isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them?
Abstract
1. Multi-Layered miR Gene Control as a Source of Plentiful miRNA Sequence Variations
1.1. Canonical miRNA Biogenesis Pathway
1.2. Non-Canonical miRNA Biogenesis Pathways
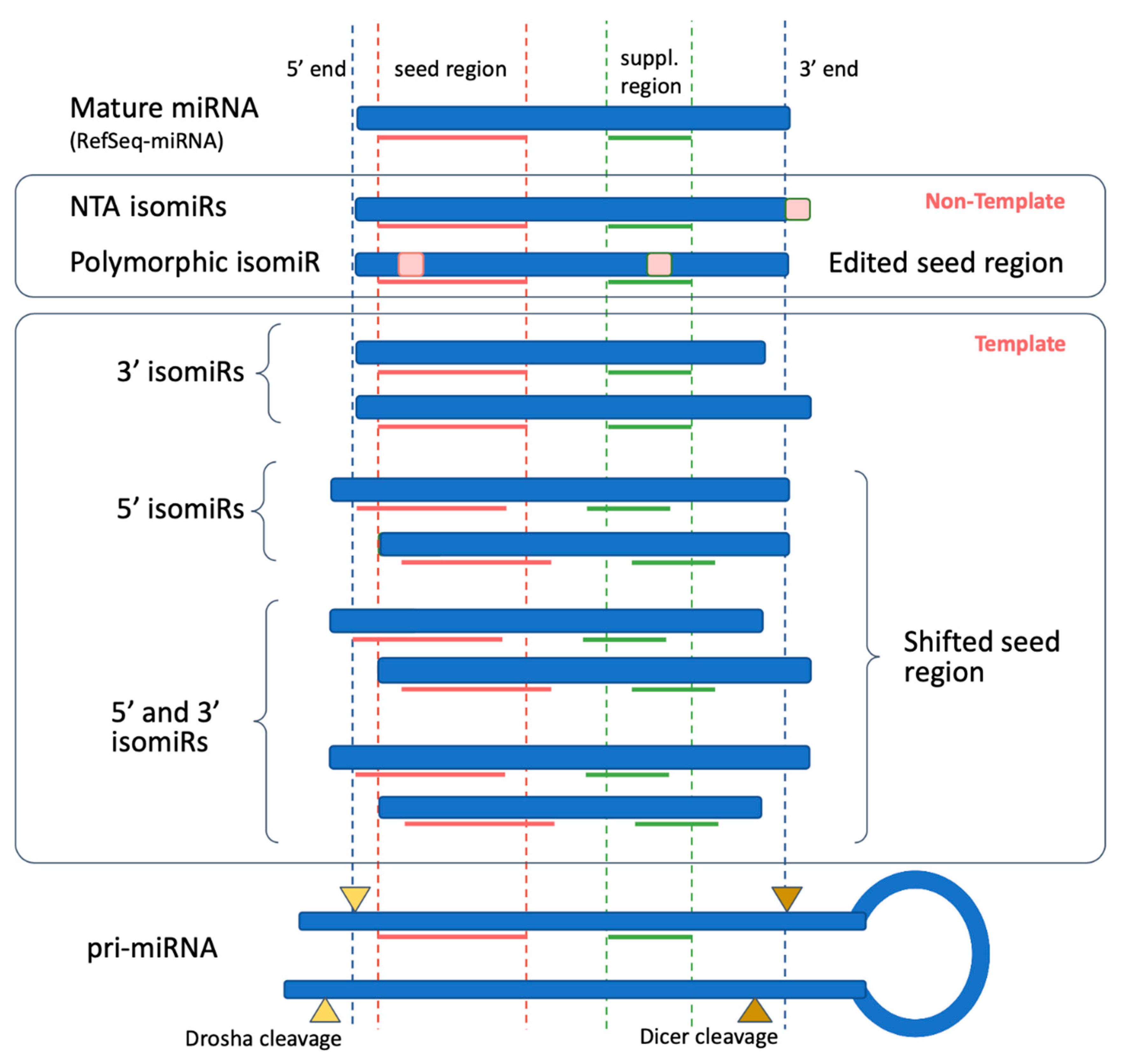
1.3. IsomiR Biogenesis Pathways
1.4. Mode of miRNA Action (miRNA Mediated Gene Expression Regulation)
1.5. IsomiR Biological Implication
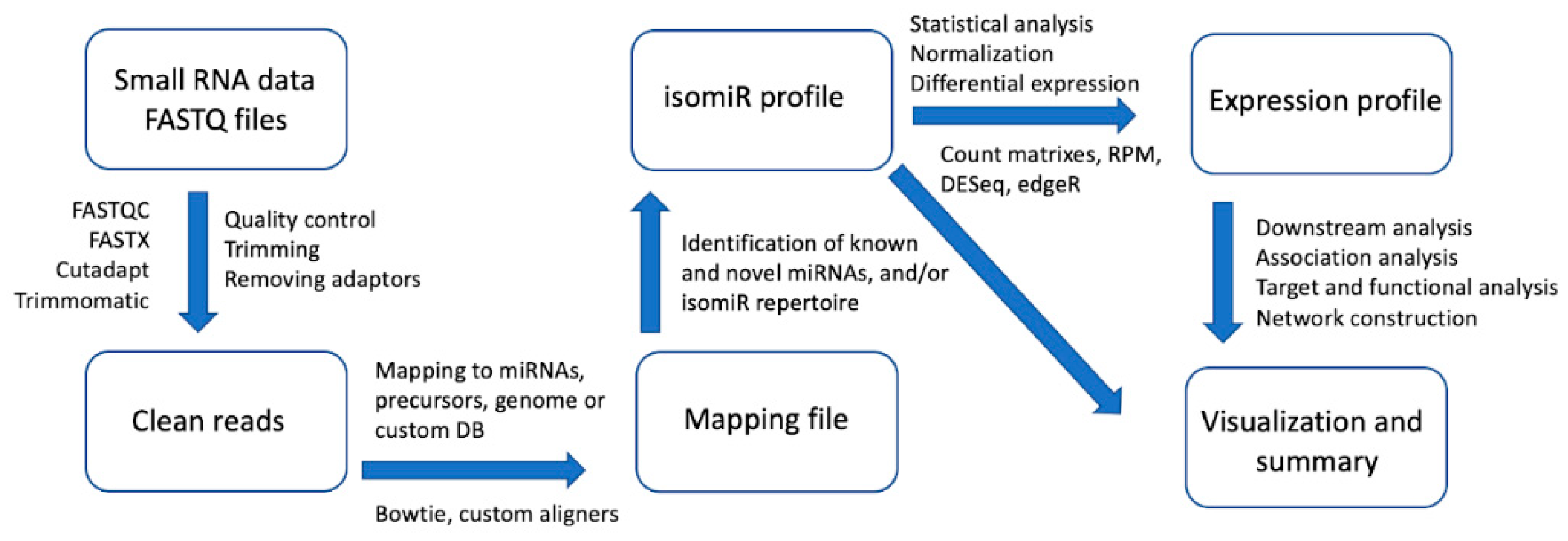
2. Bioinformatics Approaches of Identifying and Analyzing miRNA and isomiR Molecules
3. IsomiRs—A Limitless Source of Potentially Novel Biomarkers
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Rna, S.T.; Hutvágner, G.; Mclachlan, J.; Pasquinelli, A.E.; Bálint, É.; Zamore, P.D.; Rock, K.; Hutvagner, G.; Mclachlan, J.; Pasquinelli, A.E.; et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Mi, R.S.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Du, T.; Zamore, P.D. microPrimer: The biogenesis and function of microRNA. Development 2005, 132, 4645–4652. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Erratum: Functional siRNAs and miRNAs Exhibit Strand Bias (Cell 115 (209-216)). Cell 2003, 115, 505. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Katsura, A.; Yasuda, T.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol. 2015, 22, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Reichholf, B.; Herzog, V.A.; Fasching, N.; Manzenreither, R.A.; Sowemimo, I.; Ameres, S.L. Time-Resolved Small RNA Sequencing Unravels the Molecular Principles of MicroRNA Homeostasis. Mol. Cell 2019, 75, 756–768.e7. [Google Scholar] [CrossRef]
- Yang, J.S.; Maurin, T.; Lai, E.C. Functional parameters of Dicer-independent microRNA biogenesis. RNA 2012, 18, 945–957. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of dicer requires argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Chung, W.J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian Mirtron Genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.L.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs—The overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.; Ellis, P.; Langford, C.F.; et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. MiRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015, 31, 613–626. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, 1–6. [Google Scholar] [CrossRef]
- Loher, P.; Londin, E.R.; Rigoutsos, I. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget 2014, 5, 8790–8802. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Jing, Y.; Londin, E.; Rigoutsos, I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015, 43, 9158–9175. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005, 24, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ye, C.; Ramirez, D.; Manjunath, N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Jeong, K.; Kim, V.N. Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Mol. Cell 2017, 66, 258–269.e5. [Google Scholar] [CrossRef]
- Wyman, S.K.; Knouf, E.C.; Parkin, R.K.; Fritz, B.R.; Lin, D.W.; Dennis, L.M.; Krouse, M.A.; Webster, P.J.; Tewari, M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011, 21, 1450–1461. [Google Scholar] [CrossRef]
