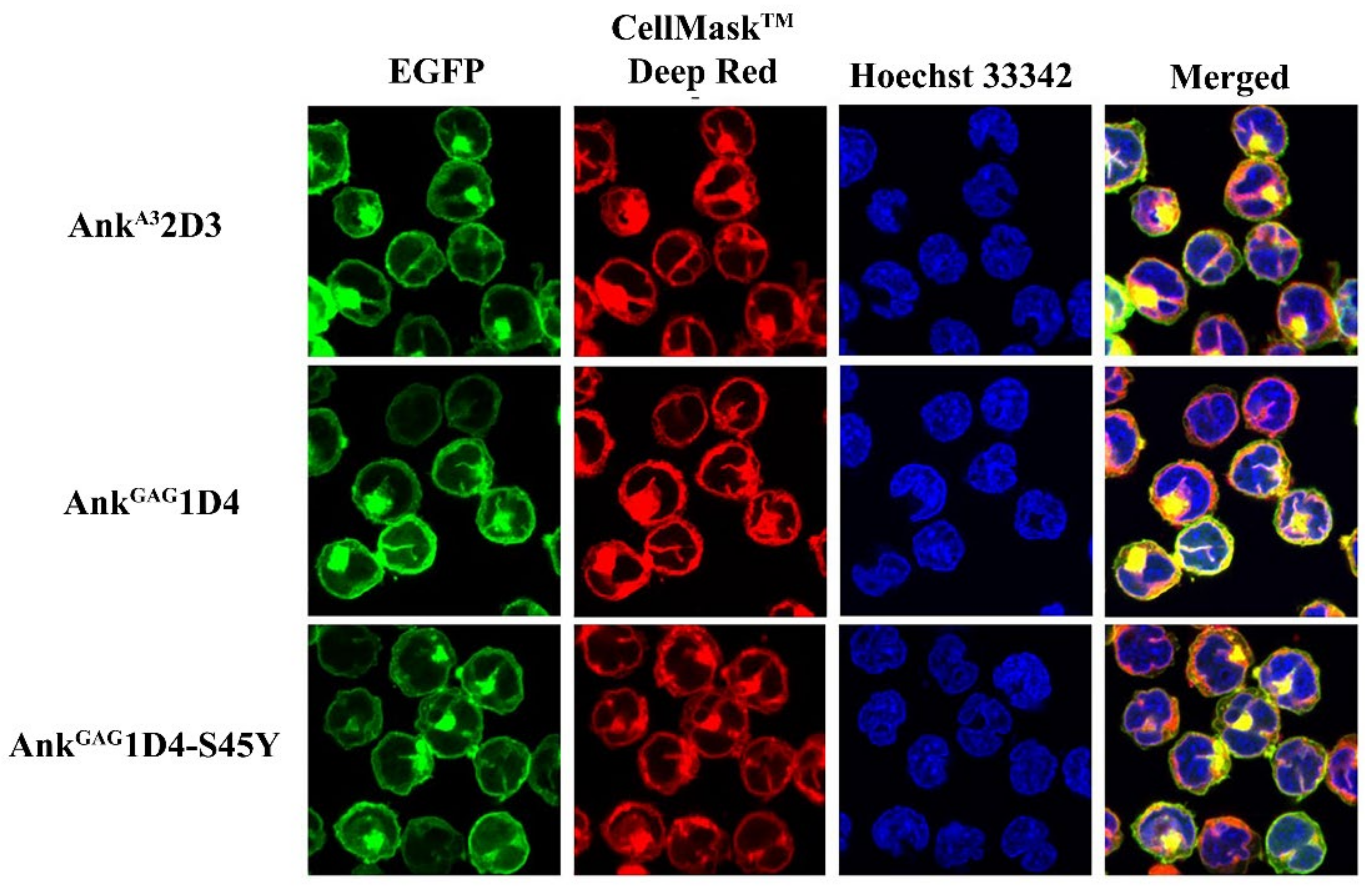
2.5. Determination of Subcellular Localization of Ankyrin Proteins in SupT1 Cells
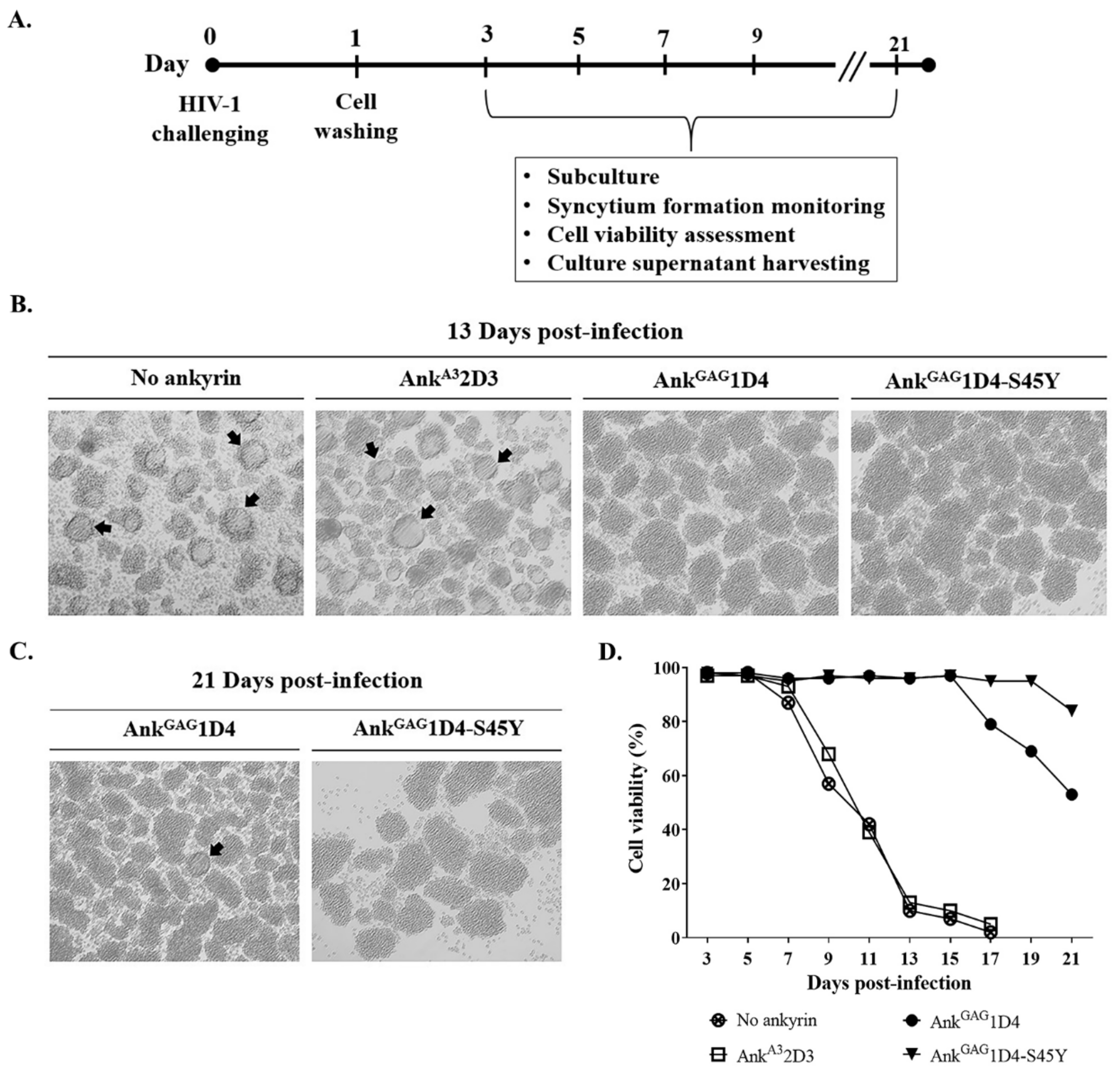
2.6. HIV-1 Challenge
2.7. Evaluation of HIV-1 p24 and Viral Load
2.8. Analysis of HIV-1 Capsid Sequence
2.9. Statistical Analysis
3. Results
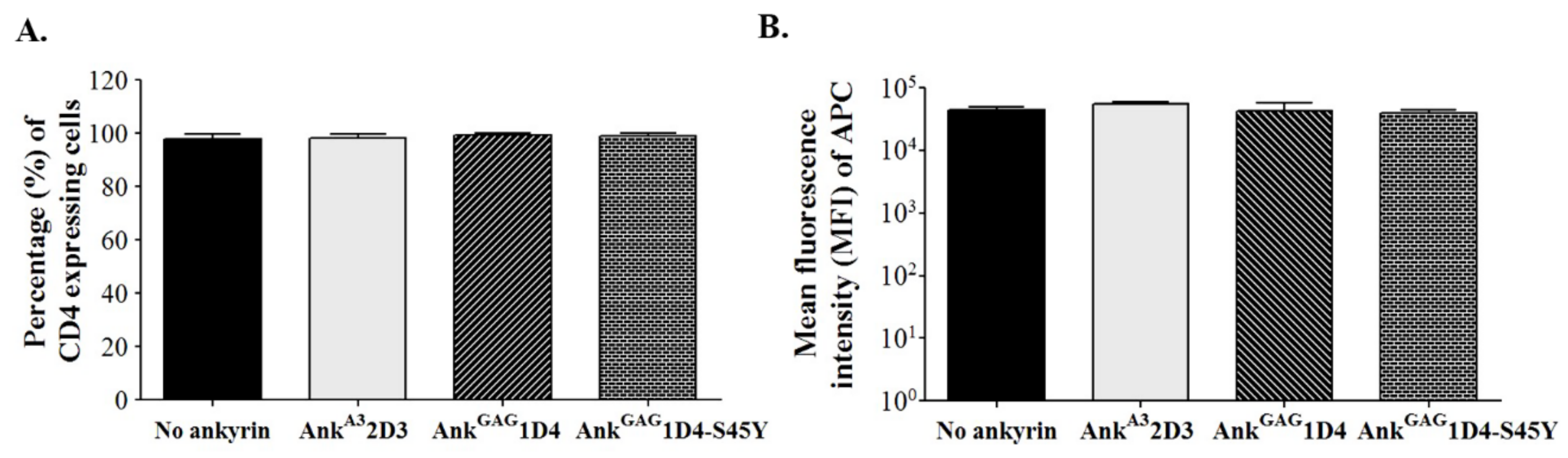
3.1. Expression of Ankyrin Protein Did Not Interfere with Cell-Surface CD4
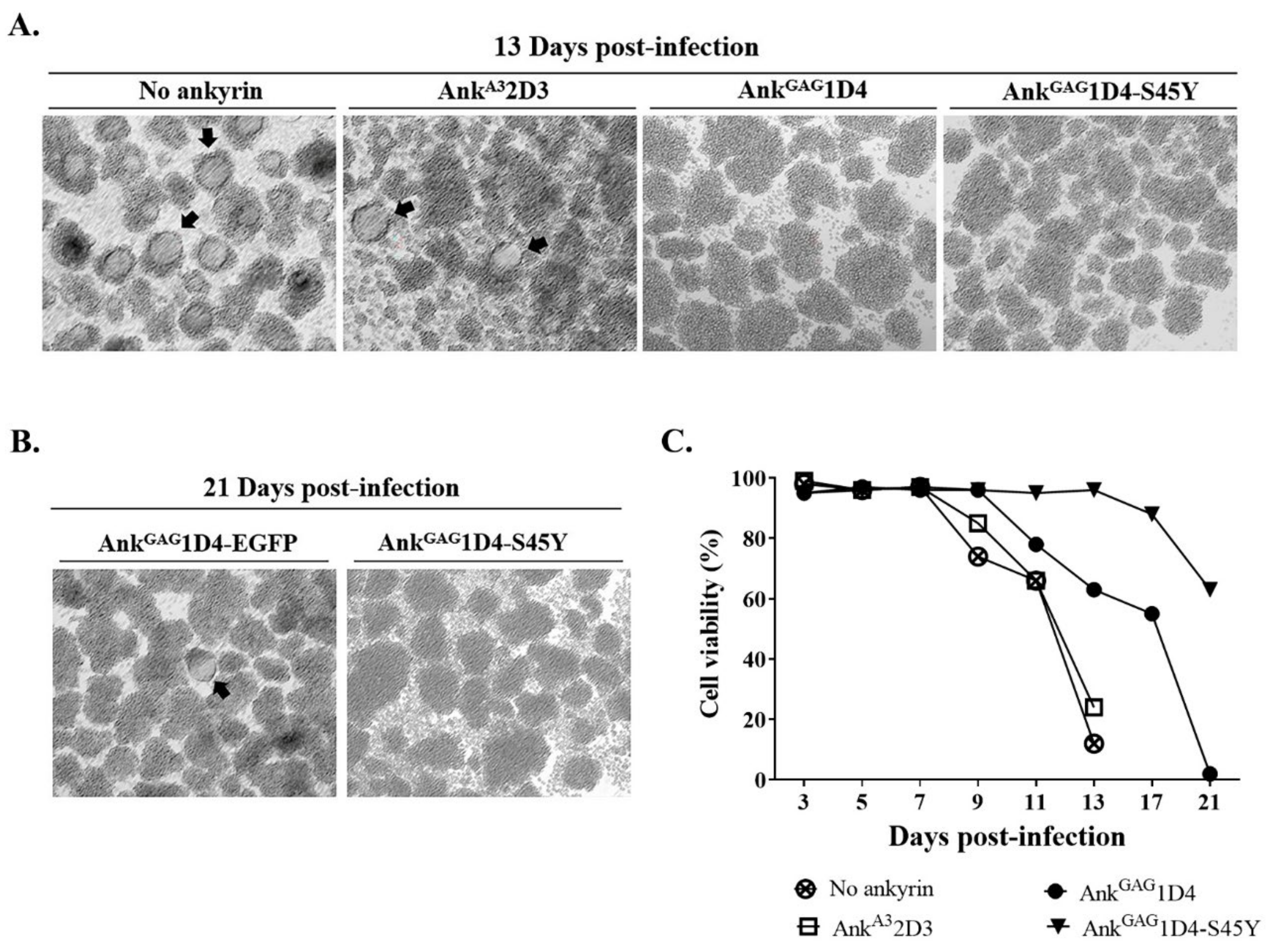
3.2. AnkGAG1D4-S45Y Provides More Protection against HIV-1-Mediated Cell Death
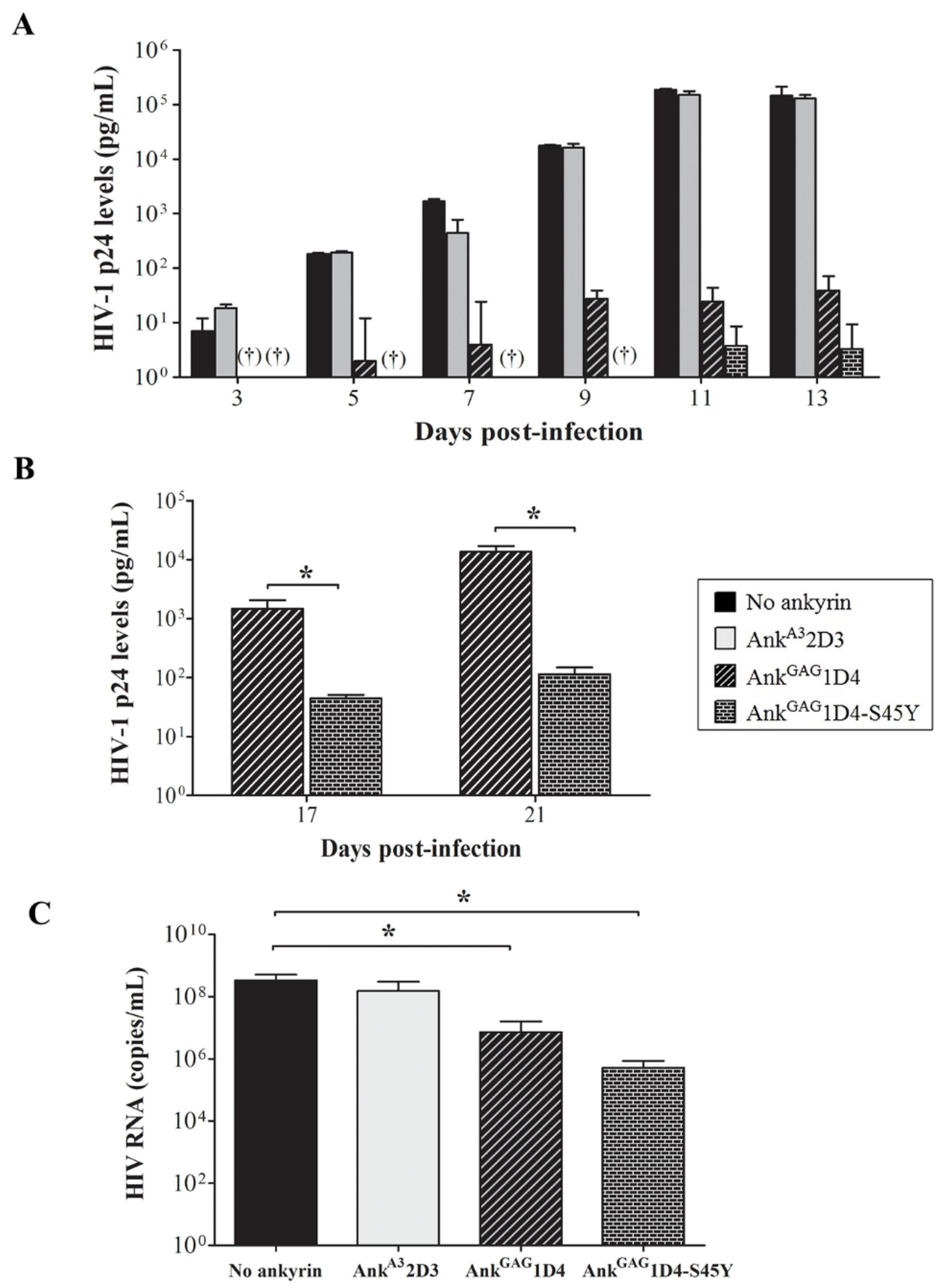
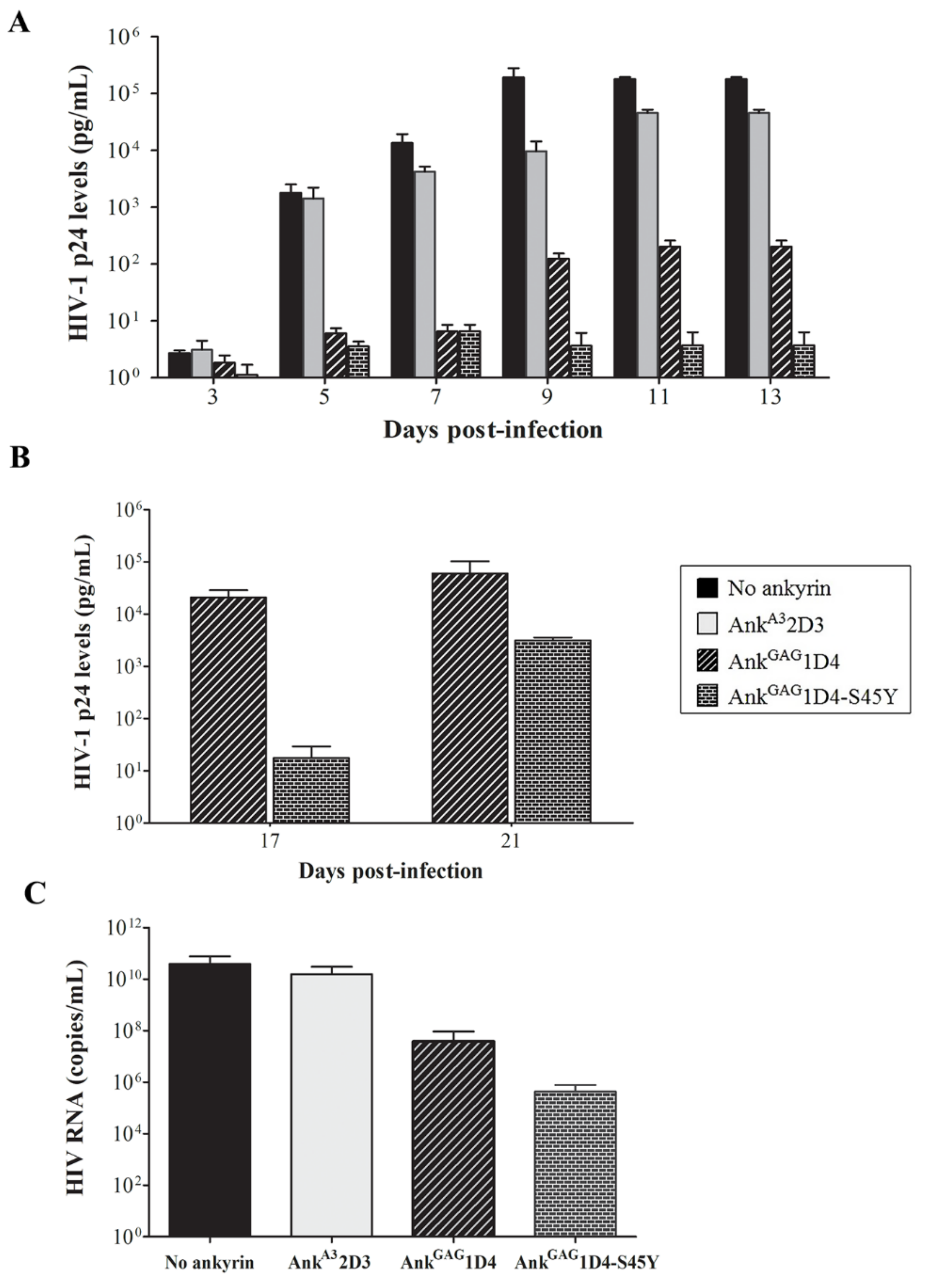
3.3. AnkGAG1D4-S45Y Improves Antiviral Activity Than Parental Ankyrin in HIV-1-Infected SupT1 Cells
3.4. Anti-HIV-1 Ankyrins Do Not Drive Mutation in Amino Acid Sequence of HIV-1 Capsid
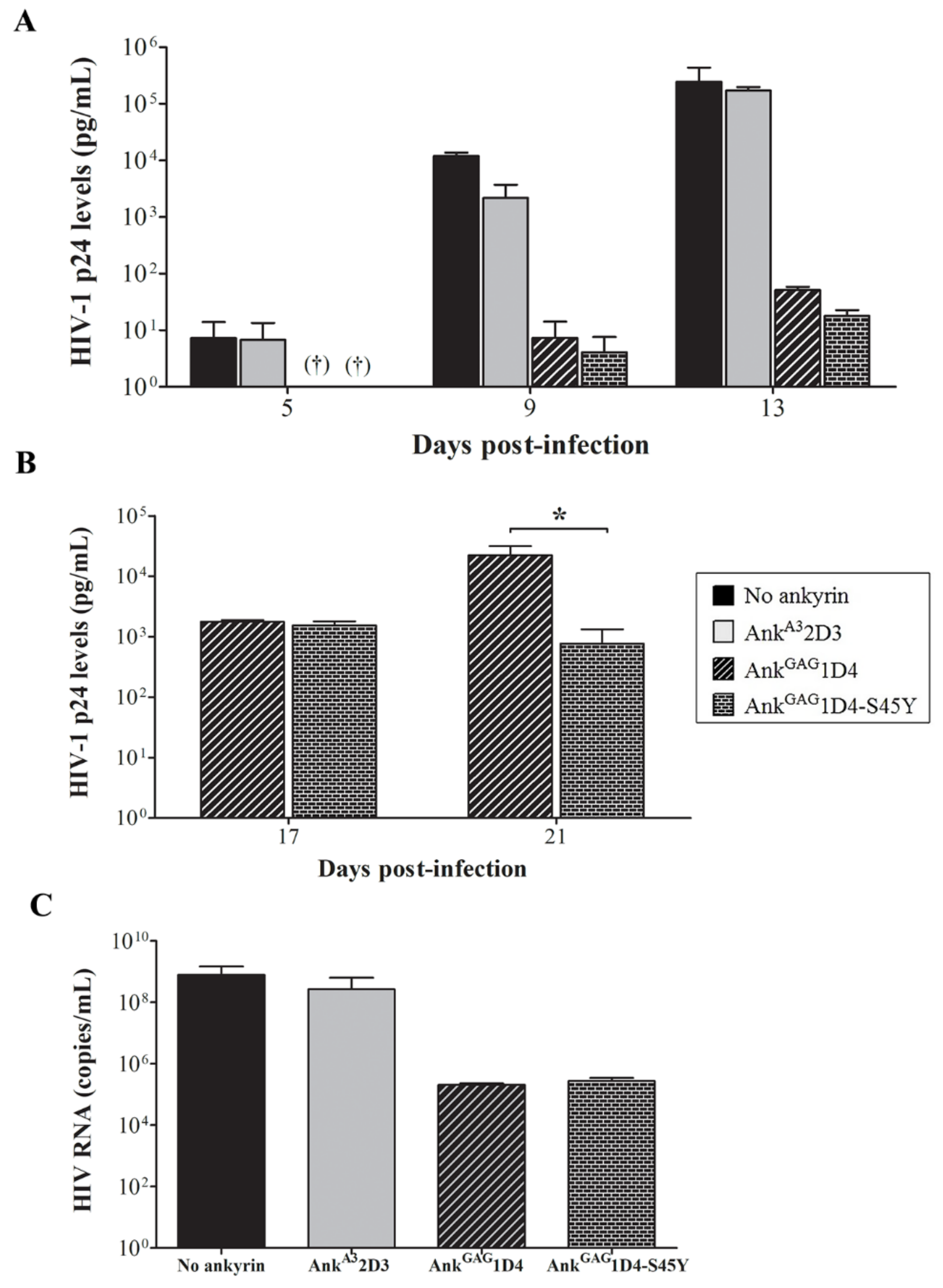
3.5. Binding Affinity-Enhanced Ankyrin Provides Antiviral Effects on HIV-1 Maturation Inhibitor Resistant Virus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reust, C.E. Common adverse effects of antiretroviral therapy for HIV disease. Am. Fam. Physician 2011, 83, 1443–1451. [Google Scholar] [PubMed]
- Carr, A. Toxicity of antiretroviral therapy and implications for drug development. Nat. Rev. Drug Discov. 2003, 2, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Chupradit, K.; Moonmuang, S.; Nangola, S.; Kitidee, K.; Yasamut, U.; Mougel, M.; Tayapiwatana, C. Current peptide and protein candidates challenging HIV therapy beyond the vaccine era. Viruses 2017, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Legastelois, I.; Desgranges, C. Design and intracellular activity of a human single-chain antibody to human immunodeficiency virus type 1 conserved gp41 epitope. J. Virol. 2000, 74, 5712–5715. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Wang, W.; Cheng, L.; Li, G.; Wen, M.; Wang, Q.; Zhang, Q.; Li, D.; Zhou, P.; Su, L. Glycosylphosphatidylinositol-anchored anti-HIV scFv efficiently protects CD4 T cells from HIV-1 infection and deletion in hu-PBL mice. J. Virol. 2017, 91, e01389–e01416. [Google Scholar] [CrossRef]
