Seminal Plasma Modulates miRNA Expression by Sow Genital Tract Lining Explants
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Ethics Statement
2.3. Reagents and Media
2.4. Boars, Ejaculates and Seminal Plasma
2.5. Sows, Tissue Collection and Explant Preparation
2.6. Explant Culture
2.7. RNA Isolation
2.8. miRNA Microarray Analysis
2.9. Bioinformatics of Microarray Data
2.10. Target Gene Prediction and Functional Analysis
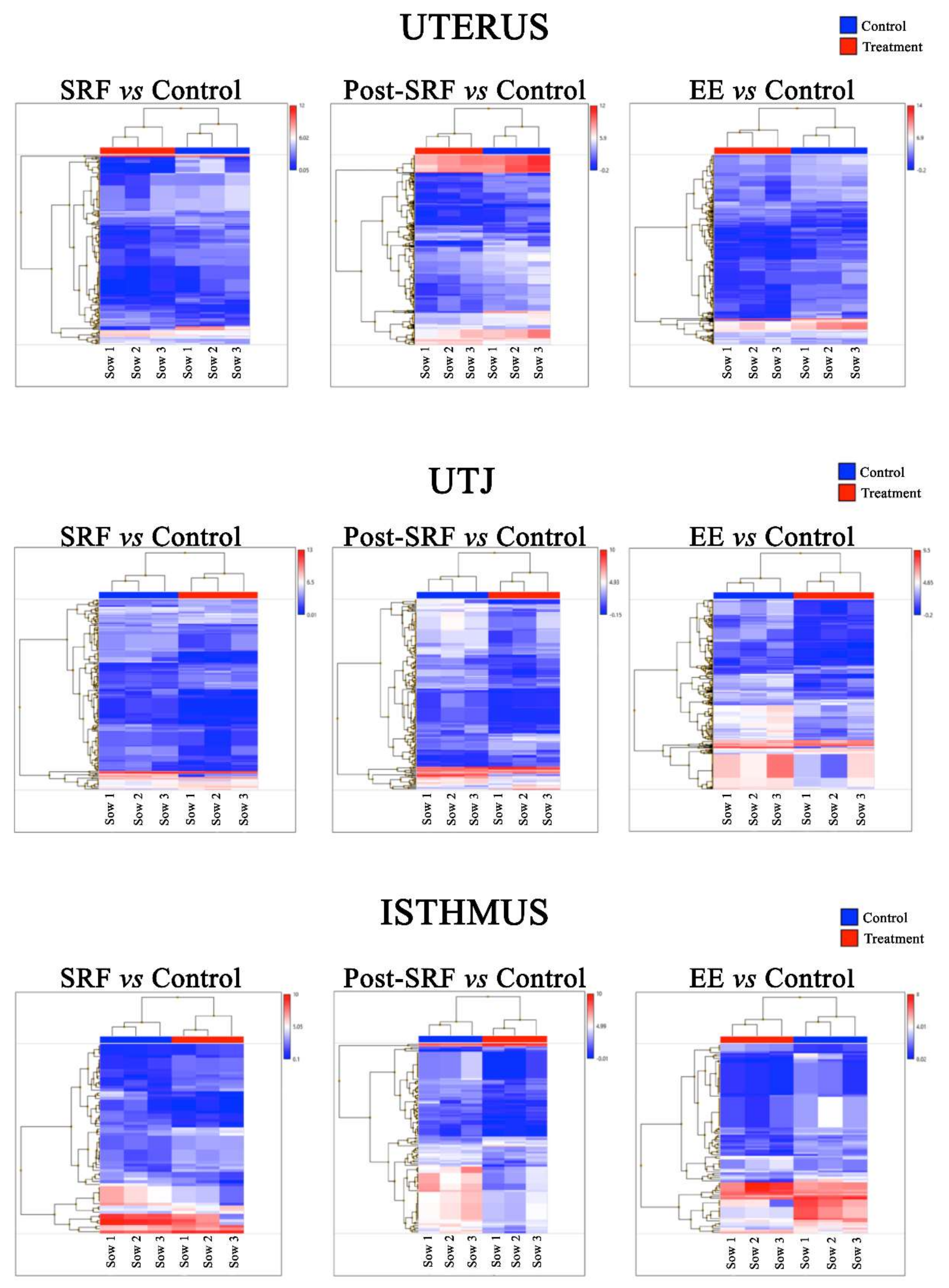
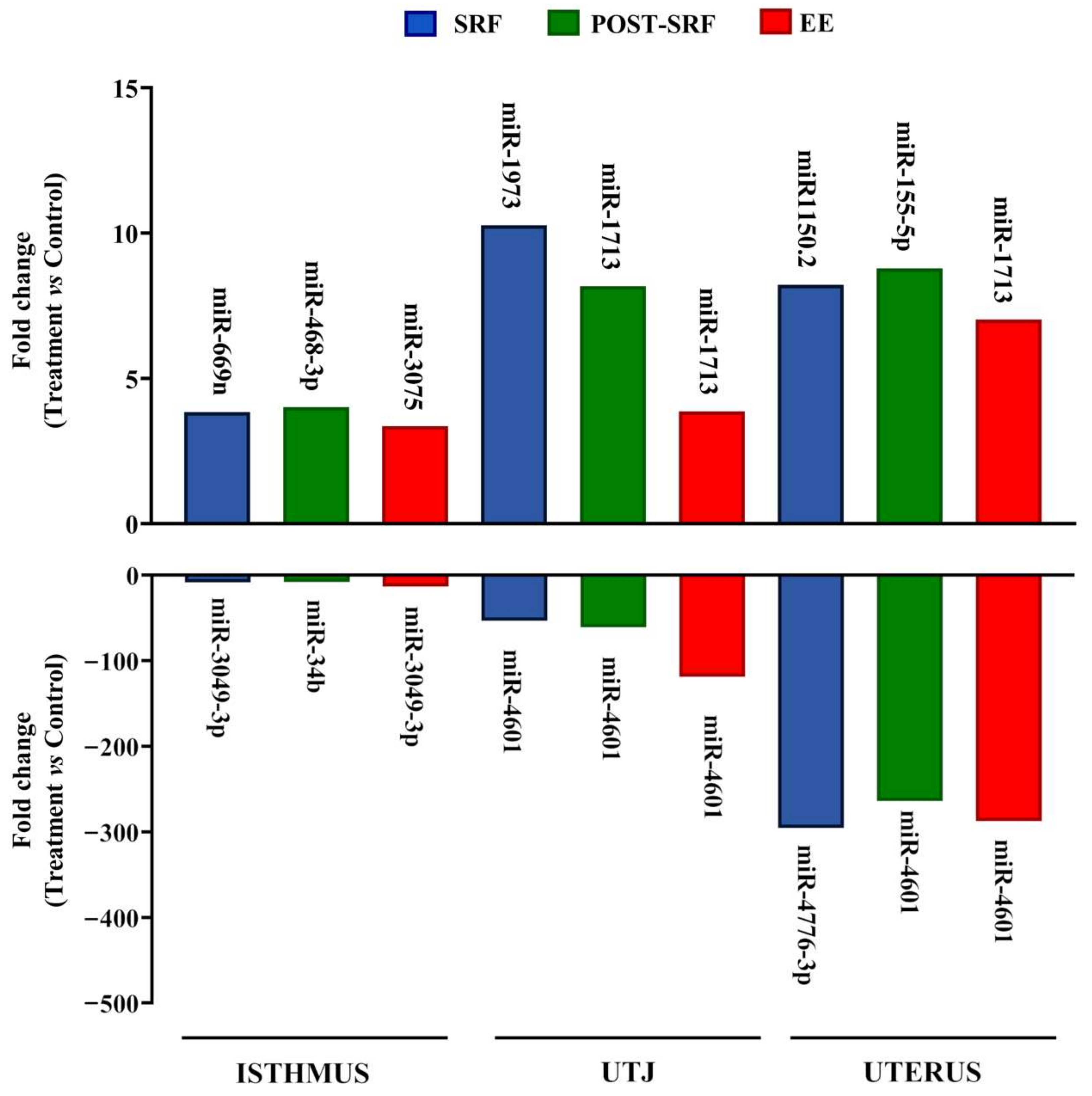
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bromfield, J.J. Seminal fluid and reproduction: Much more than previously thought. J. Assist. Reprod. Genet. 2014, 31, 627–636. [Google Scholar] [CrossRef]
- Recuero, S.; Fernandez-Fuertes, B.; Bonet, S.; Barranco, I.; Yeste, M. Potential of seminal plasma to improve the fertility of frozen-thawed boar spermatozoa. Theriogenology 2019, 137, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Szczykutowicz, J.; Kaluza, A.; Kazmierowska-Niemczuk, M.; Ferens-Sieczkowska, M. The Potential Role of Seminal Plasma in the Fertilization Outcomes. Biomed. Res. Int. 2019, 2019, 5397804. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.A. Seminal plasma and male factor signalling in the female reproductive tract. Cell Tissue Res. 2005, 322, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Martinez, H.; Kvist, U.; Ernerudh, J.; Sanz, L.; Calvete, J.J. Seminal plasma proteins: What role do they play? Am. J. Reprod. Immunol. 2011, 66 (Suppl. 1), 11–22. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.L.; Watkins, A.J. The influence of seminal plasma on offspring development and health. Semin. Cell Dev. Biol. 2020, 97, 131–137. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, S.; Jasper, M.J.; Warnes, G.M.; Armstrong, D.T.; Robertson, S.A. Seminal plasma regulates endometrial cytokine expression, leukocyte recruitment and embryo development in the pig. Reproduction 2004, 128, 237–247. [Google Scholar] [CrossRef]
- Sharkey, D.J.; Tremellen, K.P.; Jasper, M.J.; Gemzell-Danielsson, K.; Robertson, S.A. Seminal fluid induces leukocyte recruitment and cytokine and chemokine mRNA expression in the human cervix after coitus. J. Immunol. 2012, 188, 2445–2454. [Google Scholar] [CrossRef]
- Schjenken, J.E.; Robertson, S.A. Seminal Fluid Signalling in the Female Reproductive Tract: Implications for Reproductive Success and Offspring Health. Adv. Exp. Med. Biol. 2015, 868, 127–158. [Google Scholar] [PubMed]
- Alvarez-Rodriguez, M.; Atikuzzaman, M.; Venhoranta, H.; Wright, D.; Rodriguez-Martinez, H. Expression of Immune Regulatory Genes in the Porcine Internal Genital Tract Is Differentially Triggered by Spermatozoa and Seminal Plasma. Int. J. Mol. Sci. 2019, 20, 513. [Google Scholar] [CrossRef]
