A Computational Approach to Explore the Interaction of Semisynthetic Nitrogenous Heterocyclic Compounds with the SARS-CoV-2 Main Protease
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Receptor and Ligand Structures
2.2. Molecular Docking Simulations
3. Results and Discussion
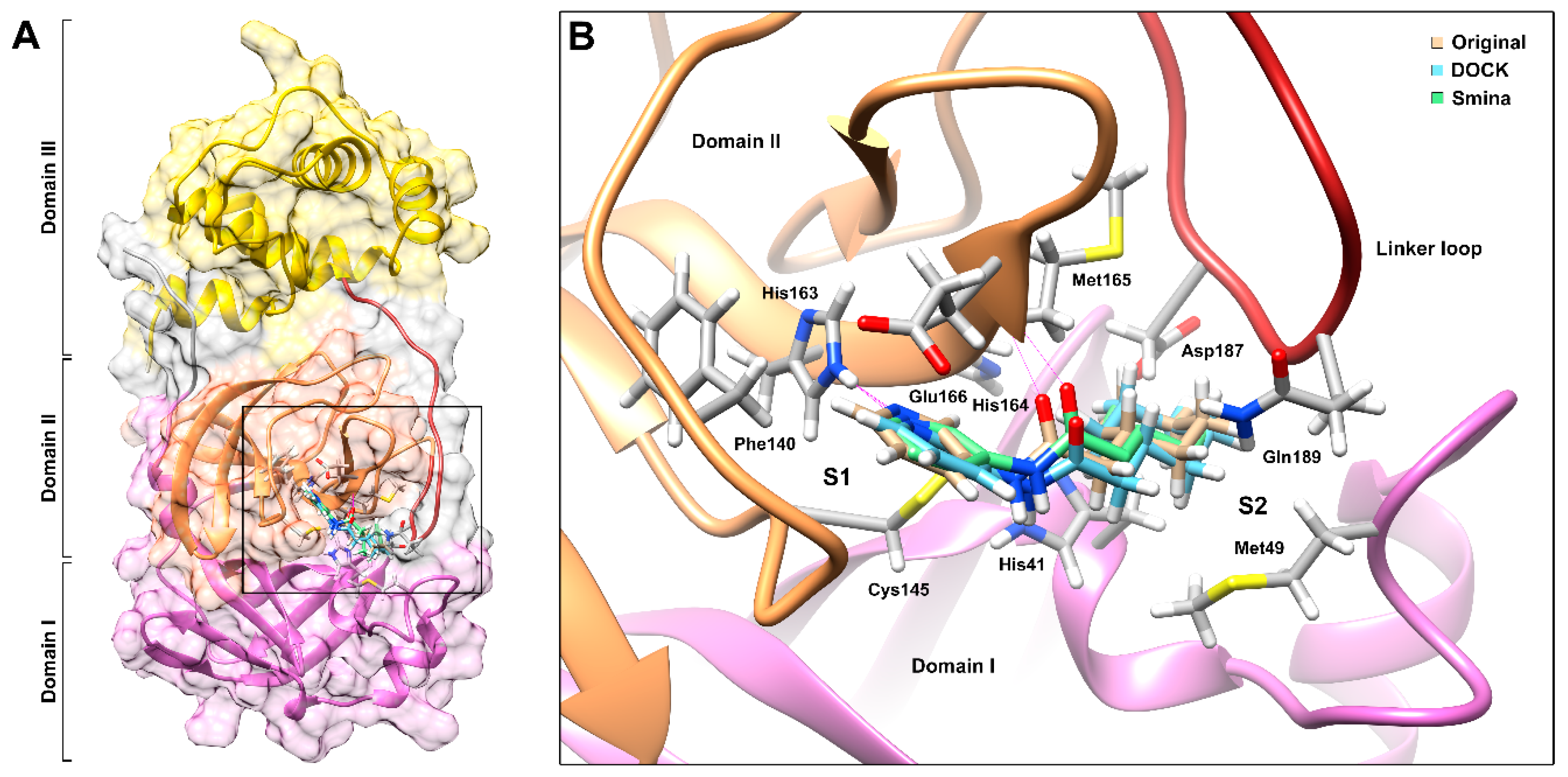
3.1. Validation of Molecular Docking Parameters and Criteria for Selection
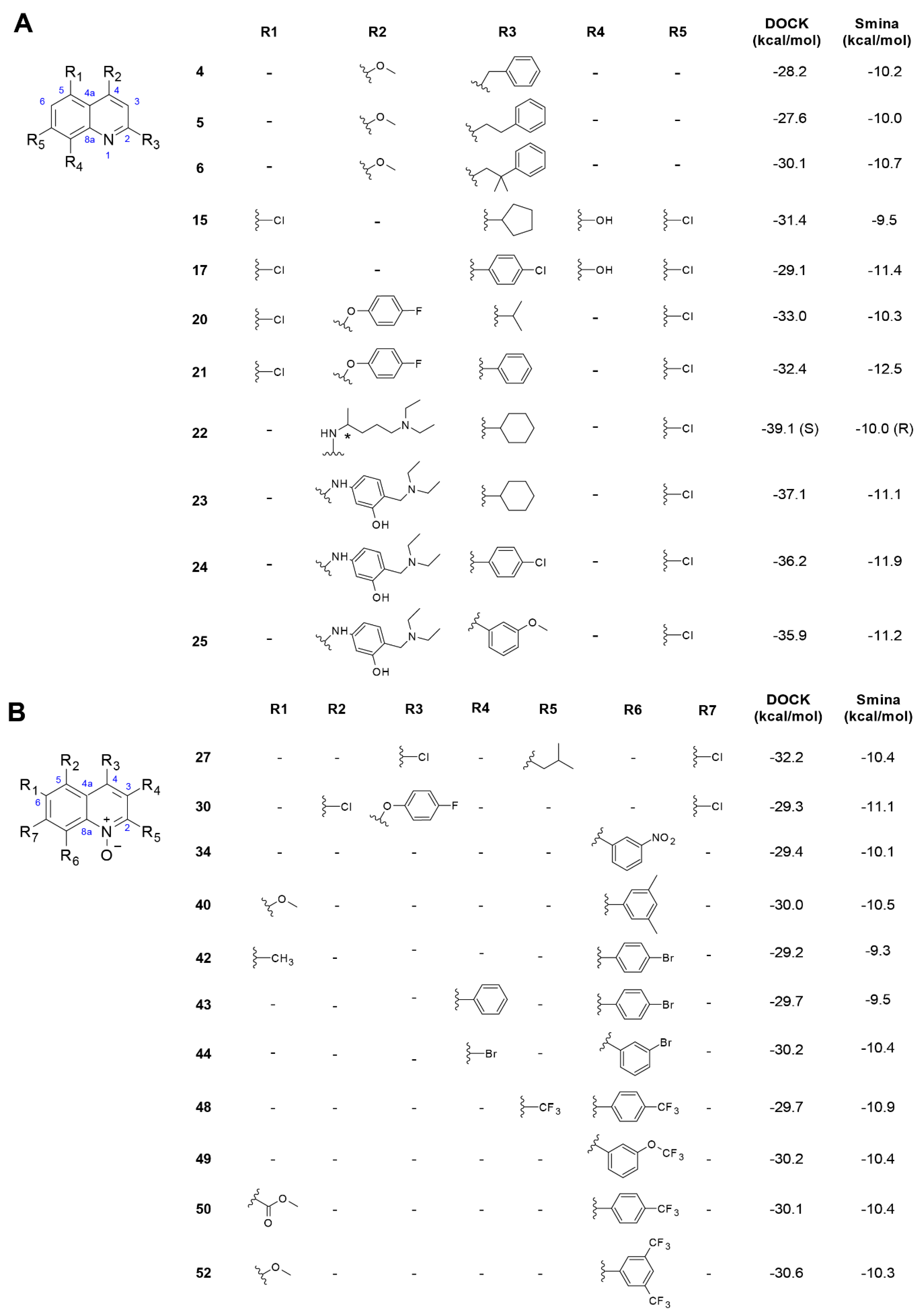
3.2. Quinoline and Quinoline N-Oxide Derivatives
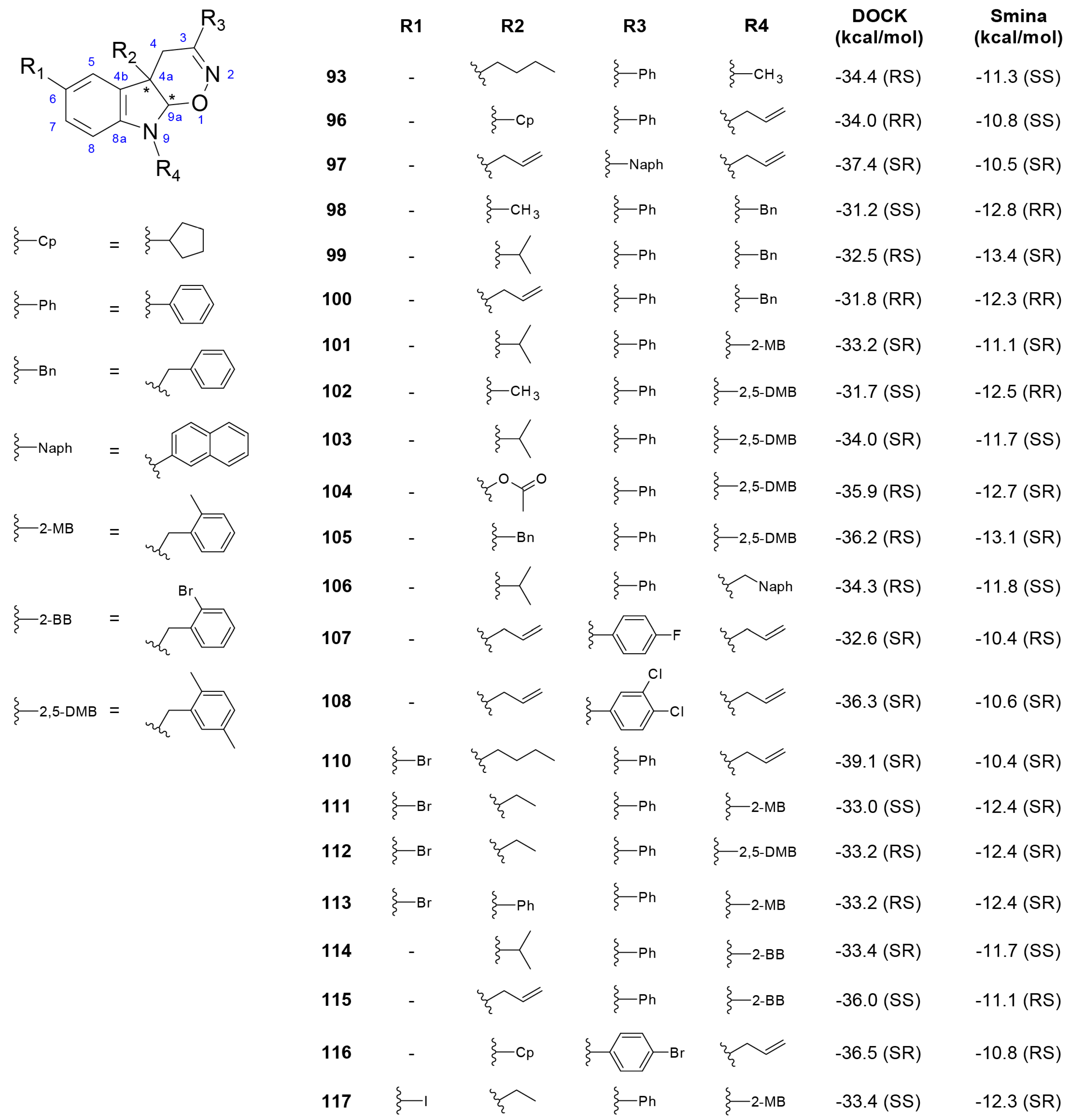
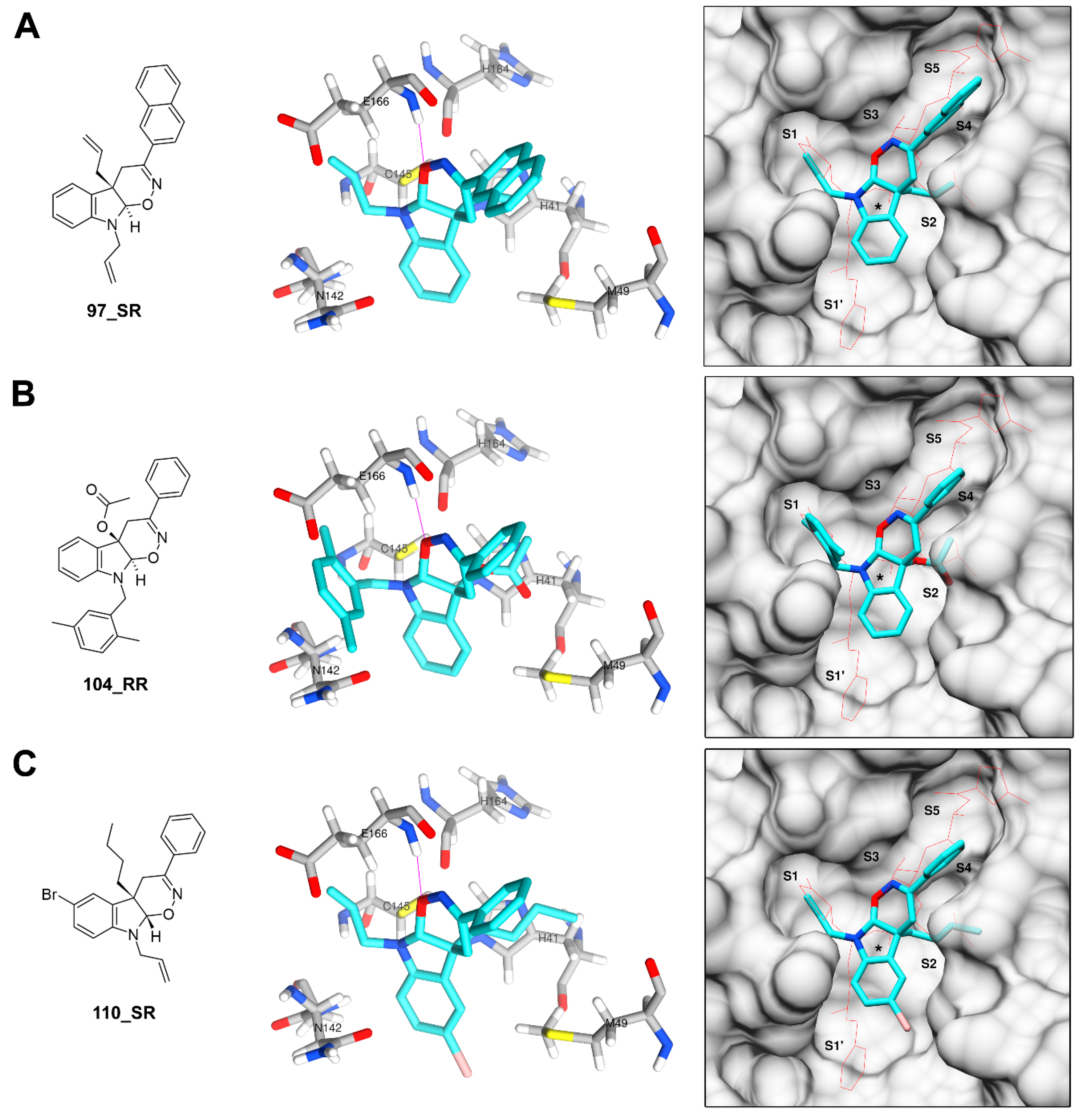
3.3. Derivatives of Hexahydropyrrolo[2,3-b]indole (HPI) Natural Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.-H. An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; et al. Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. J. Virol. 2008, 82, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xie, W.; Xue, X.; Yang, K.; Ma, J.; Liang, W.; Zhao, Q.; Zhou, Z.; Pei, D.; Ziebuhr, J.; et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005, 3, e324. [Google Scholar]
- Wang, J. Fast Identification of Possible Drug Treatment of Coronavirus Disease-19 (COVID-19) through Computational Drug Repurposing Study. J. Chem. Inf. Model. 2020, 60, 3277–3286. [Google Scholar] [CrossRef]
- Chtita, S.; Belhassan, A.; Aouidate, A.; Belaidi, S.; Bouachrine, M.; Lakhlifi, T. Discovery of Potent SARS-CoV-2 Inhibitors from Approved Antiviral Drugs via Docking Screening. Comb. Chem. High Throughput Screen. 2020, 23. [Google Scholar] [CrossRef]
