Transcriptome Based Profiling of the Immune Cell Gene Signature in Rat Experimental Colitis and Human IBD Tissue Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vivo Rat Model and Sample Collection
2.2. IBD Patients
2.3. Extraction of Total RNA
2.4. Reverse Transcription and Quantitative Real-Time PCR (QPCR)
2.5. Whole Transcriptome Analysis by RNA-Seq and Bioinformatics
2.6. Pathway Analysis
2.7. Translation of Homologous Immune Signature Gene IDs
2.8. Immune Signature Analysis
2.9. Statistical Analysis, Data Representation and Data Availability
3. Results and Discussion
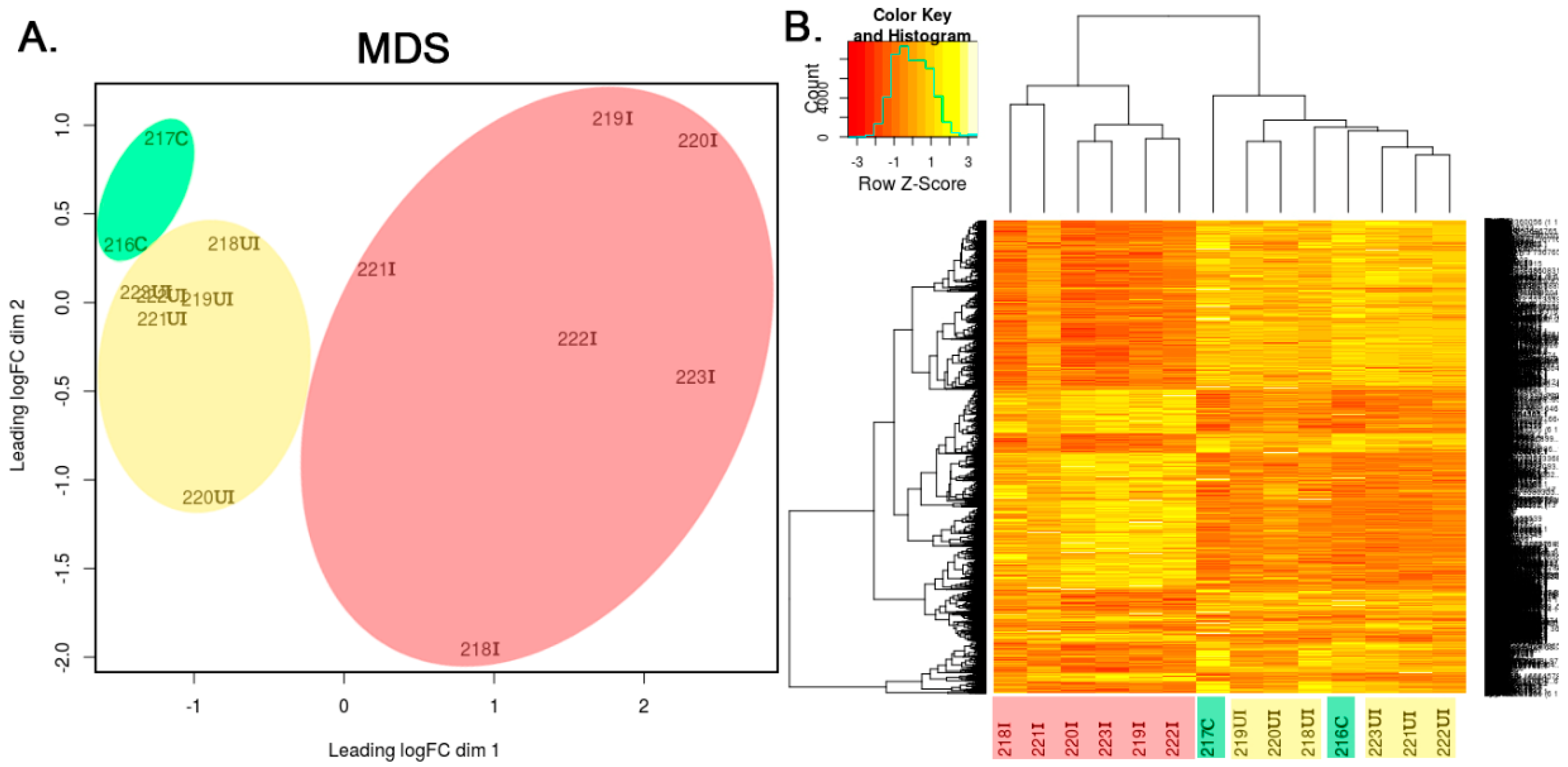
3.1. Global Gene Expression Pattern of In Vivo Model of Colitis
3.2. Correlation between Murine and Human Samples
3.3. In Silico Analysis of Transcriptomic Data
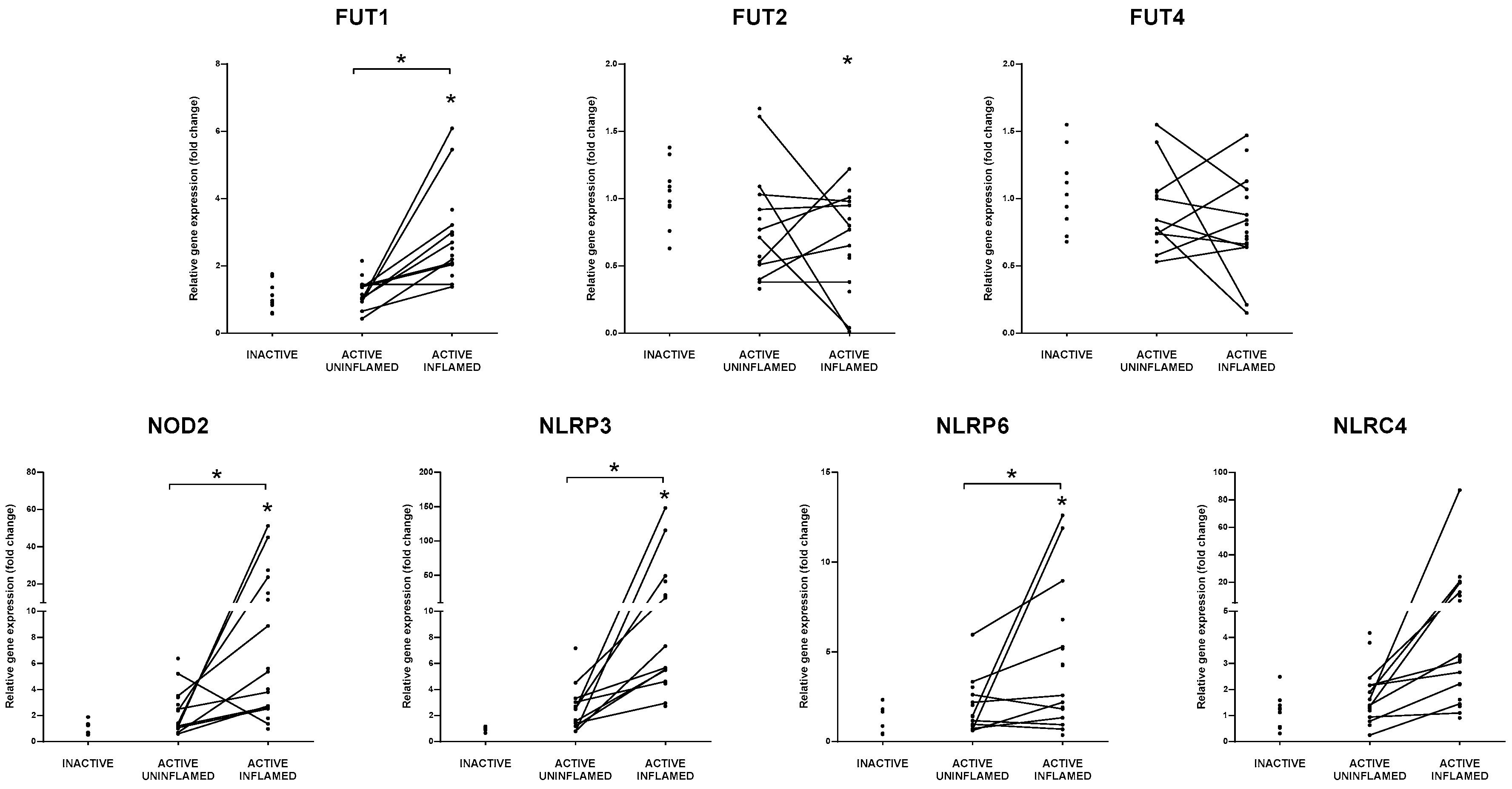
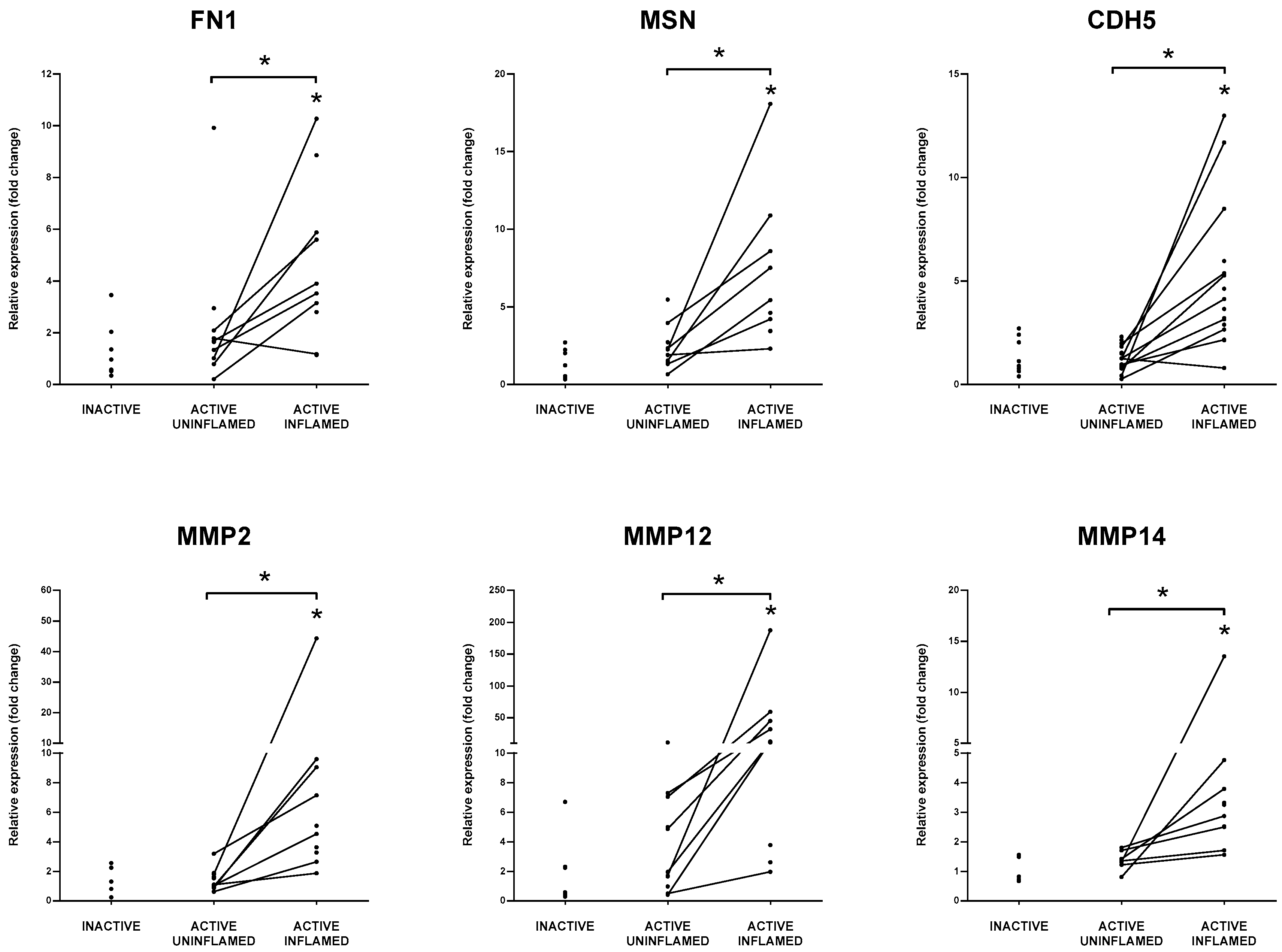
3.4. Genes Influencing Host-Microbiome Relationship during Inflammation Have Altered Expression in IBD Patients
3.5. Balancing between Danger and Tolerance
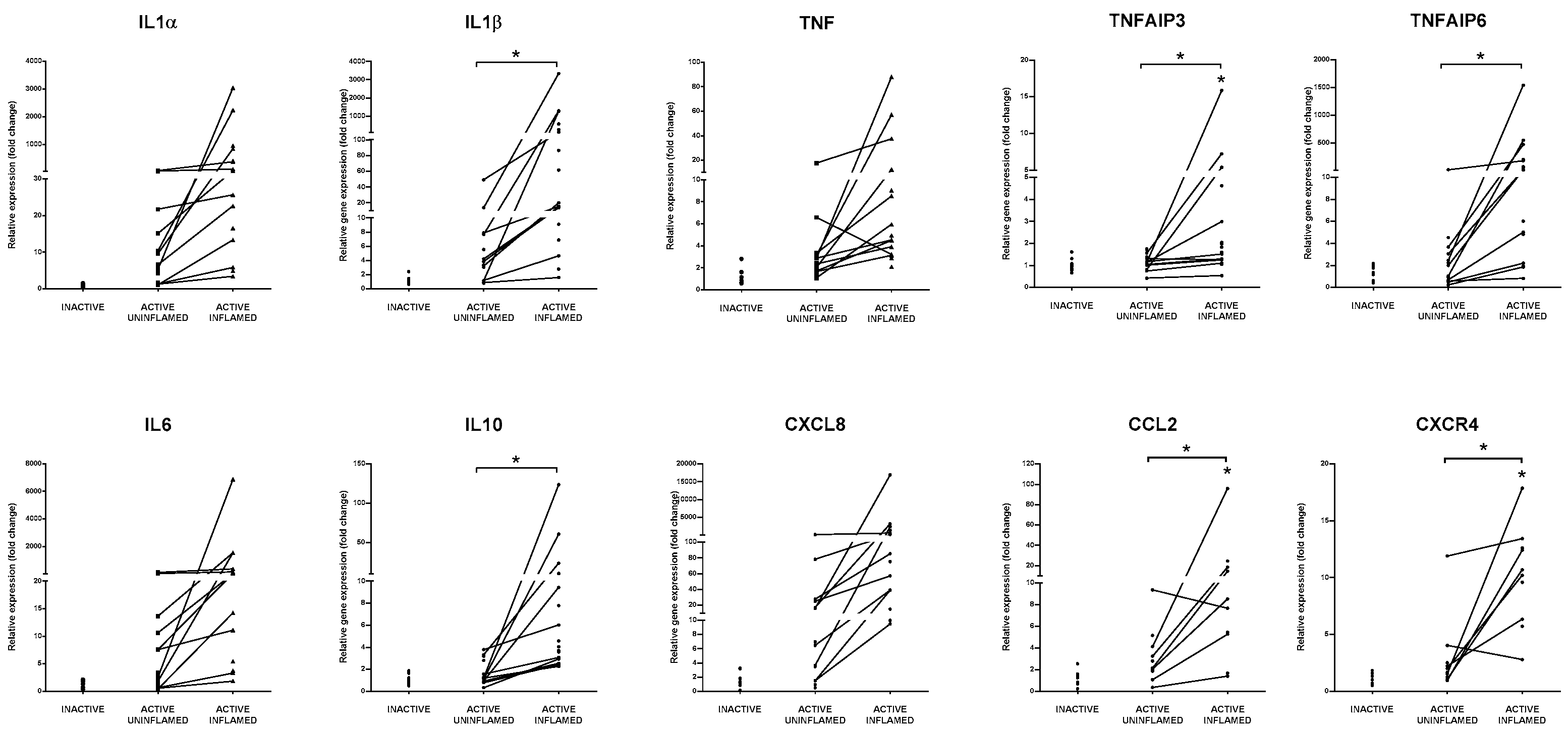
3.6. Burst of Cytokine and Chemokine Expression
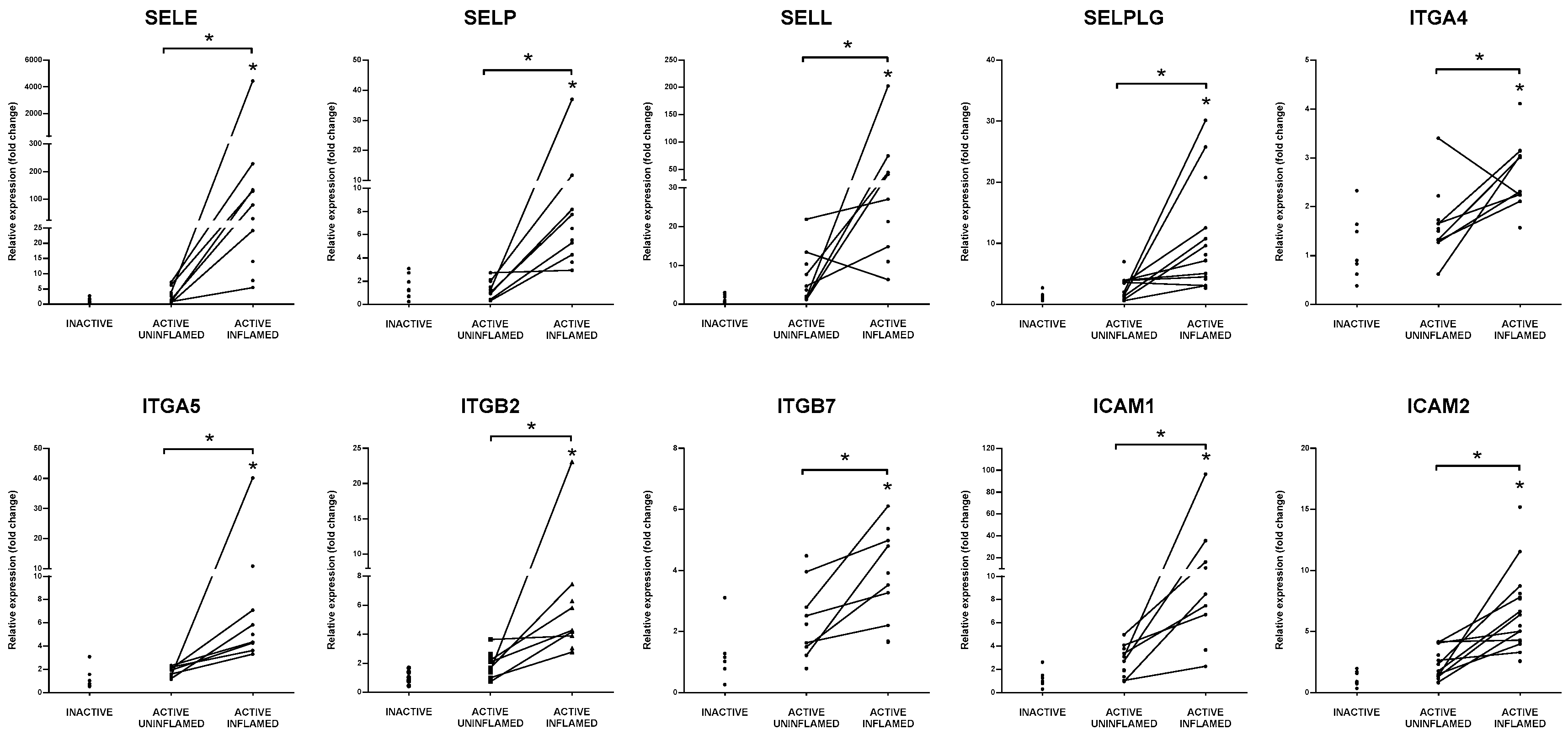
3.7. Support of Immune Cell Trafficking
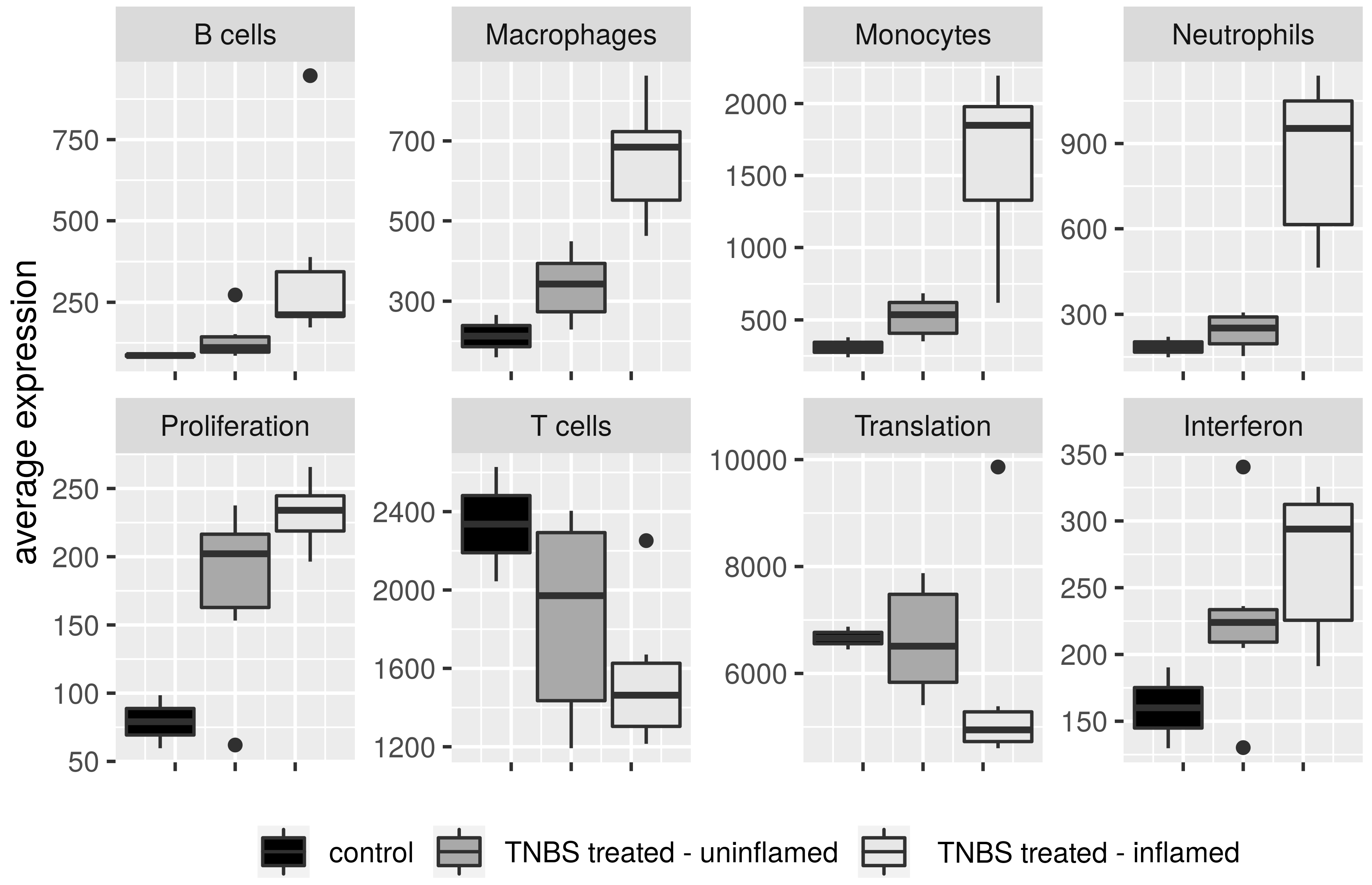
3.8. Definition of Immune Cell Signature
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Podolsky, D.K. Inflammatory bowel disease (1). N. Engl. J. Med. 1991, 325, 928–937. [Google Scholar] [CrossRef]
- Burisch, J.; Munkholm, P. The epidemiology of inflammatory bowel disease. Scand. J. Gastroenterol. 2015, 50, 942–951. [Google Scholar] [CrossRef]
- Borren, N.Z.; van der Woude, C.J.; Ananthakrishnan, A.N. Fatigue in IBD: Epidemiology, pathophysiology and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.C.; Stappenbeck, T.S. Genetics and Pathogenesis of Inflammatory Bowel Disease. Annu. Rev. Pathol. 2016, 11, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Ek, W.E.; D’Amato, M.; Halfvarson, J. The history of genetics in inflammatory bowel disease. Ann. Gastroenterol. 2014, 27, 294–303. [Google Scholar]
- De Lange, K.M.; Moutsianas, L.; Lee, J.C.; Lamb, C.A.; Luo, Y.; Kennedy, N.A.; Jostins, L.; Rice, D.L.; Gutierrez-Achury, J.; Ji, S.G.; et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017, 49, 256–261. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef]
- Knox, N.C.; Forbes, J.D.; Peterson, C.L.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiome in Inflammatory Bowel Disease: Lessons Learned From Other Immune-Mediated Inflammatory Diseases. Am. J. Gastroenterol. 2019, 114, 1051–1070. [Google Scholar] [CrossRef]
- Sun, Y.; Li, L.; Xia, Y.; Li, W.; Wang, K.; Wang, L.; Miao, Y.; Ma, S. The gut microbiota heterogeneity and assembly changes associated with the IBD. Sci. Rep. 2019, 9, 440. [Google Scholar] [CrossRef]