- Martinez, C.A.; Cambra, J.M.; Parrilla, I.; Roca, J.; Ferreira-Dias, G.; Pallares, F.J.; Lucas, X.; Vazquez, J.M.; Martinez, E.A.; Gil, M.A.; et al. Seminal Plasma Modifies the Transcriptional Pattern of the Endometrium and Advances Embryo Development in Pigs. Front. Vet. Sci. 2019, 6, 465. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Jin, D.; Choi, C.-H.; Lee, H. Integration of MicroRNA, mRNA, and Protein Expression Data for the Identification of Cancer-Related MicroRNAs. PLoS ONE 2017, 12, e0168412. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Liang, J.; Wang, S.; Wang, Z. Role of microRNAs in embryo implantation. Reprod. Biol. Endocrinol. 2017, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.A.; Zhang, B.; Chan, H.; Sharkey, D.J.; Barry, S.C.; Fullston, T.; Schjenken, J.E. MicroRNA regulation of immune events at conception. Mol. Reprod. Dev. 2017, 84, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; James, E.R.; Aston, K.I.; Jenkins, T.G.; Carrell, D.T.; Yeste, M. The Expression of miRNAs in Human Ovaries, Oocytes, Extracellular Vesicles, and Early Embryos: A Systematic Review. Cells 2019, 8, 1564. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; James, E.R.; Aston, K.I.; Carrell, D.T.; Jenkins, T.G.; Yeste, M. The role of miRNAs in male human reproduction: A systematic review. Andrology 2020, 8, 7–26. [Google Scholar] [CrossRef] [PubMed]
- Nothnick, W.B. The role of micro-RNAs in the female reproductive tract. Reproduction 2012, 143, 559–576. [Google Scholar] [CrossRef]
- Pan, Q.; Chegini, N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin. Reprod. Med. 2008, 26, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Bidarimath, M.; Khalaj, K.; Wessels, J.M.; Tayade, C. MicroRNAs, immune cells and pregnancy. Cell. Mol. Immunol. 2014, 11, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Liu, R.; Cheng, W.; Zhu, M.; Li, X.; Zhao, S.; Yu, M. Expression patterns of microRNAs in porcine endometrium and their potential roles in embryo implantation and placentation. PLoS ONE 2014, 9, e87867. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xi, Y.; Xue, S.; Wang, Y.; Wu, L.; Liu, H.; Lei, M. Sequence analysis of microRNAs during pre-implantation between Meishan and Yorkshire pigs. Gene 2018, 646, 20–27. [Google Scholar] [CrossRef]
- Hong, L.; Liu, R.; Qiao, X.; Wang, X.; Wang, S.; Li, J.; Wu, Z.; Zhang, H. Differential microRNA Expression in Porcine Endometrium Involved in Remodeling and Angiogenesis That Contributes to Embryonic Implantation. Front. Genet. 2019, 10, 661. [Google Scholar] [CrossRef]
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef]
- Gracias, D.T.; Katsikis, P.D. MicroRNAs: Key components of immune regulation. Adv. Exp. Med. Biol. 2011, 780, 15–26. [Google Scholar]
- Schjenken, J.E.; Zhang, B.; Chan, H.Y.; Sharkey, D.J.; Fullston, T.; Robertson, S.A. Mi RNA Regulation of Immune Tolerance in Early Pregnancy. Am. J. Reprod. Immunol. 2016, 75, 272–280. [Google Scholar] [CrossRef]