- Peele, K.A.; Potla Durthi, C.; Srihansa, T.; Krupanidhi, S.; Ayyagari, V.S.; Babu, D.J.; Indira, M.; Reddy, A.R.; Venkateswarulu, T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Inform. Med. Unlocked 2020, 19, 100345. [Google Scholar] [CrossRef]
- Mittal, L.; Kumari, A.; Srivastava, M.; Singh, M.; Asthana, S. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Kandeel, M.; Al-Nazawi, M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020, 251, 117627. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Sellner, M.; Neranjan, S.; Smieško, M.; Lill, M.A. Potential Inhibitors for Novel Coronavirus Protease Identified by Virtual Screening of 606 Million Compounds. Int. J. Mol. Sci. 2020, 21, 3626. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, M. Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Open Bio 2020, 10, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.U.; Froeyen, M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharm. Anal. 2020, 10, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, Q.; Wang, Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics 2020, 36, 3295–3298. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, A.A.; Kremer, F.S.; Dos Santos, J.C.; Souza, J.D.; Pinto, L.D.S.; McBride, A.J.A. Discovery of Novel Leptospirosis Vaccine Candidates Using Reverse and Structural Vaccinology. Front. Immunol. 2017, 8, 463. [Google Scholar] [CrossRef]
- Zhang, H.; Saravanan, K.M.; Yang, Y.; Hossain, M.T.; Li, J.; Ren, X.; Pan, Y.; Wei, Y. Deep Learning Based Drug Screening for Novel Coronavirus 2019-nCov. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 368–376. [Google Scholar] [CrossRef]
- Berry, M.; Fielding, B.C.; Gamieldien, J. Potential Broad Spectrum Inhibitors of the Coronavirus 3CLpro: A Virtual Screening and Structure-Based Drug Design Study. Viruses 2015, 7, 6642–6660. [Google Scholar] [CrossRef]
- Lu, I.-L.; Mahindroo, N.; Liang, P.-H.; Peng, Y.-H.; Kuo, C.-J.; Tsai, K.-C.; Hsieh, H.-P.; Chao, Y.-S.; Wu, S.-Y. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J. Med. Chem. 2006, 49, 5154–5161. [Google Scholar] [CrossRef]
- Blanchard, J.E.; Elowe, N.H.; Huitema, C.; Fortin, P.D.; Cechetto, J.D.; Eltis, L.D.; Brown, E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004, 11, 1445–1453. [Google Scholar] [CrossRef]
- Ton, A.-T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 2020, 39, e2000028. [Google Scholar] [CrossRef] [PubMed]
- Vitaku, E.; Smith, D.T.; Njardarson, J.T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014, 57, 10257–10274. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.; Meurice, N.; Kaiser, C.; Maggiora, G. Softening the Rule of Five—where to draw the line? Bioorg. Med. Chem. 2012, 20, 5343–5351. [Google Scholar] [CrossRef] [PubMed]
- Larionov, O.V.; Stephens, D.; Mfuh, A.; Chavez, G. Direct, catalytic, and regioselective synthesis of 2-alkyl-, aryl-, and alkenyl-substituted N -Heterocycles from n -oxides. Org. Lett. 2014, 16, 864–867. [Google Scholar] [CrossRef]
