Autocrine TGF-β1 Maintains the Stability of Foxp3+ Regulatory T Cells via IL-12Rβ2 Downregulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Mice
2.3. Flow Cytometric Analysis
2.4. In Vitro Differentiation
2.5. In Vitro Suppression Assay
2.6. In Vivo Adoptive T Cell Transfer
2.7. In Vitro Treg Cell Conversion Assay
2.8. Quantitative RT-PCR
2.9. Statistical Analysis
3. Results
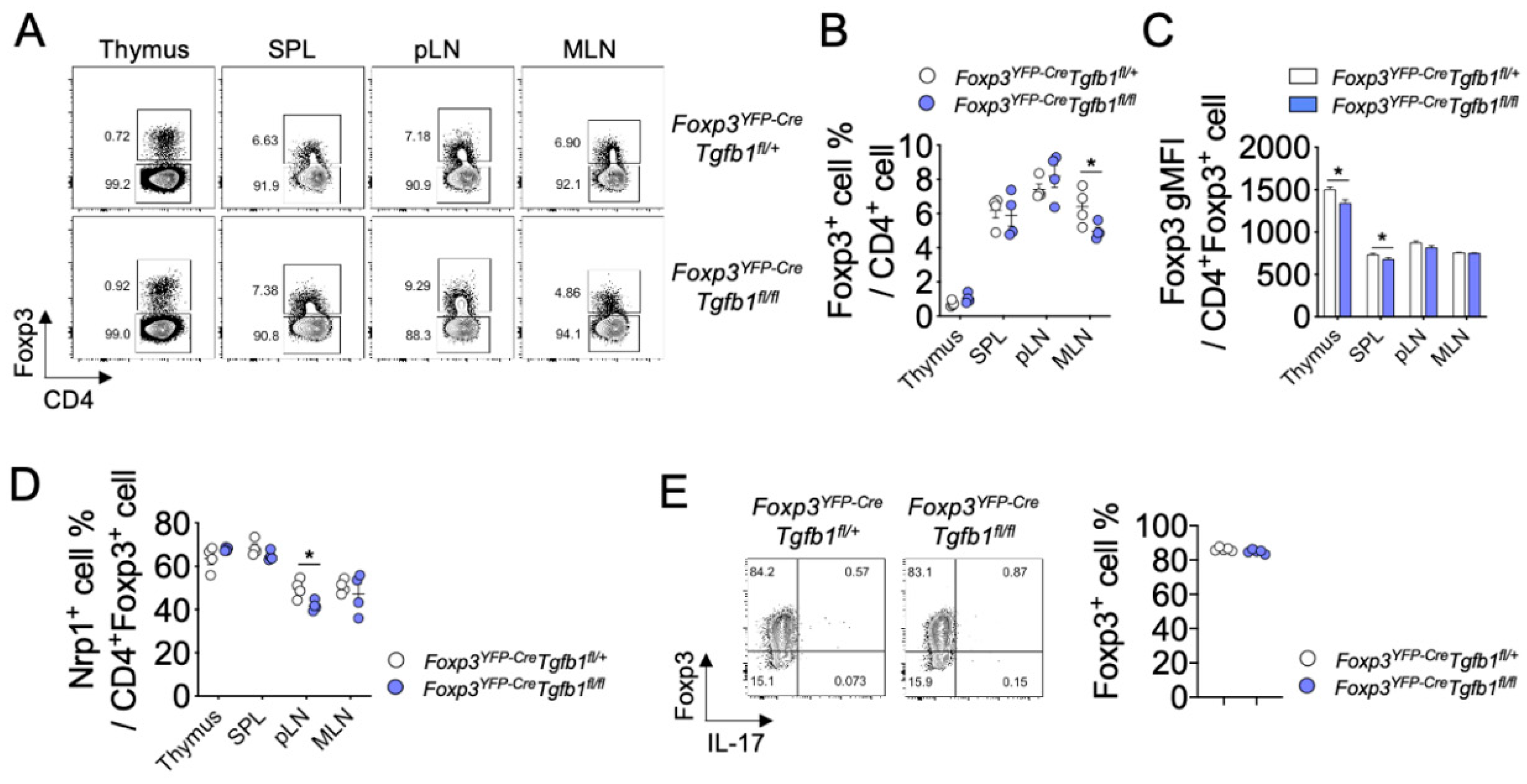
3.1. The Role of Autocrine TGF-β1 in the Development of Thymic and Peripheral Treg Cells
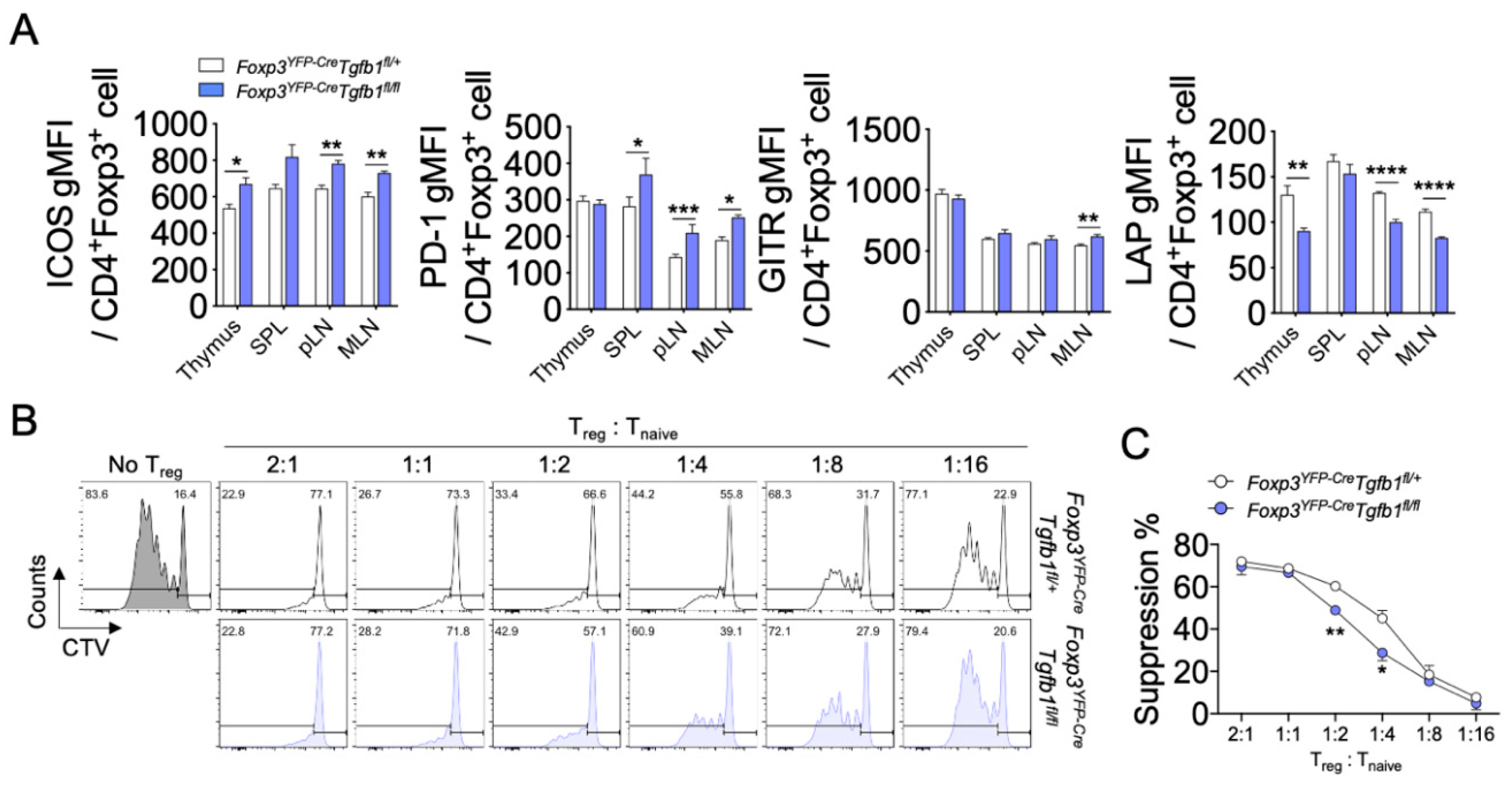
3.2. The Role of Autocrine TGF-β1 in the Phenotype and Suppressive Function of Treg Cells
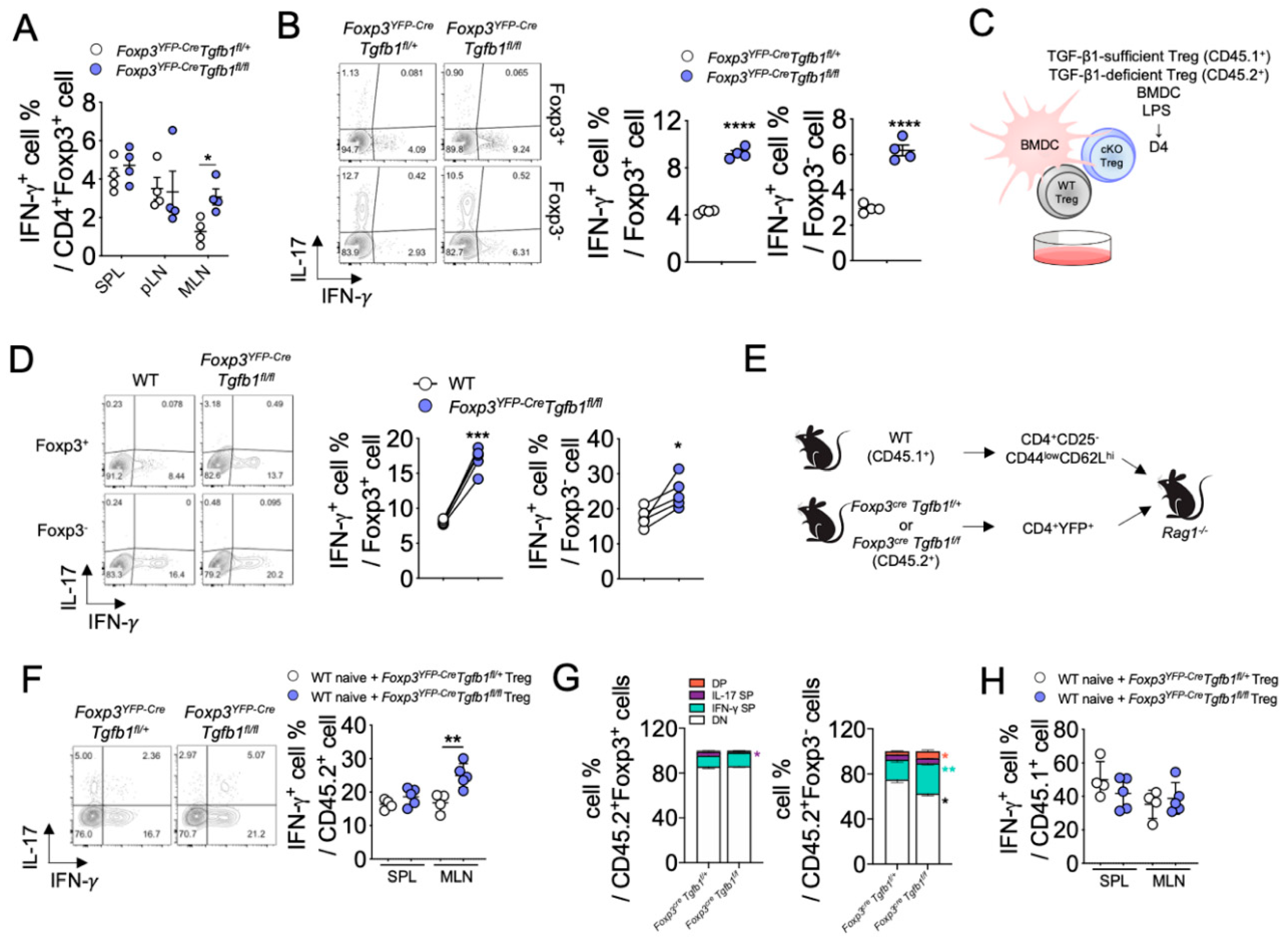