- Katoh, T.; Sakaguchi, Y.; Miyauchi, K.; Suzuki, T.; Suzuki, T.; Kashiwabara, S.I.; Baba, T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009, 23, 433–438. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Ando, Y.; De Hoon, M.J.L.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010, 20, 1398–1410. [Google Scholar] [CrossRef]
- Boele, J.; Persson, H.; Shin, J.W.; Ishizu, Y.; Newie, I.S.; Søkilde, R.; Hawkins, S.M.; Coarfa, C.; Ikeda, K.; Takayama, K.I.; et al. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11467–11472. [Google Scholar] [CrossRef]
- Katoh, T.; Hojo, H.; Suzuki, T. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015, 43, 7521–7534. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Kim, Y.K.; Ha, M.; Yoon, M.J.; Cho, J.; Yeom, K.H.; Han, J.; Kim, V.N. TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef]
- Hagan, J.P.; Piskounova, E.; Gregory, R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009, 16, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.J.; Park, J.E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Du, P.; Jing, L.; Sjekloca, L.; Lin, S.; Grossi, E.; Sliz, P.; Zon, L.I.; Gregory, R.I. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 2014, 42, 11777–11791. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 2007, 315, 1137–1140. [Google Scholar] [CrossRef]
- Bass, B.L.; Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988, 55, 1089–1098. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, V.N. MicroRNA Factory: RISC Assembly from Precursor MicroRNAs. Mol. Cell 2012, 46, 384–386. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Bukhari, S.I.A.; Truesdell, S.S.; Lee, S.; Kollu, S.; Classon, A.; Boukhali, M.; Jain, E.; Mortensen, R.D.; Yanagiya, A.; Sadreyev, R.I.; et al. A Specialized Mechanism of Translation Mediated by FXR1a-Associated MicroRNP in Cellular Quiescence. Mol. Cell 2016, 61, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Truesdell, S.S.; Mortensen, R.D.; Seo, M.; Schroeder, J.C.; Lee, J.H.; Letonqueze, O.; Vasudevan, S.V. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep. 2012, 2, 842. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein mRNAs and Enhances Their Translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Valverde, S.L.; Taft, R.J.; Mattick, J.S. Dynamic isomiR regulation in Drosophila development. Rna 2010, 16, 1881–1888. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.M.; D’Ambrogio, A.; Nottrott, S.; Richter, J.D. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 2011, 473, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Khudayberdiev, S.A.; Zampa, F.; Rajman, M.; Schratt, G. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front. Mol. Neurosci. 2013, 6, 1–19. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef]