- Mfuh, A.M.; Larionov, O. V Heterocyclic N-Oxides - An Emerging Class of Therapeutic Agents. Curr. Med. Chem. 2015, 22, 2819–2857. [Google Scholar] [CrossRef]
- Larionov, O.V.; Stephens, D.; Mfuh, A.M.; Arman, H.D.; Naumova, A.S.; Chavez, G.; Skenderi, B. Insights into the mechanistic and synthetic aspects of the Mo/P-catalyzed oxidation of N-heterocycles. Org. Biomol. Chem. 2014, 12, 3026–3036. [Google Scholar] [CrossRef]
- Ruiz-Sanchis, P.; Savina, S.A.; Albericio, F.; Álvarez, M. Structure, bioactivity and synthesis of natural products with hexahydropyrrolo[2,3-b]indole. Chemistry 2011, 17, 1388–1408. [Google Scholar] [CrossRef]
- Mfuh, A.M.; Zhang, Y.; Stephens, D.E.; Vo, A.X.T.; Arman, H.D.; Larionov, O.V. Concise Total Synthesis of Trichodermamides A, B, and C Enabled by an Efficient Construction of the 1,2-Oxazadecaline Core. J. Am. Chem. Soc. 2015, 137, 8050–8053. [Google Scholar] [CrossRef]
- Jans, P.E.; Mfuh, A.M.; Arman, H.D.; Shaffer, C.V.; Larionov, O.V.; Mooberry, S.L. Cytotoxicity and Mechanism of Action of the Marine-Derived Fungal Metabolite Trichodermamide B and Synthetic Analogues. J. Nat. Prod. 2017, 80, 676–683. [Google Scholar] [CrossRef]
- Zhang, Y.; Stephens, D.; Hernandez, G.; Mendoza, R.; Larionov, O.V. Catalytic diastereo- and enantioselective annulations between transient nitrosoalkenes and indoles. Chemistry 2012, 18, 16612–16615. [Google Scholar] [CrossRef]