- Szalai, Z.; Szasz, A.; Nagy, I.; Puskas, L.G.; Kupai, K.; Kiraly, A.; Berko, A.M.; Posa, A.; Strifler, G.; Barath, Z.; et al. Anti-inflammatory effect of recreational exercise in TNBS-induced colitis in rats: Role of NOS/HO/MPO system. Oxidative Med. Cell. Longev. 2014, 2014, 925981. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.P.; Beck, P.L.; Herridge, M.S.; Depew, W.T.; Szewczuk, M.R.; Wallace, J.L. Hapten-induced model of chronic inflammation and ulceration in the rat colon. Gastroenterology 1989, 96, 795–803. [Google Scholar] [CrossRef]
- Boros, E.; Kellermayer, Z.; Balogh, P.; Strifler, G.; Voros, A.; Sarlos, P.; Vincze, A.; Varga, C.; Nagy, I. Elevated Expression of AXL May Contribute to the Epithelial-to-Mesenchymal Transition in Inflammatory Bowel Disease Patients. Mediat. Inflamm. 2018, 2018, 3241406. [Google Scholar] [CrossRef] [PubMed]
- Magro, F.; Langner, C.; Driessen, A.; Ensari, A.; Geboes, K.; Mantzaris, G.J.; Villanacci, V.; Becheanu, G.; Borralho Nunes, P.; Cathomas, G.; et al. European consensus on the histopathology of inflammatory bowel disease. J. Crohns Colitis 2013, 7, 827–851. [Google Scholar] [CrossRef]
- Boros, E.; Csatari, M.; Varga, C.; Balint, B.; Nagy, I. Specific Gene- and MicroRNA-Expression Pattern Contributes to the Epithelial to Mesenchymal Transition in a Rat Model of Experimental Colitis. Mediat. Inflamm. 2017, 2017, 5257378. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database, G. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef] [PubMed]
- Nirmal, A.J.; Regan, T.; Shih, B.B.; Hume, D.A.; Sims, A.H.; Freeman, T.C. Immune Cell Gene Signatures for Profiling the Microenvironment of Solid Tumors. Cancer Immunol. Res. 2018, 6, 1388–1400. [Google Scholar] [CrossRef] [PubMed]
- Goyal, N.; Rana, A.; Ahlawat, A.; Bijjem, K.R.; Kumar, P. Animal models of inflammatory bowel disease: A review. Inflammopharmacology 2014, 22, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Boros, E.; Nagy, I. The Role of MicroRNAs upon Epithelial-to-Mesenchymal Transition in Inflammatory Bowel Disease. Cells 2019, 8, 1461. [Google Scholar] [CrossRef] [PubMed]
- Lovisa, S.; Genovese, G.; Danese, S. Role of Epithelial-to-Mesenchymal Transition in Inflammatory Bowel Disease. J. Crohns Colitis 2019, 13, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Corfield, A.P. Mucins: A biologically relevant glycan barrier in mucosal protection. BBA-Gen. Subj. 2015, 1850, 236–252. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Larsson, J.M.H.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.M.H.; Karlsson, H.; Crespo, J.G.; Johansson, M.E.V.; Eklund, L.; Sjovall, H.; Hansson, G.C. Altered O-glycosylation Profile of MUC2 Mucin Occurs in Active Ulcerative Colitis and Is Associated with Increased Inflammation. Inflamm. Bowel Dis. 2011, 17, 2299–2307. [Google Scholar] [CrossRef] [PubMed]
- Arike, L.; Holmen-Larsson, J.; Hansson, G.C. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 2017, 27, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Theodoratou, E.; Campbell, H.; Ventham, N.T.; Kolarich, D.; Pucic-Bakovic, M.; Zoldos, V.; Fernandes, D.; Pemberton, I.K.; Rudan, I.; Kennedy, N.A.; et al. The role of glycosylation in IBD. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 588–600. [Google Scholar] [CrossRef]