- Schjenken, J.E.; Moldenhauer, L.M.; Zhang, B.; Care, A.S.; Groome, H.M.; Chan, H.-Y.; Hope, C.M.; Barry, S.C.; Robertson, S.A. MicroRNA miR-155 is required for expansion of regulatory T cells to mediate robust pregnancy tolerance in mice. Mucosal Immunol. 2020, 1–17. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef] [PubMed]
- Parks, J.C.; Patton, A.L.; McCallie, B.; Griffin, D.K.; Schoolcraft, W.B.; Katz-Jaffe, M. Secreted endometrial microRNA gene regulators are critical for successful embryo implantation. Fertil. Steril. 2015, 104, e339. [Google Scholar] [CrossRef]
- Vilella, F.; Moreno-Moya, J.M.; Balaguer, N.; Grasso, A.; Herrero, M.; Martinez, S.; Marcilla, A.; Simon, C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 2015, 142, 3210–3221. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Martinez, H.; Kvist, U.; Saravia, F.; Wallgren, M.; Johannisson, A.; Sanz, L.; Pena, F.J.; Martinez, E.A.; Roca, J.; Vazquez, J.M.; et al. The physiological roles of the boar ejaculate. Soc. Reprod. Fertil. Suppl. 2009, 66, 1–21. [Google Scholar]
- Rodriguez-Martinez, H.; Saravia, F.; Wallgren, M.; Martinez, E.A.; Sanz, L.; Roca, J.; Vazquez, J.M.; Calvete, J.J. Spermadhesin PSP-I/PSP-II heterodimer induces migration of polymorphonuclear neutrophils into the uterine cavity of the sow. J. Reprod. Immunol. 2010, 84, 57–65. [Google Scholar] [CrossRef]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef]
- Hall, J.S.; Taylor, J.; Valentine, H.R.; Irlam, J.J.; Eustace, A.; Hoskin, P.J.; Miller, C.J.; West, C.M.L. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br. J. Cancer 2012, 107, 684–694. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. MiRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Alvarez-Rodriguez, M.; Martinez, C.; Wright, D.; Barranco, I.; Roca, J.; Rodriguez-Martinez, H. The Transcriptome of Pig Spermatozoa, and Its Role in Fertility. Int. J. Mol. Sci. 2020, 21, 1572. [Google Scholar] [CrossRef]
- Robertson, S.A.; Guerin, L.R.; Moldenhauer, L.M.; Hayball, J.D. Activating T regulatory cells for tolerance in early pregnancy-the contribution of seminal fluid. J. Reprod. Immunol. 2009, 83, 109–116. [Google Scholar] [CrossRef]
- Quenby, S.; Bates, M.; Doig, T.; Brewster, J.; Lewis-Jones, D.I.; Johnson, P.M.; Vince, G. Pre-implantation endometrial leukocytes in women with recurrent miscarriage. Hum. Reprod. 1999, 14, 2386–2391. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Jana, S.K.; Pasricha, P.; Ghosh, S.; Chakravarty, B.; Chaudhury, K. Proinflammatory cytokines induced altered expression of cyclooxygenase-2 gene results in unreceptive endometrium in women with idiopathic recurrent spontaneous miscarriage. Fertil. Steril. 2013, 99, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Petrunkina, A.M.; Gehlhaar, R.; Drommer, W.; Waberski, D.; Topfer-Petersen, E. Selective sperm binding to pig oviductal epithelium in vitro. Reproduction 2001, 121, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Bersinger, N.A.; Genewein, E.M.; Müller, O.; Altermatt, H.J.; McKinnon, B.; Mueller, M.D. Morphology of human endometrial explants and secretion of stromal marker proteins in short- and long-term cultures. Gynecol. Surg. 2010, 7, 75–80. [Google Scholar] [CrossRef]