- Herrera, L.; Stephens, D.E.; D’Avila, A.; George, K.G.; Arman, H.; Zhang, Y.; Perry, G.; Lleonart, R.; Larionov, O.V.; Fernández, P.L. Insights into the structural patterns of the antileishmanial activity of bi- and tricyclic N-heterocycles. Org. Biomol. Chem. 2016, 14, 7053–7060. [Google Scholar] [CrossRef] [PubMed]
- Doens, D.; Valdés-Tresanco, M.E.; Vasquez, V.; Carreira, M.B.; De La Guardia, Y.; Stephens, D.E.; Nguyen, V.D.; Nguyen, V.T.; Gu, J.; Hegde, M.L.; et al. Hexahydropyrrolo[2,3-b]indole Compounds as Potential Therapeutics for Alzheimer’s Disease. ACS Chem. Neurosci. 2019, 10, 4250–4263. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J Comput Chem 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Allen, W.J.; Balius, T.E.; Mukherjee, S.; Brozell, S.R.; Moustakas, D.T.; Lang, P.T.; Case, D.A.; Kuntz, I.D.; Rizzo, R.C. DOCK 6: Impact of new features and current docking performance. J. Comput. Chem. 2015, 36, 1132–1156. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Gendrot, M.; Andreani, J.; Boxberger, M.; Jardot, P.; Fonta, I.; Le Bideau, M.; Duflot, I.; Mosnier, J.; Rolland, C.; Bogreau, H.; et al. Antimalarial drugs inhibit the replication of SARS-CoV-2: An in vitro evaluation. Travel Med. Infect. Dis. 2020, 37, 101873. [Google Scholar] [CrossRef]
- Ianevski, A.; Yao, R.; Fenstad, M.H.; Biza, S.; Zusinaite, E.; Reisberg, T.; Lysvand, H.; Løseth, K.; Landsem, V.M.; Malmring, J.F.; et al. Potential antiviral options against SARS-CoV-2 infection. Viruses 2020, 12, 642. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.-M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef]