- Yamane, D.; Selitsky, S.R.; Shimakami, T.; Li, Y.; Zhou, M.; Honda, M.; Sethupathy, P.; Lemon, S.M. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3′ variants. Nucleic Acids Res. 2017, 45, 4743–4755. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Humphreys, D.T.; Hynes, C.J.; Patel, H.R.; Wei, G.H.; Cannon, L.; Fatkin, D.; Suter, C.M.; Clancy, J.L.; Preiss, T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Salem, O.; Erdem, N.; Jung, J.; Münstermann, E.; Wörner, A.; Wilhelm, H.; Wiemann, S.; Körner, C. The highly expressed 5′isomiR of hsa-miR-140-3p contributes to the tumor-suppressive effects of miR-140 by reducing breast cancer proliferation and migration. BMC Genom. 2016, 17, 566. [Google Scholar] [CrossRef] [PubMed]
- Mercey, O.; Popa, A.; Cavard, A.; Paquet, A.; Chevalier, B.; Pons, N.; Magnone, V.; Zangari, J.; Brest, P.; Zaragosi, L.E.; et al. Characterizing isomiR variants within the microRNA-34/449 family. FEBS Lett. 2017, 591, 693–705. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Sdassi, N.; Silveri, L.; Laubier, J.; Tilly, G.; Costa, J.; Layani, S.; Vilotte, J.L.; Le Provost, F. Identification and characterization of new miRNAs cloned from normal mouse mammary gland. BMC Genom. 2009, 10, 149. [Google Scholar] [CrossRef]
- Lee, L.W.; Zhang, S.; Etheridge, A.; Ma, L.; Martin, D.; Galas, D.; Wang, K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 2010, 16, 2170–2180. [Google Scholar] [CrossRef]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011, 12. [Google Scholar] [CrossRef]
- Ziemann, M.; Kaspi, A.; El-Osta, A. Evaluation of microRNA alignment techniques. RNA 2016, 22, 1120–1138. [Google Scholar] [CrossRef]
- Fehlmann, T.; Backes, C.; Kahraman, M.; Haas, J.; Ludwig, N.; Posch, A.E.; Würstle, M.L.; Hübenthal, M.; Franke, A.; Meder, B.; et al. Web-based NGS data analysis using miRMaster: A large-scale meta-analysis of human miRNAs. Nucleic Acids Res. 2017, 45, 8731–8744. [Google Scholar] [CrossRef]
- Zhao, S.; Gordon, W.; Du, S.; Zhang, C.; He, W.; Xi, L.; Mathur, S.; Agostino, M.; Paradis, T.; von Schack, D.; et al. QuickMIRSeq: A pipeline for quick and accurate quantification of both known miRNAs and isomiRs by jointly processing multiple samples from microRNA sequencing. BMC Bioinform. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Giurato, G.; De Filippo, M.R.; Rinaldi, A.; Hashim, A.; Nassa, G.; Ravo, M.; Rizzo, F.; Tarallo, R.; Weisz, A. iMir: An integrated pipeline for high-throughput analysis of small non-coding RNA data obtained by smallRNA-Seq. BMC Bioinform. 2013, 14, 362. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yu, J.; Liang, T.; Zou, Q. MIR-isomiRExp: A web-server for the analysis of expression of miRNA at the miRNA/isomiR levels. Sci. Rep. 2016, 6, 23700. [Google Scholar] [CrossRef]
- Aparicio, E.L.; Rueda, A.; Fromm, B.; Gómez-Martín, C.; Lebrón, R.; Oliver, J.L.; Marchal, J.A.; Kotsyfakis, M.; Hackenberg, M. MiRNAgFree: Prediction and profiling of novel microRNAs without genome assembly. bioRxiv 2017. [Google Scholar] [CrossRef]
- De Oliveira, L.F.V.; Christoff, A.P.; Margis, R. isomiRID: A framework to identify microRNA isoforms. Bioinformatics 2013, 29, 2521–2523. [Google Scholar] [CrossRef]