- Omar, M.T.C. Expression of Functional Anti-p24 scFv 183-H12-5C in HEK293T and Jurkat T Cells. Adv. Pharm. Bull. 2017, 7, 299–312. [Google Scholar] [CrossRef]
- Hadpech, S.; Nangola, S.; Chupradit, K.; Fanhchaksai, K.; Furnon, W.; Urvoas, A.; Valerio-Lepiniec, M.; Minard, P.; Boulanger, P.; Hong, S.-S.; et al. Alpha-helicoidal HEAT-like repeat proteins (αRep) selected as interactors of HIV-1 nucleocapsid negatively interfere with viral genome packaging and virus maturation. Sci. Rep. 2017, 7, 16335. [Google Scholar] [CrossRef]
- Hadpech, S.; Peerakam, N.; Chupradit, K.; Tayapiwatana, C. Occupation of a thermoresistant-scaffold (αRep) at SP1-NC cleavage site disturbs the function of HIV-1 protease. Biosci. Rep. 2020, 40, BSR20201131. [Google Scholar] [CrossRef]
- Shilova, O.N.; Deyev, S.M. DARPins: Promising scaffolds for theranostics. Acta Nat. 2019, 11, 42–53. [Google Scholar] [CrossRef]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.-Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef]
- Cunha, S.R.; Mohler, P.J. Ankyrin protein networks in membrane formation and stabilization. J. Cell. Mol. Med. 2009, 13, 4364–4376. [Google Scholar] [CrossRef]
- Cunha, S.R.; Mohler, P.J. Ankyrin-based cellular pathways for cardiac ion channel and transporter targeting and regulation. Semin. Cell Dev. Biol. 2011, 22, 166–170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zahnd, C.; Wyler, E.; Schwenk, J.M.; Steiner, D.; Lawrence, M.C.; McKern, N.M.; Pecorari, F.; Ward, C.W.; Joos, T.O.; Plückthun, A. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J. Mol. Biol. 2007, 369, 1015–1028. [Google Scholar] [CrossRef]