- Ma, B.; Simala-Grant, J.L.; Taylor, D.E. Fucosylation in prokaryotes and eukaryotes. Glycobiology 2006, 16, 158R–184R. [Google Scholar] [CrossRef]
- Goto, Y.; Obata, T.; Kunisawa, J.; Sato, S.; Ivanov, I.I.; Lamichhane, A.; Takeyama, N.; Kamioka, M.; Sakamoto, M.; Matsuki, T.; et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014, 345, 1310. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Ellinghaus, E.; Nair, R.P.; Stuart, P.E.; Esko, T.; Metspalu, A.; Debrus, S.; Raelson, J.V.; Tejasvi, T.; Belouchi, M.; et al. Combined Analysis of Genome-wide Association Studies for Crohn Disease and Psoriasis Identifies Seven Shared Susceptibility Loci. Am. J. Hum. Genet. 2012, 90, 636–647. [Google Scholar] [CrossRef]
- Ting, J.P.Y.; Lovering, R.C.; Alnemri, E.S.; Bertin, J.; Boss, J.M.; Davis, B.K.; Flavell, R.A.; Girardin, S.E.; Godzik, A.; Harton, J.A.; et al. The NLR gene family: A standard nomenclature. Immunity 2008, 28, 285–287. [Google Scholar] [CrossRef]
- Elinav, E.; Strowig, T.; Henao-Mejia, J.; Flavell, R.A. Regulation of the antimicrobial response by NLR proteins. Immunity 2011, 34, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Elinav, E.; Strowig, T.; Kau, A.L.; Henao-Mejia, J.; Thaiss, C.A.; Booth, C.J.; Peaper, D.R.; Bertin, J.; Eisenbarth, S.C.; Gordon, J.I.; et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011, 145, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.B.; Bergstralh, D.T.; Duncan, J.A.; Lei, Y.; Morrison, T.E.; Zimmermann, A.G.; Accavitti-Loper, M.A.; Madden, V.J.; Sun, L.J.; Ye, Z.M.; et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 2008, 451, U573–U578. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cezard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.K.; Kanneganti, T.D. NLRP6 in infection and inflammation. Microbes Infect. 2013, 15, 661–668. [Google Scholar] [CrossRef]
- Prochnicki, T.; Latz, E. Inflammasomes on the Crossroads of Innate Immune Recognition and Metabolic Control. Cell Metab. 2017, 26, 71–93. [Google Scholar] [CrossRef]
- Simard, J.C.; Cesaro, A.; Chapeton-Montes, J.; Tardif, M.; Antoine, F.; Girard, D.; Tessier, P.A. S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-kappaB(1.). PLoS ONE 2013, 8, e72138. [Google Scholar] [CrossRef]
- Anand, P.K.; Malireddi, R.K.; Lukens, J.R.; Vogel, P.; Bertin, J.; Lamkanfi, M.; Kanneganti, T.D. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 2012, 488, 389–393. [Google Scholar] [CrossRef]
- Lage, S.L.; Longo, C.; Branco, L.M.; da Costa, T.B.; Buzzo, C.D.; Bortoluci, K.R. Emerging concepts about NAIP/NLIRC4 inflammasomes. Front. Immunol. 2014, 5, 309. [Google Scholar] [CrossRef] [PubMed]
- Feghali, C.A.; Wright, T.M. Cytokines in acute and chronic inflammation. Front. Biosci. 1997, 2, d12–d26. [Google Scholar] [PubMed]