- Borges, A.M.; Healey, G.D.; Sheldon, I.M. Explants of intact endometrium to model bovine innate immunity and inflammation ex vivo. Am. J. Reprod. Immunol. 2012, 67, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Skowronska, A.; Mlotkowska, P.; Nielsen, S.; Skowronski, M.T. Difference in expression between AQP1 and AQP5 in porcine endometrium and myometrium in response to steroid hormones, oxytocin, arachidonic acid, forskolin and cAMP during the mid-luteal phase of the estrous cycle and luteolysis. Reprod. Biol. Endocrinol. 2015, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fuertes, B.; Sanchez, J.M.; Bages-Arnal, S.; McDonald, M.; Yeste, M.; Lonergan, P. Species-specific and collection method-dependent differences in endometrial susceptibility to seminal plasma-induced RNA degradation. Sci. Rep. 2019, 9, 15072. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.A.; Vazquez, J.M.; Roca, J.; Lucas, X.; Gil, M.A.; Parrilla, I.; Vazquez, J.L.; Day, B.N. Minimum number of spermatozoa required for normal fertility after deep intrauterine insemination in non-sedated sows. Reproduction 2002, 123, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Kaeoket, K.; Persson, E.; Dalin, A.-M. Influence of pre-ovulatory insemination and early pregnancy on the infiltration by cells of the immune system in the sow endometrium. Anim. Reprod. Sci. 2003, 75, 55–71. [Google Scholar] [CrossRef]
- Ibrahim, L.A.; Rizo, J.A.; Fontes, P.L.P.; Lamb, G.C.; Bromfield, J.J. Seminal plasma modulates expression of endometrial inflammatory meditators in the bovinedagger. Biol. Reprod. 2019, 100, 660–671. [Google Scholar] [CrossRef]
- Einarsson, S.; Jones, B.; Larsson, K.; Viring, S. Distribution of small- and medium-sized molecules within the genital tract of artificially inseminated gilts. J. Reprod. Fertil. 1980, 59, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, Y.; Juma, C.A.; Yang, C.; Huang, J.; Zhang, X.; Zeng, Y. Differential Inhibition of Target Gene Expression by Human microRNAs. Cells 2019, 8, 791. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Hebles, M.; Dorado, M.; Gallardo, M.; Gonzalez-Martinez, M.; Sanchez-Martin, P. Seminal quality in the first fraction of ejaculate. Syst. Biol. Reprod. Med. 2015, 61, 113–116. [Google Scholar] [CrossRef]
- Kareskoski, M.; Sankari, S.; Johannisson, A.; Kindahl, H.; Andersson, M.; Katila, T. The association of the presence of seminal plasma and its components with sperm longevity in fractionated stallion ejaculates. Reprod. Domest. Anim. 2011, 46, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Perez-Patino, C.; Barranco, I.; Parrilla, I.; Valero, M.L.; Martinez, E.A.; Rodriguez-Martinez, H.; Roca, J. Characterization of the porcine seminal plasma proteome comparing ejaculate portions. J. Proteom. 2016, 142, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Barranco, I.; Ruber, M.; Perez-Patino, C.; Atikuzzaman, M.; Martinez, E.A.; Roca, J.; Rodriguez-Martinez, H. The Seminal Plasma of the Boar is Rich in Cytokines, with Significant Individual and Intra-Ejaculate Variation. Am. J. Reprod. Immunol. 2015, 74, 523–532. [Google Scholar] [CrossRef]