- Boersma, Y.L.; Chao, G.; Steiner, D.; Wittrup, K.D.; Plückthun, A. Bispecific designed ankyrin repeat proteins (DARPins) targeting epidermal growth factor receptor inhibit A431 cell proliferation and receptor recycling. J. Biol. Chem. 2011, 286, 41273–41285. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.A.; Mason, M.; Christie, L.-A.; Hansen, C.; Hernandez, L.M.; Burke, J.; Luhrs, K.A.; Hohman, T.C. Functional characterization of abicipar-pegol, an anti-VEGF DARPin therapeutic that potently inhibits angiogenesis and vascular permeability. Invest. Ophthalmol. Vis. Sci. 2018, 59, 5836–5846. [Google Scholar] [CrossRef]
- Yant, S.R.; Mulato, A.; Hansen, D.; Tse, W.C.; Niedziela-Majka, A.; Zhang, J.R.; Stepan, G.J.; Jin, D.; Wong, M.H.; Perreira, J.M.; et al. A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 2019, 25, 1377–1384. [Google Scholar] [CrossRef]
- Schweizer, A.; Rusert, P.; Berlinger, L.; Ruprecht, C.R.; Mann, A.; Corthésy, S.; Turville, S.G.; Aravantinou, M.; Fischer, M.; Robbiani, M.; et al. CD4-specific designed ankyrin repeat proteins are novel potent HIV entry inhibitors with unique characteristics. PLoS Pathog. 2008, 4, e1000109. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Friedrich, N.; Krarup, A.; Weber, J.; Stiegeler, E.; Dreier, B.; Pugach, P.; Robbiani, M.; Riedel, T.; Moehle, K.; et al. Conformation-dependent recognition of HIV gp120 by designed ankyrin repeat proteins provides access to novel HIV entry inhibitors. J. Virol. 2013, 87, 5868. [Google Scholar] [CrossRef]
- Pugach, P.; Krarup, A.; Gettie, A.; Kuroda, M.; Blanchard, J.; Piatak, M., Jr.; Lifson, J.D.; Trkola, A.; Robbiani, M. In vivo binding and retention of CD4-specific DARPin 57.2 in macaques. PLoS ONE 2010, 5, e12455. [Google Scholar] [CrossRef]