- Mantovani, A.; Dinarello, C.A.; Molgora, M.; Garlanda, C. Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity 2019, 50, 778–795. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.A.; O’Callaghan, A.; Corr, S.C. IL-33 and IL-18 in Inflammatory Bowel Disease Etiology and Microbial Interactions. Front. Immunol. 2019, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, N.C.; Shayakhmetov, D.M. Interleukin 1alpha and the inflammatory process. Nat. Immunol. 2016, 17, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Bersudsky, M.; Luski, L.; Fishman, D.; White, R.M.; Ziv-Sokolovskaya, N.; Dotan, S.; Rider, P.; Kaplanov, I.; Aychek, T.; Dinarello, C.A.; et al. Non-redundant properties of IL-1alpha and IL-1beta during acute colon inflammation in mice. Gut 2014, 63, 598–609. [Google Scholar] [CrossRef]
- Kalliolias, G.D.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef]
- Day, A.J.; Milner, C.M. TSG-6: A multifunctional protein with anti-inflammatory and tissue-protective properties. Matrix Biol. 2019, 78–79, 60–83. [Google Scholar] [CrossRef]
- Das, T.; Chen, Z.; Hendriks, R.W.; Kool, M. A20/Tumor Necrosis Factor alpha-Induced Protein 3 in Immune Cells Controls Development of Autoinflammation and Autoimmunity: Lessons from Mouse Models. Front. Immunol. 2018, 9, 104. [Google Scholar] [CrossRef]
- Beigel, F.; Friedrich, M.; Probst, C.; Sotlar, K.; Goke, B.; Diegelmann, J.; Brand, S. Oncostatin M mediates STAT3-dependent intestinal epithelial restitution via increased cell proliferation, decreased apoptosis and upregulation of SERPIN family members. PLoS ONE 2014, 9, e93498. [Google Scholar] [CrossRef]
- Hunter, C.A.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Hegazy, A.N.; Owens, B.M.J.; Bullers, S.J.; Linggi, B.; Buonocore, S.; Coccia, M.; Gortz, D.; This, S.; Stockenhuber, K.; et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017, 23, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; O’Garra, A. IL-10 Family Cytokines IL-10 and IL-22: From Basic Science to Clinical Translation. Immunity 2019, 50, 871–891. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef]
- Hol, J.; Wilhelmsen, L.; Haraldsen, G. The murine IL-8 homologues KC, MIP-2, and LIX are found in endothelial cytoplasmic granules but not in Weibel-Palade bodies. J. Leukoc. Biol. 2010, 87, 501–508. [Google Scholar] [CrossRef]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. BBA-Mol. Cell. Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef]
- Khan, W.I.; Motomura, Y.; Wang, H.; El-Sharkawy, R.T.; Verdu, E.F.; Verma-Gandhu, M.; Rollins, B.J.; Collins, S.M. Critical role of MCP-1 in the pathogenesis of experimental colitis in the context of immune and enterochromaffin cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 291, G803–G811. [Google Scholar] [CrossRef]
- Werner, L.; Elad, H.; Brazowski, E.; Tulchinsky, H.; Vigodman, S.; Kopylov, U.; Halpern, Z.; Guzner-Gur, H.; Dotan, I. Reciprocal regulation of CXCR4 and CXCR7 in intestinal mucosal homeostasis and inflammatory bowel disease. J. Leukoc. Biol. 2011, 90, 583–590. [Google Scholar] [CrossRef]