- Kuenne, C.; Preussner, J.; Herzog, M.; Braun, T.; Looso, M. MIRPIPE: Quantification of microRNAs in niche model organisms. Bioinformatics 2014, 30, 3412–3413. [Google Scholar] [CrossRef]
- Wu, C.W.; Evans, J.M.; Huang, S.; Mahoney, D.W.; Dukek, B.A.; Taylor, W.R.; Yab, T.C.; Smyrk, T.C.; Jen, J.; Kisiel, J.B.; et al. A Comprehensive Approach to Sequence-oriented IsomiR annotation (CASMIR): Demonstration with IsomiR profiling in colorectal neoplasia. BMC Genom. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Urgese, G.; Paciello, G.; Acquaviva, A.; Ficarra, E. IsomiR-SEA: An RNA-Seq analysis tool for miRNAs/isomiRs expression level profiling and miRNA-mRNA interaction sites evaluation. BMC Bioinform. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zang, Q.; Zhang, H.; Ban, R.; Yang, Y.; Iqbal, F.; Li, A.; Shi, Q. DeAnnIso: A tool for online detection and annotation of isomiRs from small RNA sequencing data. Nucleic Acids Res. 2016, 44, W166–W175. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.; Marzi, M.J.; Nicassio, F. IsomiRage: From Functional Classification to Differential Expression of miRNA Isoforms. Front. Bioeng. Biotechnol. 2014, 2, 38. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Chen, K.; Chen, S.; Tesic, N.; Randjelovic, D.; Skundric, N.; Nesic, S.; Varjacic, V.; Williams, E.H.; Malhotra, R.; et al. QuagmiR: A cloud-based application for isomiR big data analytics. Bioinformatics 2019, 35, 1576–1578. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Pla, A.; Rayner, S. Jasmine: A Java pipeline for isomiR characterization in miRNA-Seq data. Bioinformatics 2020, 36, 1933–1936. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, R.K.; Chiesa, M.; Bellazzi, R.; Colombo, G.I. CBS-miRSeq: A comprehensive tool for accurate and extensive analyses of microRNA-sequencing data. Comput. Biol. Med. 2019, 110, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Rueda, A.; Barturen, G.; Lebrón, R.; Gómez-Martín, C.; Alganza, Á.; Oliver, J.L.; Hackenberg, M. SRNAtoolbox: An integrated collection of small RNA research tools. Nucleic Acids Res. 2015, 43, W467–W473. [Google Scholar] [CrossRef]
- Wu, X.; Kim, T.K.; Baxter, D.; Scherler, K.; Gordon, A.; Fong, O.; Etheridge, A.; Galas, D.J.; Wang, K. SRNAnalyzer-A flexible and customizable small RNA sequencing data analysis pipeline. Nucleic Acids Res. 2017, 45, 12140–12151. [Google Scholar] [CrossRef]
- Shi, J.; Dong, M.; Li, L.; Liu, L.; Luz-Madrigal, A.; Tsonis, P.A.; Del Rio-Tsonis, K.; Liang, C. MirPRo-a novel standalone program for differential expression and variation analysis of miRNAs. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Baras, A.S.; Mitchell, C.J.; Myers, J.R.; Gupta, S.; Weng, L.C.; Ashton, J.M.; Cornish, T.C.; Pandey, A.; Halushka, M.K. MiRge—A multiplexed method of processing small RNA-seq data to determine MicroRNA entropy. PLoS ONE 2015, 10, e0143066. [Google Scholar] [CrossRef]
- Zhang, H.; e Silva, B.V.R.; Cui, J. miRDis: A Web tool for endogenous and exogenous microRNA discovery based on deep-sequencing data analysis. Brief. Bioinform. 2018, 19, 415–424. [Google Scholar] [CrossRef]