- Hermann, F.G.; Egerer, L.; Brauer, F.; Gerum, C.; Schwalbe, H.; Dietrich, U.; von Laer, D. Mutations in gp120 contribute to the resistance of human immunodeficiency virus type 1 to membrane-anchored C-peptide maC46. J. Virol. 2009, 83, 4844–4853. [Google Scholar] [CrossRef] [PubMed]
- Praditwongwan, W.; Chuankhayan, P.; Saoin, S.; Wisitponchai, T.; Lee, V.S.; Nangola, S.; Hong, S.S.; Minard, P.; Boulanger, P.; Chen, C.-J.; et al. Crystal structure of an antiviral ankyrin targeting the HIV-1 capsid and molecular modeling of the ankyrin-capsid complex. J. Comput. Aided Mol. Des. 2014, 28, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Nangola, S.; Urvoas, A.; Valerio-Lepiniec, M.; Khamaikawin, W.; Sakkhachornphop, S.; Hong, S.S.; Boulanger, P.; Minard, P.; Tayapiwatana, C. Antiviral activity of recombinant ankyrin targeted to the capsid domain of HIV-1 Gag polyprotein. Retrovirology 2012, 9, 17. [Google Scholar] [CrossRef]
- Sakkhachornphop, S.; Hadpech, S.; Wisitponchai, T.; Panto, C.; Kantamala, D.; Utaipat, U.; Praparattanapan, J.; Kotarathitithum, W.; Taejaroenkul, S.; Yasamut, U.; et al. Broad-spectrum antiviral activity of an ankyrin repeat protein on viral assembly against chimeric NL4-3 viruses carrying Gag/PR derived from circulating strains among Northern Thai patients. Viruses 2018, 10, 625. [Google Scholar] [CrossRef]
- Khamaikawin, W.; Saoin, S.; Nangola, S.; Chupradit, K.; Sakkhachornphop, S.; Hadpech, S.; Onlamoon, N.; Ansari, A.A.; Byrareddy, S.N.; Boulanger, P.; et al. Combined antiviral therapy using designed molecular scaffolds targeting two distinct viral functions, HIV-1 genome integration and capsid assembly. Mol. Ther. Nucleic Acids 2015, 4, e249. [Google Scholar] [CrossRef]