- Friedrich, M.; Pohin, M.; Powrie, F. Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 2019, 50, 992–1006. [Google Scholar] [CrossRef]
- Sala, E.; Genua, M.; Petti, L.; Anselmo, A.; Arena, V.; Cibella, J.; Zanotti, L.; D’Alessio, S.; Scaldaferri, F.; Luca, G.; et al. Mesenchymal Stem Cells Reduce Colitis in Mice via Release of TSG6, Independently of Their Localization to the Intestine. Gastroenterology 2015, 149, 163–176.e120. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Vestweber, D. How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 2015, 15, 692–704. [Google Scholar] [CrossRef] [PubMed]
- Lishko, V.K.; Yakubenko, V.P.; Ugarova, T.P. The interplay between integrins alpha(M)beta(2) and alpha(5)beta(1), during cell migration to fibronectin. Exp. Cell Res. 2003, 283, 116–126. [Google Scholar] [CrossRef]
- Cauwe, B.; Van den Steen, P.E.; Opdenakker, G. The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 113–185. [Google Scholar] [CrossRef]
- Schonbeck, U.; Mach, F.; Libby, P. Generation of biologically active IL-1 beta by matrix metalloproteinases: A novel caspase-1-independent pathway of IL-1 beta processing. J. Immunol. 1998, 161, 3340–3346. [Google Scholar]
- Van Lint, P.; Libert, C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol. 2007, 82, 1375–1381. [Google Scholar] [CrossRef]
- Parks, W.C.; Wilson, C.L.; Lopez-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef]
- Nakayama, F.; Nishihara, S.; Iwasaki, H.; Kudo, T.; Okubo, R.; Kaneko, M.; Nakamura, M.; Karube, M.; Sasaki, K.; Narimatsu, H. CD15 expression in mature granulocytes is determined by alpha 1,3-fucosyltransferase IX, but in promyelocytes and monocytes by alpha 1,3-fucosyltransferase IV. J. Biol. Chem. 2001, 276, 16100–16106. [Google Scholar] [CrossRef]
- Buffone, A.; Mondal, N.; Gupta, R.; McHugh, K.P.; Lau, J.T.Y.; Neelamegham, S. Silencing alpha 1,3-Fucosyltransferases in Human Leukocytes Reveals a Role for FUT9 Enzyme during E-selectin-mediated Cell Adhesion. J. Biol. Chem. 2013, 288, 1620–1633. [Google Scholar] [CrossRef]
- Kannagi, R. Molecular mechanism for cancer-associated induction of sialyl Lewis X and sialyl Lewis A expression—The Warburg effect revisited. Glycoconj. J. 2003, 20, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, S.; Yan, Q. Role of fucosyltransferase IV in epithelial-mesenchymal transition in breast cancer cells. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Petretti, T.; Kemmner, W.; Schulze, B.; Schlag, P.M. Altered mRNA expression of glycosyltransferases in human colorectal carcinomas and liver metastases. Gut 2000, 46, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Auslander, N.; Cunningham, C.E.; Toosi, B.M.; McEwen, E.J.; Yizhak, K.; Vizeacoumar, F.S.; Parameswaran, S.; Gonen, N.; Freywald, T.; Bhanumathy, K.K.; et al. An integrated computational and experimental study uncovers FUT9 as a metabolic driver of colorectal cancer. Mol. Syst. Biol. 2017, 13, 956. [Google Scholar] [CrossRef] [PubMed]