- Kaushik, A.; Saraf, S.; Mukherjee, S.K.; Gupta, D. miRMOD: A tool for identification and analysis of 5′ and 3′ miRNA modifications in Next Generation Sequencing small RNA data. PeerJ 2015, 2015. [Google Scholar] [CrossRef][Green Version]
- Wu, J.; Liu, Q.; Wang, X.; Zheng, J.; Wang, T.; You, M.; Sun, Z.S.; Shi, Q. MirTools 2.0 for non-coding RNA discovery, profiling and functional annotation based on high-throughput sequencing. RNA Biol. 2013, 10, 1087–1092. [Google Scholar] [CrossRef]
- Vitsios, D.M.; Enright, A.J. Chimira: Analysis of small RNA sequencing data and microRNA modifications. Bioinformatics 2015, 31, 3365–3367. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Loher, P.; Eilbeck, K.; Ma, J.; Urgese, G.; Fromm, B.; Sydes, J.; Aparicio-Puerta, E.; Barrera, V.; Espín, R.; et al. Unification of miRNA and isomiR research: The mirGFF3 format and the mirtop API. bioRxiv 2018, 36, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, T.; Batzel, P.; Sydes, J.; Eames, B.F.; Postlethwait, J.H. miRNA analysis with Prost! reveals evolutionary conservation of organ-enriched expression and post-transcriptional modifications in three-spined stickleback and zebrafish. Sci. Rep. 2019, 9, 3913. [Google Scholar] [CrossRef] [PubMed]
- Thibord, F.; Perret, C.; Roux, M.; Suchon, P.; Germain, M.; Deleuze, J.F.; Morange, P.E.; Trégouët, D.A. OPTIMIR, a novel algorithm for integrating available genome-wide genotype data into miRNA sequence alignment analysis. RNA 2019, 25, 657–668. [Google Scholar] [CrossRef]
- Pantano, L.; Estivill, X.; Martí, E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2009, 38. [Google Scholar] [CrossRef]
- Guo, L.; Liang, T.; Yu, J.; Zou, Q. A comprehensive analysis of miRNA/isomiR expression with gender difference. PLoS ONE 2016, 11, e0154955. [Google Scholar] [CrossRef]
- Martí, E.; Pantano, L.; Bañez-Coronel, M.; Llorens, F.; Miñones-Moyano, E.; Porta, S.; Sumoy, L.; Ferrer, I.; Estivill, X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010, 38, 7219–7235. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Y.; Li, M.; Tu, J.; Lu, Z. Dysregulation of miRNA isoform level at 5′ end in Alzheimer’s disease. Gene 2016, 584, 167–172. [Google Scholar] [CrossRef]
- Haseeb, A.; Makki, M.S.; Khan, N.M.; Ahmad, I.; Haqqi, T.M. Deep sequencing and analyses of miRNAs, isomiRs and miRNA induced silencing complex (miRISC)-associated miRNome in primary human chondrocytes. Sci. Rep. 2017, 7, 15178. [Google Scholar] [CrossRef]
- van der Kwast, R.V.C.T.; Woudenberg, T.; Quax, P.H.A.; Nossent, A.Y. MicroRNA-411 and Its 5′-IsomiR Have Distinct Targets and Functions and Are Differentially Regulated in the Vasculature under Ischemia. Mol. Ther. 2019, 28, 157–170. [Google Scholar] [CrossRef]
- Ormseth, M.J.; Solus, J.F.; Sheng, Q.; Ye, F.; Wu, Q.; Guo, Y.; Oeser, A.M.; Allen, R.M.; Vickers, K.C.; Stein, C.M. Development and validation of a MicroRNA panel to differentiate between patients with rheumatoid arthritis or systemic lupus erythematosus and controls. J. Rheumatol. 2020, 47, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Siddle, K.J.; Tailleux, L.; Deschamps, M.; Loh, Y.H.E.; Deluen, C.; Gicquel, B.; Antoniewski, C.; Barreiro, L.B.; Farinelli, L.; Quintana-Murci, L. Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs. PLoS Genet. 2015, 11, e1005064. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef] [PubMed]