- Saoin, S.; Wisitponchai, T.; Intachai, K.; Chupradit, K.; Moonmuang, S.; Nangola, S.; Kitidee, K.; Fanhchaksai, K.; Lee, V.S.; Hong, S.S.; et al. Deciphering critical amino acid residues to modify and enhance the binding affinity of ankyrin scaffold specific to capsid protein of human immunodeficiency virus type 1. Asian Pac. J. Allergy Immunol. 2018, 36, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Zazzi, M.; Hu, H.; Prosperi, M. The global burden of HIV-1 drug resistance in the past 20 years. PeerJ 2018, 6, e4848. [Google Scholar] [CrossRef]
- Wensing, A.M.; Calvez, V.; Ceccherini-Silberstein, F.; Charpentier, C.; Günthard, H.F.; Paredes, R.; Shafer, R.W.; Richman, D.D. 2019 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 2019, 27, 111. [Google Scholar]
- Zhang, X. Anti-retroviral drugs: Current state and development in the next decade. Acta Pharm. Sin. B 2018, 8, 131–136. [Google Scholar] [CrossRef]
- Pak, A.J.; Grime, J.M.A.; Yu, A.; Voth, G.A. Off-pathway assembly: A broad-spectrum mechanism of action for drugs that undermine controlled HIV-1 viral capsid formation. J. Am. Chem. Soc. 2019, 141, 10214–10224. [Google Scholar] [CrossRef]
- Carnes, S.K.; Sheehan, J.H.; Aiken, C. Inhibitors of the HIV-1 capsid, a target of opportunity. Curr. Opin. HIV AIDS 2018, 13, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Salzwedel, K.; Allaway, G.P. Bevirimat: A novel maturation inhibitor for the treatment of HIV-1 infection. Antivir. Chem. Chemother. 2008, 19, 107–113. [Google Scholar] [CrossRef]
- Urano, E.; Ablan, S.D.; Mandt, R.; Pauly, G.T.; Sigano, D.M.; Schneider, J.P.; Martin, D.E.; Nitz, T.J.; Wild, C.T.; Freed, E.O. Alkyl amine bevirimat derivatives are potent and broadly active HIV-1 maturation inhibitors. Antimicrob. Agents Chemother. 2016, 60, 190–197. [Google Scholar] [CrossRef]
- Waki, K.; Durell, S.R.; Soheilian, F.; Nagashima, K.; Butler, S.L.; Freed, E.O. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS Pathog. 2012, 8, e1002997. [Google Scholar] [CrossRef]