- Robertson, S.A.; Mau, V.J.; Hudson, S.N.; Tremellen, K.P. Cytokine-leukocyte networks and the establishment of pregnancy. Am. J. Reprod. Immunol. 1997, 37, 438–442. [Google Scholar] [CrossRef]
- Salvi, V.; Gianello, V.; Tiberio, L.; Sozzani, S.; Bosisio, D. Cytokine Targeting by miRNAs in Autoimmune Diseases. Front. Immunol. 2019, 10, 15. [Google Scholar] [CrossRef]
- Burney, R.O.; Hamilton, A.E.; Aghajanova, L.; Vo, K.C.; Nezhat, C.N.; Lessey, B.A.; Giudice, L.C. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol. Hum. Reprod. 2009, 15, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Hiroki, E.; Suzuki, F.; Akahira, J.-I.; Nagase, S.; Ito, K.; Sugawara, J.-I.; Miki, Y.; Suzuki, T.; Sasano, H.; Yaegashi, N. MicroRNA-34b functions as a potential tumor suppressor in endometrial serous adenocarcinoma. Int. J. Cancer 2012, 131, E395–E404. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Gu, C.; Ye, M.; Zhang, Z.; Li, L.; Fan, W.; Meng, Y. Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod. Biol. Endocrinol. 2018, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Herington, J.L.; Bruner-Tran, K.L.; Lucas, J.A.; Osteen, K.G. Immune interactions in endometriosis. Expert Rev. Clin. Immunol. 2011, 7, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Jasinski-Bergner, S.; Mandelboim, O.; Seliger, B. The role of microRNAs in the control of innate immune response in cancer. J. Natl. Cancer Inst. 2014, 106, dju257. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Seo, H.; Choi, Y.; Shim, J.; Choi, Y.; Ka, H. Regulatory mechanism for expression of IL1B receptors in the uterine endometrium and effects of IL1B on prostaglandin synthetic enzymes during the implantation period in pigs. Biol. Reprod. 2012, 87, 31. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Ibrahim, S.; Gebremedhn, S.; Tesfaye, D.; Heppelmann, M.; Bollwein, H.; Pfarrer, C.; Tholen, E.; Neuhoff, C.; Schellander, K.; et al. Clinical and subclinical endometritis induced alterations in bovine endometrial transcriptome and miRNome profile. BMC Genom. 2016, 17, 218. [Google Scholar] [CrossRef]
- de Kouchkovsky, D.; Esensten, J.H.; Rosenthal, W.L.; Morar, M.M.; Bluestone, J.A.; Jeker, L.T. MicroRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J. Immunol. 2013, 191, 1594–1605. [Google Scholar] [CrossRef]
- Aluvihare, V.R.; Kallikourdis, M.; Betz, A.G. Regulatory T cells mediate maternal tolerance to the fetus. Nat. Immunol. 2004, 5, 266–271. [Google Scholar] [CrossRef]