- Parafioriti, A.; Cifola, I.; Gissi, C.; Pinatel, E.; Vilardo, L.; Armiraglio, E.; Di Bernardo, A.; Daolio, P.A.; Felsani, A.; D’Agnano, I.; et al. Expression profiling of microRNAs and isomiRs in conventional central chondrosarcoma. Cell Death Discov. 2020, 6. [Google Scholar] [CrossRef]
- Li, S.C.; Liao, Y.L.; Ho, M.R.; Tsai, K.W.; Lai, C.H.; Lin, W.C. MiRNA arm selection and isomiR distribution in gastric cancer. Ser. Adv. Bioinforma. Comput. Biol. 2012, 13, S13. [Google Scholar] [CrossRef]
- Babapoor, S.; Fleming, E.; Wu, R.; Dadras, S.S. A novel mir-451a isomir, associated with amelanotypic phenotype, acts as a tumor suppressor in melanoma by retarding cell migration and invasion. PLoS ONE 2014, 9, e120. [Google Scholar] [CrossRef]
- Ni, H.; Dai, X.; Leng, X.; Deng, M.; Qin, Y.; Ji, Q.; Xu, C.; Li, J.; Liu, Y. Higher variety and quantity of microRNA-139-5p isoforms confer suppressive role in hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 6806–6813. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Z.; Chen, P.; Wu, M. Tumor classification and biomarker discovery based on the 5′isomiR expression level. BMC Cancer 2019, 19, 127. [Google Scholar] [CrossRef]
- Koi, Y.; Tsutani, Y.; Nishiyama, Y.; Ueda, D.; Ibuki, Y.; Sasada, S.; Akita, T.; Masumoto, N.; Kadoya, T.; Yamamoto, Y.; et al. Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci. 2020, 111, 2104–2115. [Google Scholar] [CrossRef]
- Ibuki, Y.; Nishiyama, Y.; Tsutani, Y.; Emi, M.; Hamai, Y.; Okada, M.; Tahara, H. Circulating microRNA/isomiRs as novel biomarkers of esophageal squamous cell carcinoma. PLoS ONE 2020, 15, e0231116. [Google Scholar] [CrossRef]
- Londin, E.; Magee, R.; Shields, C.L.; Lally, S.E.; Sato, T.; Rigoutsos, I. IsomiRs and tRNA-derived fragments are associated with metastasis and patient survival in uveal melanoma. Pigment Cell Melanoma Res. 2020, 33, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Li, D.; Wang, X.; Li, L.; Zou, Q. Cancer Diagnosis Through IsomiR Expression with Machine Learning Method. Curr. Bioinform. 2016, 13, 57–63. [Google Scholar] [CrossRef]
- Plé, H.; Landry, P.; Benham, A.; Coarfa, C.; Gunaratne, P.H.; Provost, P. The Repertoire and Features of Human Platelet microRNAs. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Geekiyanage, H.; Rayatpisheh, S.; Wohlschlegel, J.A.; Brown, R.; Ambros, V. Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc. Natl. Acad. Sci. USA 2020, 117, 24213–24223. [Google Scholar] [CrossRef] [PubMed]
- Lunavat, T.R.; Cheng, L.; Kim, D.K.; Bhadury, J.; Jang, S.C.; Lässer, C.; Sharples, R.A.; López, M.D.; Nilsson, J.; Gho, Y.S.; et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–Evidence of unique microRNA cargos. RNA Biol. 2015, 12, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Menezes, R. De Non-invasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016, 7, 22566–22578. [Google Scholar] [CrossRef]