- Blair, W.S.; Cao, J.; Fok-Seang, J.; Griffin, P.; Isaacson, J.; Jackson, R.L.; Murray, E.; Patick, A.K.; Peng, Q.; Perros, M.; et al. New small-molecule inhibitor class targeting human immunodeficiency virus type 1 virion maturation. Antimicrob. Agents Chemother. 2009, 53, 5080–5087. [Google Scholar] [CrossRef]
- Ghimire, D.; Timilsina, U.; Srivastava, T.P.; Gaur, R. Insights into the activity of maturation inhibitor PF-46396 on HIV-1 clade C. Sci. Rep. 2017, 7, 43711. [Google Scholar] [CrossRef] [PubMed]
- Sakkhachornphop, S.; Jiranusornkul, S.; Kodchakorn, K.; Nangola, S.; Sirisanthana, T.; Tayapiwatana, C. Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions. Protein Sci. 2009, 18, 2219–2230. [Google Scholar] [CrossRef]
- Yildirim, O.; Gottwald, M.; Schüler, P.; Michel, M.C. Opportunities and challenges for drug development: Public–private partnerships, adaptive designs and big data. Front. Pharmacol. 2016, 7, 461. [Google Scholar] [CrossRef]
- Simeon, R.; Chen, Z. In vitro-engineered non-antibody protein therapeutics. Protein Cell 2018, 9, 3–14. [Google Scholar] [CrossRef]
- Stumpp, M.T.; Dawson, K.M.; Binz, H.K. Beyond antibodies: The DARPin® drug platform. BioDrugs 2020, 34, 423–433. [Google Scholar] [CrossRef]
- Takemura, T.; Murakami, T. Functional constraints on HIV-1 capsid: Their impacts on the viral immune escape potency. Front. Microbiol. 2012, 3, 369. [Google Scholar] [CrossRef]
- Ramalho, R.; Rankovic, S.; Zhou, J.; Aiken, C.; Rousso, I. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology 2016, 13, 17. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Alam, S.L.; Fricke, T.; Zadrozny, K.; Sedzicki, J.; Taylor, A.B.; Demeler, B.; Pornillos, O.; Ganser-Pornillos, B.K.; Diaz-Griffero, F.; et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 2014, 111, 18625–18630. [Google Scholar] [CrossRef]