- Robertson, S.A.; Prins, J.R.; Sharkey, D.J.; Moldenhauer, L.M. Seminal fluid and the generation of regulatory T cells for embryo implantation. Am. J. Reprod. Immunol. 2013, 69, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, S.; Balcells, I.; Castello, A.; Ovilo, C.; Noguera, J.L.; Timoneda, O.; Sanchez, A. Endometrial gene expression profile of pregnant sows with extreme phenotypes for reproductive efficiency. Sci. Rep. 2015, 5, 14416. [Google Scholar] [CrossRef]
- Karaayvaz, M.; Zhang, C.; Liang, S.; Shroyer, K.R.; Ju, J. Prognostic significance of miR-205 in endometrial cancer. PLoS ONE 2012, 7, e35158. [Google Scholar] [CrossRef]
- Dong, Y.; Si, J.-W.; Li, W.-T.; Liang, L.; Zhao, J.; Zhou, M.; Li, D.; Li, T. MiR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int. J. Clin. Exp. Pathol. 2015, 8, 2864–2875. [Google Scholar] [PubMed]
- Wilczynski, M.; Danielska, J.; Dzieniecka, M.; Szymanska, B.; Wojciechowski, M.; Malinowski, A. Prognostic and Clinical Significance of miRNA-205 in Endometrioid Endometrial Cancer. PLoS ONE 2016, 11, e0164687. [Google Scholar] [CrossRef]
- Peng, P.; Li, Z.; Liu, X. Reduced Expression of miR-23a Suppresses A20 in TLR-stimulated Macrophages. Inflammation 2015, 38, 1787–1793. [Google Scholar] [CrossRef]
- Si, X.; Cao, D.; Chen, J.; Nie, Y.; Jiang, Z.; Chen, M.-Y.; Wu, J.-F.; Guan, X.-D. MiR23a downregulation modulates the inflammatory response by targeting ATG12mediated autophagy. Mol. Med. Rep. 2018, 18, 1524–1530. [Google Scholar] [PubMed]
- Gu, X.; Gao, Y.; Mu, D.-G.; Fu, E.-Q. MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-kappaB pathway by targeting TLR2. Exp. Cell Res. 2017, 354, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Nunez-Hernandez, F.; Perez, L.J.; Munoz, M.; Vera, G.; Tomas, A.; Egea, R.; Cordoba, S.; Segales, J.; Sanchez, A.; Nunez, J.I. Identification of microRNAs in PCV2 subclinically infected pigs by high throughput sequencing. Vet. Res. 2015, 46, 18. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Lin, H.; Qi, J.; Zhu, C.; Gao, Z.; Wang, H. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction 2012, 143, 389–397. [Google Scholar] [CrossRef]
- Yang, Q.; Gu, W.-W.; Gu, Y.; Yan, N.-N.; Mao, Y.-Y.; Zhen, X.-X.; Wang, J.-M.; Yang, J.; Shi, H.-J.; Zhang, X.; et al. Association of the peripheral blood levels of circulating microRNAs with both recurrent miscarriage and the outcomes of embryo transfer in an in vitro fertilization process. J. Transl. Med. 2018, 16, 186. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-L.; Sun, J.-J.; Ma, H.; Liu, S.-J.; Li, N.; Guo, S.-J.; Shi, Y.; Xu, Y.-Y.; Qi, Z.-Y.; Wang, Y.-Q.; et al. MicroRNA-23a inhibits endometrial cancer cell development by targeting SIX1. Oncol. Lett. 2019, 18, 3792–3802. [Google Scholar] [CrossRef] [PubMed]




| miRNA | No. of Predicted Target Genes | Names of Predicted Target Genes |
|---|---|---|
| miR-23a-5p | 12 | GIPC3, ABCA1, MTMR4, VSIG1, GOLGA6L1, GOLGA6L6, MLIP, LOC100132813, SSMEM1, USH2A, TMEM127, MYEOV |