- WHO Solidarity Trial Consortium; Pan, H.; Peto, R.; Henao-Restrepo, A.-M.; Preziosi, M.-P.; Sathiyamoorthy, V.; Abdool Karim, Q.; Alejandria, M.M.; Hernández García, C.; Kieny, M.-P.; et al. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llanes, A.; Cruz, H.; Nguyen, V.D.; Larionov, O.V.; Fernández, P.L. A Computational Approach to Explore the Interaction of Semisynthetic Nitrogenous Heterocyclic Compounds with the SARS-CoV-2 Main Protease. Biomolecules 2021, 11, 18. https://doi.org/10.3390/biom11010018
Llanes A, Cruz H, Nguyen VD, Larionov OV, Fernández PL. A Computational Approach to Explore the Interaction of Semisynthetic Nitrogenous Heterocyclic Compounds with the SARS-CoV-2 Main Protease. Biomolecules. 2021; 11(1):18. https://doi.org/10.3390/biom11010018
Chicago/Turabian StyleLlanes, Alejandro, Héctor Cruz, Viet D. Nguyen, Oleg V. Larionov, and Patricia L. Fernández. 2021. "A Computational Approach to Explore the Interaction of Semisynthetic Nitrogenous Heterocyclic Compounds with the SARS-CoV-2 Main Protease" Biomolecules 11, no. 1: 18. https://doi.org/10.3390/biom11010018
APA StyleLlanes, A., Cruz, H., Nguyen, V. D., Larionov, O. V., & Fernández, P. L. (2021). A Computational Approach to Explore the Interaction of Semisynthetic Nitrogenous Heterocyclic Compounds with the SARS-CoV-2 Main Protease. Biomolecules, 11(1), 18. https://doi.org/10.3390/biom11010018