- Karlsen, T.A.; Aae, T.F.; Brinchmann, J.E. Robust profiling of microRNAs and isomiRs in human plasma exosomes across 46 individuals. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Vaz, C.; Ahmad, H.M.; Bharti, R.; Pandey, P.; Kumar, L.; Kulshreshtha, R.; Bhattacharya, A. Analysis of the microRNA transcriptome and expression of different isomiRs in human peripheral blood mononuclear cells. BMC Res. Notes 2013, 6, 1. [Google Scholar] [CrossRef]
| Tool Name | Type | Tool Features | Year | Authors | Journal/URL | Reference (DOI Number) |
|---|---|---|---|---|---|---|
| miRMaster | Web |
| 2017 | Tobias Fehlmann et al. | Nucleic Acids Research Open Access https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5587802/ | 10.1093/nar/gkx595 [80] |
| QuickMIRSeq | Linux |
| 2017 | Shanrong Zhao et al. | BMC Bioinformatics Open Access https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5359966/ | 10.1186/s12859-017-1601-4 [81] |
| iMir | Linux Mac |
| 2013 | Giorgio Giurato et al. | BMC Bioinformatics Open access https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-14-362 | 10.1186/1471-2105-14-362 [82] |
| MIR-isomiRExp | Web |
| 2016 | Li Guo et al. | Scientific Reports Open Access https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4806314/ | 10.1038/srep23700 [83] |
| miRNAgFree | Linux Windows |
| 2017 | EL Aparicio et al. | BioRxiv https://www.biorxiv.org/content/10.1101/193094v1.full | 10.1101/193094 [84] |
| isomiRID | Linux Mac |
| 2013 | Luiz Felipe Valter de Oliveira et al. | Bioinformatics https://academic.oup.com/bioinformatics/article/29/20/2521/276800 | 10.1093/bioinformatics/btt424 [85] |
| MIRPIPE | Web |
| 2014 | Carsten Kuenne et al. | Bioinformatics https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4816158/ | 10.1093/bioinformatics/btu573 [86] |
| CASMIR | Standalone, Linux |
| 2018 | Chung Wah Wu et al. | BMC Genomics Open Access https://www.ncbi.nlm.nih.gov/pubmed/29801434 | 10.1186/s12864-018-4794-7 [87] |
| IsomiR-SEA | Linux Windows Mac |
| 2016 | Gianvito Urgese et al. | BMC Bioinformatics https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-0958-0 | 10.1186/s12859-016-0958-0 [88] |
| DeAnnIso | Web |
| 2016 | Yuanwei Zhang et al. | Nucleic Acids Research https://www.ncbi.nlm.nih.gov/pubmed/27179030 | 10.1093/nar/gkw427 [89] |
| IsomiRage | Windows Mac |
| 2014 | Muller Heiko et al. | Frontiers in Bioengineering and Biotechnology https://www.frontiersin.org/articles/10.3389/fbioe.2014.00038/full | 10.3389/fbioe.2014.00038 [90] |
| QuagmiR | Standalone, Web |
| 2018 | Xavier Bofill-De Ros et al. | Bioimformatics https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6499244/ | 10.1093/bioinformatics/bty843 [91] |
| Jasmine | R + Java |
| 2019 | Xiangfu Zhong et al. | Bioinformatics https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btz806/5612093 | 10.1093/bioinformatics/btz806 [92] |
| CBS-miRSeq | Linux, Docker VM |