- Zundler, S.; Schillinger, D.; Fischer, A.; Atreya, R.; Lopez-Posadas, R.; Watson, A.; Neufert, C.; Atreya, I.; Neurath, M.F. Blockade of alphaEbeta7 integrin suppresses accumulation of CD8(+) and Th9 lymphocytes from patients with IBD in the inflamed gut in vivo. Gut 2017, 66, 1936–1948. [Google Scholar] [CrossRef]
- Neurath, M.F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 2019, 20, 970–979. [Google Scholar] [CrossRef]
- Kinchen, J.; Chen, H.H.; Parikh, K.; Antanaviciute, A.; Jagielowicz, M.; Fawkner-Corbett, D.; Ashley, N.; Cubitt, L.; Mellado-Gomez, E.; Attar, M.; et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 2018, 175, 372–386.e17. [Google Scholar] [CrossRef]
- Smids, C.; Horjus Talabur Horje, C.S.; Drylewicz, J.; Roosenboom, B.; Groenen, M.J.M.; van Koolwijk, E.; van Lochem, E.G.; Wahab, P.J. Intestinal T Cell Profiling in Inflammatory Bowel Disease: Linking T Cell Subsets to Disease Activity and Disease Course. J. Crohns Colitis 2018, 12, 465–475. [Google Scholar] [CrossRef]
- Na, Y.R.; Stakenborg, M.; Seok, S.H.; Matteoli, G. Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 531–543. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Bhan, A.K. Immunobiology of B Cells in Inflammatory Bowel Disease. In Crohn’s Disease and Ulcerative Colitis; Springer: Boston, MA, USA, 2012; pp. 161–168. [Google Scholar] [CrossRef]
- Jones, G.R.; Bain, C.C.; Fenton, T.M.; Kelly, A.; Brown, S.L.; Ivens, A.C.; Travis, M.A.; Cook, P.C.; MacDonald, A.S. Dynamics of Colon Monocyte and Macrophage Activation During Colitis. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Fournier, B.M.; Parkos, C.A. The role of neutrophils during intestinal inflammation. Mucosal Immunol. 2012, 5, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Therrien, A.; Chapuy, L.; Bsat, M.; Rubio, M.; Bernard, G.; Arslanian, E.; Orlicka, K.; Weber, A.; Panzini, B.P.; Dorais, J.; et al. Recruitment of activated neutrophils correlates with disease severity in adult Crohn’s disease. Clin. Exp. Immunol. 2019, 195, 251–264. [Google Scholar] [CrossRef] [PubMed]
| Disease State | IBD | Patient ID | Sample ID | Sex | Age, y | Biopsy Location | Tissue State | Disease Duration, y |
|---|---|---|---|---|---|---|---|---|
| inactive | CD | 1 | H1 | M | 47 | colon descendens | − | 27 |
| inactive | CD | 2 | H3 | M | 32 | colon ascendens | − | 7 |
| inactive | UC | 3 | H5 | F | 36 | colon ascendens | − | 16 |
| inactive | CD | 9 | H27 | F | 39 | colon ascendens | − | 17 |
| inactive | CD | 9 | H28 | F | 39 | colon descendens | − | 17 |
| inactive | UC | 16 | H50 | F | 72 | colon ascendens | − | 12 |
| inactive | UC | 16 | H51 | F | 72 | colon descendens | − | 12 |
| inactive | UC | 33 | H99 | M | 56 | colon ascendens | − | 21 |
| inactive | UC | 33 | H100 | M | 56 | colon transversum | − | 21 |
| inactive | UC | 33 | H101 | M | 56 | colon descendens | − | 21 |
| active | UC/CD | 4 | H10 | F | 41 | colon ascendens | uninflamed | 20 |
| active | UC/CD | 4 | H12 | F | 41 | colon transversum | uninflamed | 20 |