- Ternois, F.; Sticht, J.; Duquerroy, S.; Kräusslich, H.G.; Rey, F.A. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat. Struct. Mol. Biol. 2005, 12, 678–682. [Google Scholar] [CrossRef]
- Perrier, M.; Bertine, M.; Hingrat, Q.L.; Joly, V.; Visseaux, B.; Collin, G.; Landman, R.; Yazdanpanah, Y.; Descamps, D.; Charpentier, C. Prevalence of gag mutations associated with in vitro resistance to capsid inhibitor GS-CA1 in HIV-1 antiretroviral-naive patients. J. Antimicrob. Chemother. 2017, 72, 2954–2955. [Google Scholar] [CrossRef]
- Binz, H.K.; Stumpp, M.T.; Forrer, P.; Amstutz, P.; Plückthun, A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003, 332, 489–503. [Google Scholar] [CrossRef]
- Costin, J.M. Cytopathic mechanisms of HIV-1. Virol. J. 2007, 4, 100. [Google Scholar] [CrossRef] [PubMed]
- Koide, S.; Sidhu, S.S. The importance of being tyrosine: Lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem. Biol. 2009, 4, 325–334. [Google Scholar] [CrossRef]
- Moreira, I.S.; Martins, J.M.; Ramos, R.M.; Fernandes, P.A.; Ramos, M.J. Understanding the importance of the aromatic amino-acid residues as hot-spots. Biochim. Biophys. Acta 2013, 1834, 404–414. [Google Scholar] [CrossRef]
- Piotukh, K.; Freund, C. A novel hSH3 domain scaffold engineered to bind folded domains in CD2BP2 and HIV capsid protein. Protein Eng. Des. Sel. 2012, 25, 649–656. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schlicksup, C.J.; Zlotnick, A. Viral structural proteins as targets for antivirals. Curr. Opin. Virol. 2020, 45, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Dicker, I.; Zhang, S.; Ray, N.; Beno, B.R.; Regueiro-Ren, A.; Joshi, S.; Cockett, M.; Krystal, M.; Lataillade, M. Resistance profile of the HIV-1 maturation inhibitor GSK3532795 in vitro and in a clinical study. PLoS ONE 2019, 14, e0224076. [Google Scholar] [CrossRef] [PubMed]