| 2019 | Rupesh K. Kesharwani et al. | Computers in Biology and Medicine https://www.sciencedirect.com/science/article/pii/S001048251930188X | 10.1016/j.compbiomed.2019.05.019. [93] |
| sRNAtoolbox | Web |
| 2015 | Antonio Rueda et al. | Nucleic Acids Research https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4489306/pdf/gkv555.pdf | 10.1093/nar/gkv555 [94] |
| sRNAnalyzer | Web |
| 2017 | Xiaogang Wu et al. | Nucleic Acids Research Open Access https://www.ncbi.nlm.nih.gov/pubmed/29069500 | 10.1093/nar/gkx999 [95] |
| mirPRo | Linux |
| 2015 | Jieming Shi et al. | Scientific Reports Open Access https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4592965/ | 10.1038/srep14617 [96] |
| miRge | Standalone |
| 2015 | Alexander S. Baras et al. | Plos one Open Access https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0143066 | 10.1371/journal.pone.0143066 [97] |
| miRDis | Web |
| 2017 | Hanyuan Zhang et al. | Briefings in Bioinformatics Open Access https://www.ncbi.nlm.nih.gov/pubmed/28073746 | 10.1093/bib/bbw140 [98] |
| miRMOD | Windows |
| 2015 | Abhinav Kaushik et al. | PeerJ Open Access https://www.ncbi.nlm.nih.gov/pubmed/26623179 | 10.7717/peerj.1332 [99] |
| mirTools 2.0 | Web |
| 2013 | Jinyu Wu et al. | RNA Biology https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3849156/ | 10.4161/rna.25193 [100] |
| Chimira | Web |
| 2015 | Dimitrios M. Vitsios et al. | Bioinformatics https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4595902/ | 10.1093/bioinformatics/btv380 [101] |
| miRTOP | Standalone, Linux |
| 2019 | Thomas Desvignes et al. | Bioinformatics Open Access https://academic.oup.com/bioinformatics/article/36/3/698/5556118 | 10.1093/bioinformatics/btz675 [102] |
| Prost! | Standalone, Linux |
| 2019 | Thomas Desvignes et al. | Scientific Reports Open Access https://www.nature.com/articles/s41598-019-40361-8 | 10.1038/s41598-019-40361-8 [103] |
| OptimiR | Standalone, Linux |
| 2019 | Florian Thibord et al. | RNA Open Access https://rnajournal.cshlp.org/content/early/2019/02/28/rna.069708.118 | 10.1261/rna. 069708.118. [104] |
| isomiRs | R library |
| 2020 | Pantano L., Escaramis G. | http://bioconductor.org/packages/release/bioc/html/isomiRs.html | 10.18129/B9.bioc.isomiRs |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glogovitis, I.; Yahubyan, G.; Würdinger, T.; Koppers-Lalic, D.; Baev, V. isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them? Biomolecules 2021, 11, 41. https://doi.org/10.3390/biom11010041
Glogovitis I, Yahubyan G, Würdinger T, Koppers-Lalic D, Baev V. isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them? Biomolecules. 2021; 11(1):41. https://doi.org/10.3390/biom11010041
Chicago/Turabian StyleGlogovitis, Ilias, Galina Yahubyan, Thomas Würdinger, Danijela Koppers-Lalic, and Vesselin Baev. 2021. "isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them?" Biomolecules 11, no. 1: 41. https://doi.org/10.3390/biom11010041
APA StyleGlogovitis, I., Yahubyan, G., Würdinger, T., Koppers-Lalic, D., & Baev, V. (2021). isomiRs–Hidden Soldiers in the miRNA Regulatory Army, and How to Find Them? Biomolecules, 11(1), 41. https://doi.org/10.3390/biom11010041