| active | UC | 6 | H18 | F | 27 | colon ascendens | uninflamed | 2 |
| active | UC | 6 | H21 | F | 27 | colon descendens | uninflamed | 2 |
| active | CD | 10 | H31 | F | 43 | colon transversum | uninflamed | 9 |
| active | UC | 11 | H32 | F | 44 | colon ascendens | uninflamed | 11 |
| active | CD | 12 | H35 | M | 37 | colon ascendens | uninflamed | 3 |
| active | CD | 15 | H48 | F | 37 | colon ascendens | uninflamed | 12 |
| active | CD | 15 | H49 | F | 37 | colon descendens | uninflamed | 12 |
| active | UC | 17 | H56 | M | 51 | colon descendens | uninflamed | 14 |
| active | UC | 18 | H58 | F | 47 | colon sigmoideum | uninflamed | 17 |
| active | UC | 20 | H62 | F | 41 | colon sigmoideum | uninflamed | 12 |
| active | CD | 22 | H68 | F | 24 | colon transversum | uninflamed | 6 |
| active | UC | 28 | H86 | M | 49 | colon descendens | uninflamed | 20 |
| active | CD | 29 | H88 | M | 33 | colon ascendens | uninflamed | 11 |
| active | UC/CD | 4 | H9 | F | 41 | colon ascendens | inflamed | 20 |
| active | UC/CD | 4 | H11 | F | 41 | colon transversum | inflamed | 20 |
| active | UC | 6 | H19 | F | 27 | colon ascendens | inflamed | 2 |
| active | UC | 6 | H20 | F | 27 | colon descendens | inflamed | 2 |
| active | CD | 8 | H25 | M | 30 | colon transversum | inflamed | 14 |
| active | CD | 12 | H36 | M | 37 | colon descendens | inflamed | 3 |
| active | UC | 13 | H39 | F | 33 | colon descendens | inflamed | 11 |
| active | UC | 17 | H54 | M | 51 | colon ascendens | inflamed | 14 |
| active | UC | 17 | H55 | M | 51 | colon transversum | inflamed | 14 |
| active | UC | 18 | H57 | F | 47 | colon sigmoideum | inflamed | 17 |
| active | UC | 20 | H63 | F | 41 | colon sigmoideum | inflamed | 12 |
| active | CD | 22 | H69 | F | 24 | colon sigmoideum | inflamed | 6 |
| active | UC | 24 | H74 | M | 42 | colon transversum | inflamed | 21 |
| active | UC | 24 | H75 | M | 42 | colon sigmoideum | inflamed | 21 |
| active | UC | 28 | H87 | M | 49 | colon sigmoideum | inflamed | 20 |
| active | CD | 29 | H89 | M | 33 | colon sigmoideum | inflamed | 11 |
| Gene ID | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Cxcl1 | CATTAATATTTAACGATGTGGATGCGTTTCA | GCCTACCATCTTTAAACTGCACAAT |
| Fut1 | AGGACCCGTTTCTCAAGCTG | CTATCCGGAGCCCACTCAAC |
| Fut2 | ACTTCCACCATCATCCACCTC | CTCTGGGCTTTCTGTGTTTCC |
| Fut4 | AAGCTACAGCATGAGAGCCG | AGAGGAGGTCCGGGGTAATC |
| Fut9 | CAACAAATCCCATGCGGTCC | CAATGCCACTCTTTTGGGGG |
| Il1β | CAGGAAGGCAGTGTCACTCA | AGACAGCACGAGGCATTTTT |
| Il6 | CACTTCACAAGTCGGAGGCT | TCTGACAGTGCATCATCGCT |
| Nlrc4 | GCGAAACCTGAAGAAGATGC | AACGCTCAGCTTGACCAAAT |
| Nlrp3 | GCTGCTCAGCTCTGACCTCT | AGGTGAGGCTGCAGTTGTCT |
| Nlrp6 | TACCTGGTCATTGTGCTCCA | TCAGAGGCTGAGGATGTGTG |
| Nod2 | TCCTTGCACACAAGCAGAAC | TGATCAGCCACAACTTCAGC |
| Tnf | ATGGGCTCCCTCTCATCAGT | GCTTGGTGGTTTGCTACGAC |
| Gene ID | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| CCL2 | CCTTCATTCCCCAAGGGCTC | CTTCTTTGGGACACTTGCTGC |
| CDH5 | GAGAATGACAATGCCCCGGA | ACTGTGATGTTGGCCGTGTT |