- Urano, E.; Timilsina, U.; Kaplan, J.A.; Ablan, S.; Ghimire, D.; Pham, P.; Kuruppu, N.; Mandt, R.; Durell, S.R.; Nitz, T.J.; et al. Resistance to second-generation HIV-1 maturation inhibitors. J. Virol. 2019, 93, e02017–e02018. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, P.R.; Stone, C.; Griffin, P.; Nájera, I.; Bloor, S.; Kemp, S.; Tisdale, M.; Larder, B. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J. Infect. Dis. 2000, 181, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Fun, A.; Wensing, A.M.; Verheyen, J.; Nijhuis, M. Human Immunodeficiency Virus Gag and protease: Partners in resistance. Retrovirology 2012, 9, 63. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sornsuwan, K.; Thongkhum, W.; Pamonsupornwichit, T.; Carraway, T.S.; Soponpong, S.; Sakkhachornphop, S.; Tayapiwatana, C.; Yasamut, U. Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production. Biomolecules 2021, 11, 1437. https://doi.org/10.3390/biom11101437
Sornsuwan K, Thongkhum W, Pamonsupornwichit T, Carraway TS, Soponpong S, Sakkhachornphop S, Tayapiwatana C, Yasamut U. Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production. Biomolecules. 2021; 11(10):1437. https://doi.org/10.3390/biom11101437
Chicago/Turabian StyleSornsuwan, Kanokporn, Weeraya Thongkhum, Thanathat Pamonsupornwichit, Tanawan Samleerat Carraway, Suthinee Soponpong, Supachai Sakkhachornphop, Chatchai Tayapiwatana, and Umpa Yasamut. 2021. "Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production" Biomolecules 11, no. 10: 1437. https://doi.org/10.3390/biom11101437
APA StyleSornsuwan, K., Thongkhum, W., Pamonsupornwichit, T., Carraway, T. S., Soponpong, S., Sakkhachornphop, S., Tayapiwatana, C., & Yasamut, U. (2021). Performance of Affinity-Improved DARPin Targeting HIV Capsid Domain in Interference of Viral Progeny Production. Biomolecules, 11(10), 1437. https://doi.org/10.3390/biom11101437