| miR-34b | 87 | TENM1, INSIG1, ELMOD1, FURIN, PPP6R3, RFX3, DLL1, RAB3C, ZC4H2, CAMSAP2, MAP2, MTF2, MYSM1, NCKAP1, PLEKHA1, SETD3, SLITRK3, STK38L, THRB, ANKS1B, CLINT1, G2E3, GPATCH8, NEUROD1, PIK3C2A, PRKAR2B, SOX6, ASCL1, CAMK4, FDX1, RPRD1A, TLNRD1, YTHDC2, YWHAG, APH1A, ARID1B, ATP11C, BRINP2, CBLB, ELMSAN1, ESPL1, F2RL2, GAS1, GBP4, MYF5, RDX, THAP12, TM9SF3, ATP6V0A2, CDK19, CNTNAP1, HOXB8, HOXC8, KCNA1, KIF2A, MARVELD2, MYCBP2, NPTN, QDPR, STK39, AKTIP, APOB, CTNND2, GABRB2, MTCL1, MYC, NHSL1, PHACTR1, STMN2, TENT4B, ACTL6A, CADM2, CELF2, FKBP1B, IL1RAP, MAPK4, MTDH, NUPL2, PGRMC1, PIEZO2, PTPN4, PTPRG, RALA, RHOH, TENT4A, WIPF3, XKR6 |
| miR-34b-3p | 32 | INSIG1, PPP6R3, FURIN, NCKAP1, TENM1, SETD3, MYSM1, MTF2, SLITRK3, MAP2, CLINT1, PRKAR2B, G2E3, GPATCH8, RPRD1A, YWHAG, YTHDC2, CAMK4, TLNRD1, FDX1, F2RL2, BRINP2, ESPL1, GBP4, TM9SF3, KIF2A, NPTN, AKTIP, TENT4B, GABRB2, PGRMC1, NUPL2 |
| miR92b-5p | 1 | FN3K |
| miR-205 | 266 | RRM2B, MOSMO, MINDY2, CSGALNACT2, RAB11FIP1, PPP2R2D, DMRT1, CHN1, CDK19, C5orf24, BICC1, RBM47, PNPT1, PIK3CG, PIAS2, NLGN1, LPGAT1, LPCAT1, LIN9, DST, BTBD3, AAK1, TOPBP1, TAPT1, PTPRJ, PDLIM5, NFAT5, NANOS1, MSANTD4, MAP3K13, LRRC8B, LRRC19, ITGB8, FAM19A1, FAM122A, DSC2, CCNJ, C9orf153, AZIN1, ATRN, ZNF606, ZFYVE16, WBP2, UBE2R2, TTPAL, TNFAIP8, TMEM245, SSR3, SLC30A8, SFT2D1, RSBN1, RPRD1A, ROCK2, RBM4B, RBM39, PMEL, PLCB1, MRC1, MGRN1, LYPLA1, GALNT13, FAM49A, CPSF6, CMC1, CDH11, CAP2, CALCRL, CADM1, C11orf86, ATRX, AGFG1, XYLT1, WASHC4, VTI1B, TSPAN2, TRIM33, SLC35B3, SLC12A2, SIRT1, SH3BGRL2, RYR3, PRKCE, NUP54, NFATC3, NECAP1, MORC3, MNT, MELK, MAGI2, LRP6, LCOR, KIAA1841, HS3ST1, GABRG1, FZD3, ENTPD1, CNR1, CASC4, B4GALT6, ATXN1, ARHGAP15, APAF1, ANKRD12, AFTPH, ACTA2, YWHAB, TMEM144, SPANXN5, SLC49A4, SLC19A2, SEC62, RELCH, QKI, PTPRM, PDE3B, NEMP1, MYOC, MPP6, INO80D, HERC3, HDX, GEMIN2, EVA1C, ERBB4, DUSP7, DRG2, DLD, CWC27, CPEB2, CEP350, CDK14, CCDC80, BOLA2-SMG1P6, AMOT, ZNF644, UNC13C, TUT7, TOB1, STRBP, SLC15A2, RTN3, RMDN1, RFX3, RCN2, PLAC8, NOL4, NKD1, NFIB, LRRK2, HS3ST2, HECW1, HDAC9, GPC6, FOXF1, FAM126A, EZR, ETNK1, ERRFI1, ENAH, DUSP16, DMXL2, COL10A1, B3GNT2, ANKRD50, AFDN, URI1, TMEM132B, TMED4, TIMM8A, SLC35A1, SIPA1L1, SGMS1, SEPT4, SEMA3A, SEL1L, SATB2, RORA, RIPOR3, PMFBP1, PDE10A, P2RY1, MSI2, MARCKS, LZIC, LRP1, INPP4A, HSF5, HSD17B11, GABRA4, F5, CREB1, COBL, CLTC, CLDN11, CCDC59, CAP1, C6orf222, ATP8A1, ARMC1, AP1S1, ANKIB1, ADAM10, ZNF800, ZNF652, ZEB1, ZC3H12C, XIRP2, TTC33, TET1, RECK, RCBTB1, RASSF6, RAB21, PTTG1IP, PSD3, PDE1C, PAX9, NEK7, MIER3, MECP2, LAMC1, KPNA1, IMPAD1, IMPA1, GLRB, FUT9, ERBB3, ELF2, E2F5, DYNLT1, CSTF2, CREBZF, COL3A1, CCSER1, CARNMT1, C5, APC, ADAMTS9, ADAM7, ZNF655, ZBED4, YAP1, TP53BP2, TNKS, SUCNR1, SORBS1, SECISBP2L, RUVBL1, RBM41, PPM1D, NOTCH2, NACC2, MMP16, MAGI1, LYSMD3, KRTAP4-2, KAT2B, IVNS1ABP, FAM120B, EPPK1, COX20, CHCHD1, CCSER2, CAPN14, ALG10, ABCD1 |
| miR-574-5p | 18 | CALCOCO1, FOXI2, C11orf96, RFX4, NSUN5, DGKG, TCF20, CCK, FOXN3, DDB1, DAZL, LARP6, L2HGDH, NRN1, FOXL2NB, MS4A7, IL5RA, HLTF |