| CXCR4 | GGTGGTCTATGTTGGCGTCT | ACACAACCACCCACAAGTCA |
| FN1 | GGGCGAGGGAGAATAAGCTG | TCTCAGCTATGGGCTTGCAG |
| FUT1 | TGGCCGGTTTGGTAATCAGA | TCCGACATCCAGTCGTGAAG |
| FUT2 | TGGACCTTCTACCACCACCT | CCACCCCCTTCCACACTTTT |
| FUT4 | GCCTCGTACCTGCTTTTCCT | AGTTCCGTATGCTCTTGGGC |
| FUT9 | CCATACCTACGGGCAAGCAT | ACAACAGGTACAGAGCCAGC |
| ICAM1 | TGACCGTGAATGTGCTCTCC | TCCCTTTTTGGGCCTGTTGT |
| ICAM2 | TCTGCTGTCCAGGATCGGA | TTTCCACTGAGCCTGTTCGT |
| IL1β | AGCTGGAGAGTGTAGATCCCAA | GGGAACTGGGCAGACTCAAA |
| ITGA4 | CGCTTCAGTGATCAATCCCG | CTCCATTAGGGCTACCCAGC |
| ITGA5 | TGGCCTTCGGTTTACAGTCC | GGAGAGCCGAAAGGAAACCA |
| ITGB2 | TGGATGAGAGCCGAGAGTGT | CAGCAGGAGAATGCCGATCA |
| ITGB7 | TCGCAGCACAGAGTTTGACT | CTGTACCACGTTGCTGGAGT |
| MMP12 | CGGGCAACTGGACACATCTA | CCTCACGGTTCATGTCAGGT |
| MMP14 | GGCGGGTGAGGAATAACCAA | ACGCCTCATCAAACACCCAA |
| MMP2 | GCCCTGATGGCACCCATTTA | GTCAGGAGAGGCCCCATAGA |
| MSN | TCTGCAAGTGAAAGAGGGCA | CCTCCCACTGGTCCTTGTTG |
| NLRC4 | GGACAATAGCCGAGCCCTTA | CCACCTTCTCGCAGCAAATG |
| NLRP3 | AAGGCCGACACCTTGATATG | CCGAATGTTACAGCCAGGAT |
| NLRP6 | CAGTTCTCAAGGCACCACAA | TCACTCAGCATACGCAGTCC |
| NOD2 | CTCCATGGCTAAGCTCCTTG | CACACTGCCAATGTTGTTCC |
| SELE | GCCTGCAATGTGGTTGAGTG | GGTACACTGAAGGCTCTGGG |
| SELL | GCCTGCCACAAACTAAAGGC | TCACAAACTGACACTGGGGC |
| SELP | CCAACCTGCAAAGGCATAGC | GTGTGAATCCAGCGTTGCAG |
| SELPLG | AGAAGGAGATAAGATGGCTGGTG | ACTCATATTCGGTGGCCTGTC |
| Gene ID | Assay Number |
|---|---|
| IL1α | Hs00174092_m1 |
| IL6 | Hs00174131_m1 |
| IL8 (CXCL8) | Hs00174103_m1 |
| IL10 | Hs99999035_m1 |
| TNFAIP3 | Hs00234713_m1 |
| TNFAIP6 | Hs01113602_m1 |
| TNF | Hs00174128_m1 |
| 18S RNA | Hs99999901_s1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boros, É.; Prontvai, B.; Kellermayer, Z.; Balogh, P.; Sarlós, P.; Vincze, Á.; Varga, C.; Maróti, Z.; Bálint, B.; Nagy, I. Transcriptome Based Profiling of the Immune Cell Gene Signature in Rat Experimental Colitis and Human IBD Tissue Samples. Biomolecules 2020, 10, 974. https://doi.org/10.3390/biom10070974
Boros É, Prontvai B, Kellermayer Z, Balogh P, Sarlós P, Vincze Á, Varga C, Maróti Z, Bálint B, Nagy I. Transcriptome Based Profiling of the Immune Cell Gene Signature in Rat Experimental Colitis and Human IBD Tissue Samples. Biomolecules. 2020; 10(7):974. https://doi.org/10.3390/biom10070974
Chicago/Turabian StyleBoros, Éva, Bence Prontvai, Zoltán Kellermayer, Péter Balogh, Patrícia Sarlós, Áron Vincze, Csaba Varga, Zoltán Maróti, Balázs Bálint, and István Nagy. 2020. "Transcriptome Based Profiling of the Immune Cell Gene Signature in Rat Experimental Colitis and Human IBD Tissue Samples" Biomolecules 10, no. 7: 974. https://doi.org/10.3390/biom10070974
APA StyleBoros, É., Prontvai, B., Kellermayer, Z., Balogh, P., Sarlós, P., Vincze, Á., Varga, C., Maróti, Z., Bálint, B., & Nagy, I. (2020). Transcriptome Based Profiling of the Immune Cell Gene Signature in Rat Experimental Colitis and Human IBD Tissue Samples. Biomolecules, 10(7), 974. https://doi.org/10.3390/biom10070974