| miR-4776-3p | 56 | ZNF99, ZNF493, CXCL5, ZNF138, ZNF117, CFL2, HOOK1, LRAT, ARHGAP32, ZNF714, ZNF730, CUL3, EIF5A2, ZNF728, SLC16A7, CADM2, ZNF431, HYPK, ZNF329, ALOX15, AGO3, GGNBP2, CCDC88A, RAB3C, TBCEL, ZNF208, COL11A1, IQSEC3, GLIPR1L2, LUZP1, RNF169, PAK5, PHF6, ZNF195, SDC2, TRIM14, BICC1, ZNF107, USH2A, ZNF468, GLYR1, MTMR9, YTHDF3, PTCHD4, TRIM33, EBF1, DEPDC4, ZNF737, ESYT3, TAX1BP1, ZNF732, ZNF708, ZNF600, USP25, RP2, FBXO22 |
| miR-3944-3p | 1 | ICAM2 |
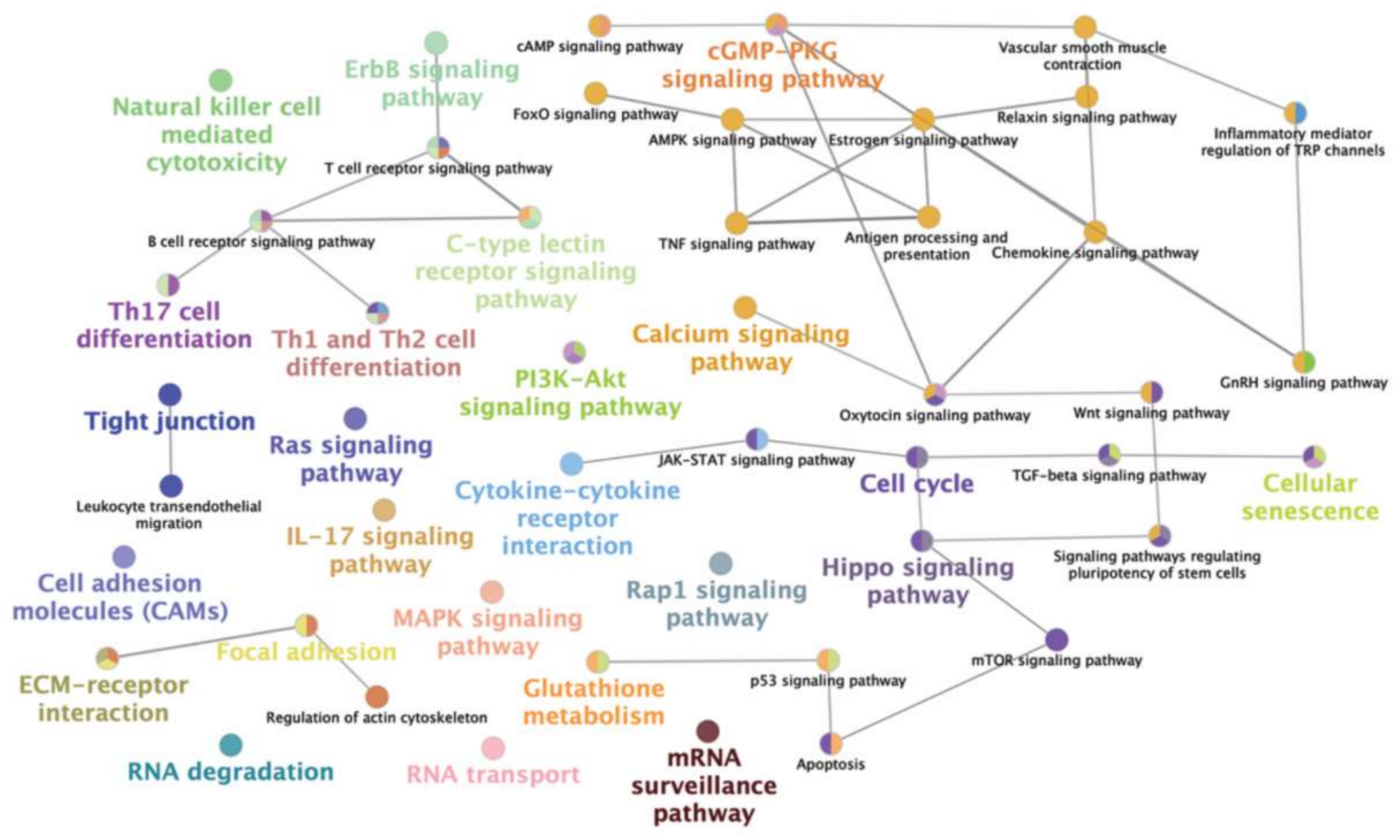
| Pathway | SP-Source | miRNAs | ||
|---|---|---|---|---|
| SRF | Post-SRF | EE | ||
| Th1 and Th2 cell differentiation | + | + | + | miR-34b, miR-205 |
| Cytokine-cytokine receptor interaction | + | + | + | miR-34b, miR-574-5p |
| T cell receptor signaling | + | + | + | miR-34b, miR-205, miR-4776-3p |
| IL-17 signaling | + | + | + | miR-34b, miR-4776-3p |
| MAPK signaling | + | + | + | miR-34b, miR-205 |
| Chemokine signaling | − | + | + | miR-205 |
| Th17 cell differentiation | + | + | + | miR-34b, miR-205 |
| TGF-beta signaling | + | + | + | miR-34b, miR-205 |
| PI3K-Akt signaling | + | + | + | miR-34b, miR-34b-3p, miR-205 |
| Cell adhesion molecules | + | + | + | miR-34b, miR-205, miR-4776-3p |
| TNF signaling | − | + | + | miR-205 |
| Wnt signaling | + | + | + | miR-34b, miR-205 |
| Focal adhesion | + | + | + | miR-205, miR-4776-3p |
| GnRH signaling | − | + | + | miR-205 |
| B cell receptor signaling | − | + | + | miR-205 |
| Oxytocin signaling | + | + | + | miR-34b, miR-34b-3p, miR-205 |
| JAK-STAT signaling | + | + | + | miR-34b, miR-205, miR-574-5p |
| Estrogen signaling | − | + | + | miR-205 |
| p53 signaling | − | + | + | miR-205 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barranco, I.; Padilla, L.; Martinez, C.A.; Alvarez-Rodriguez, M.; Parrilla, I.; Lucas, X.; Ferreira-Dias, G.; Yeste, M.; Rodriguez-Martinez, H.; Roca, J. Seminal Plasma Modulates miRNA Expression by Sow Genital Tract Lining Explants. Biomolecules 2020, 10, 933. https://doi.org/10.3390/biom10060933
Barranco I, Padilla L, Martinez CA, Alvarez-Rodriguez M, Parrilla I, Lucas X, Ferreira-Dias G, Yeste M, Rodriguez-Martinez H, Roca J. Seminal Plasma Modulates miRNA Expression by Sow Genital Tract Lining Explants. Biomolecules. 2020; 10(6):933. https://doi.org/10.3390/biom10060933
Chicago/Turabian StyleBarranco, Isabel, Lorena Padilla, Cristina A. Martinez, Manuel Alvarez-Rodriguez, Inmaculada Parrilla, Xiomara Lucas, Graça Ferreira-Dias, Marc Yeste, Heriberto Rodriguez-Martinez, and Jordi Roca. 2020. "Seminal Plasma Modulates miRNA Expression by Sow Genital Tract Lining Explants" Biomolecules 10, no. 6: 933. https://doi.org/10.3390/biom10060933
APA StyleBarranco, I., Padilla, L., Martinez, C. A., Alvarez-Rodriguez, M., Parrilla, I., Lucas, X., Ferreira-Dias, G., Yeste, M., Rodriguez-Martinez, H., & Roca, J. (2020). Seminal Plasma Modulates miRNA Expression by Sow Genital Tract Lining Explants. Biomolecules, 10(6), 933. https://doi.org/10.3390/biom10060933