3.3. Autocrine TGF-β1 Contributes to the Stability of Treg Cells
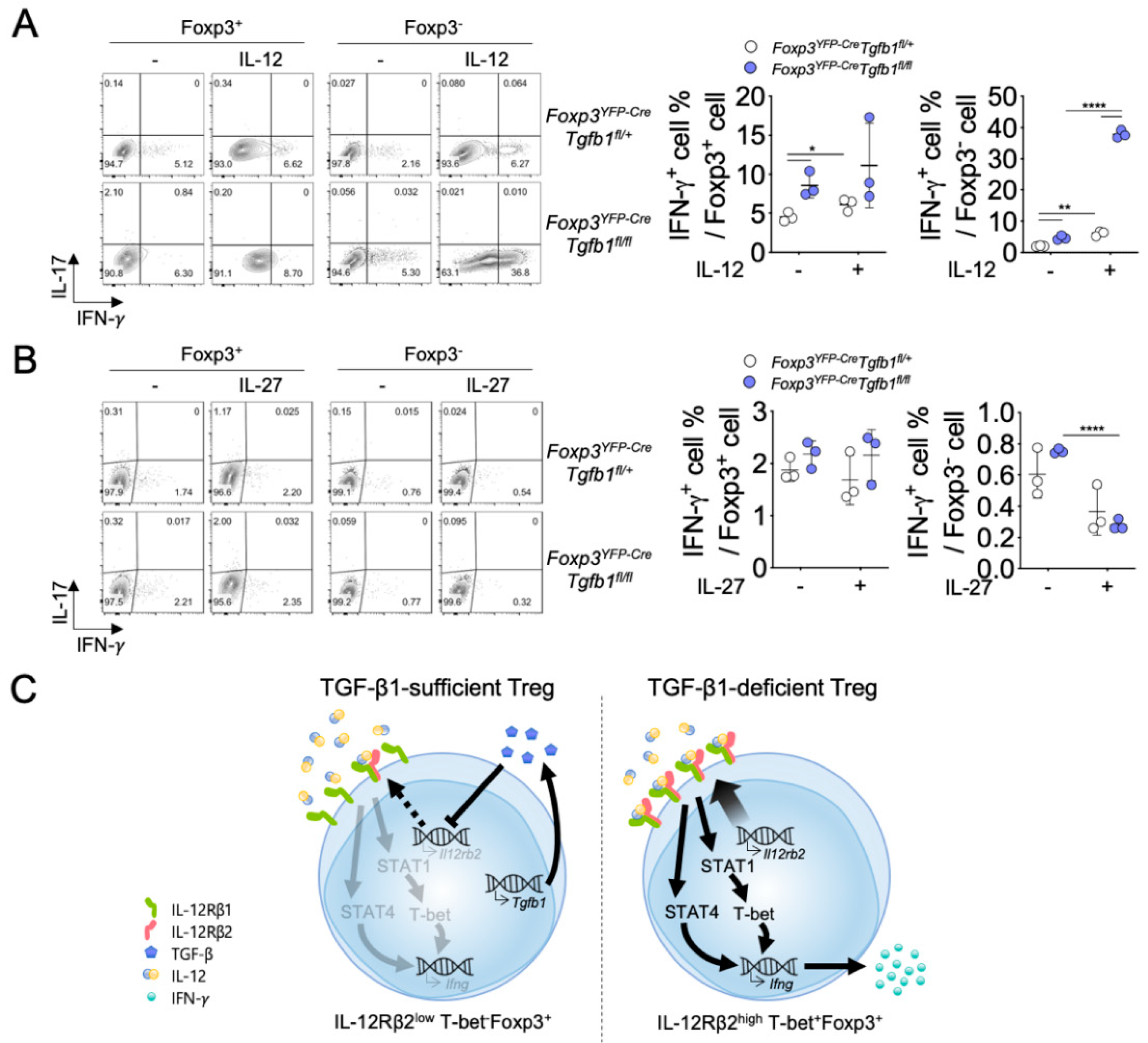
3.4. Autocrine TGF-β1 Maintains the Stability of Treg Cells by Inhibiting IL-12Rβ Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar]
- Takahashi, T.; Kuniyasu, Y.; Toda, M.; Sakaguchi, N.; Itoh, M.; Iwata, M.; Shimizu, J.; Sakaguchi, S. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: Induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 1998, 10, 1969–1980. [Google Scholar] [CrossRef] [PubMed]
- Thornton, A.M.; Shevach, E.M. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 1998, 188, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Tzavlaki, K.; Moustakas, A. TGF-beta signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Bensinger, S.J.; Bandeira, A.; Jordan, M.S.; Caton, A.J.; Laufer, T.M. Major histocompatibility complex class II-positive cortical epithelium mediates the selection of CD4(+)25(+) immunoregulatory T cells. J. Exp. Med. 2001, 194, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar] [CrossRef]
- Apostolou, I.; von Boehmer, H. In vivo instruction of suppressor commitment in naive T cells. J. Exp. Med. 2004, 199, 1401–1408. [Google Scholar] [CrossRef]
- Thornton, A.M.; Korty, P.E.; Tran, D.Q.; Wohlfert, E.A.; Murray, P.E.; Belkaid, Y.; Shevach, E.M. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010, 184, 3433–3441. [Google Scholar] [CrossRef]
- Weiss, J.M.; Bilate, A.M.; Gobert, M.; Ding, Y.; Curotto de Lafaille, M.A.; Parkhurst, C.N.; Xiong, H.; Dolpady, J.; Frey, A.B.; Ruocco, M.G.; et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 2012, 209 (Suppl. S1721), 1723–1742. [Google Scholar] [CrossRef]
- Li, M.O.; Wan, Y.Y.; Sanjabi, S.; Robertson, A.K.; Flavell, R.A. Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 2006, 24, 99–146. [Google Scholar] [CrossRef] [PubMed]
- Shull, M.M.; Ormsby, I.; Kier, A.B.; Pawlowski, S.; Diebold, R.J.; Yin, M.; Allen, R.; Sidman, C.; Proetzel, G.; Calvin, D.; et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature 1992, 359, 693–699. [Google Scholar] [CrossRef]
- Kulkarni, A.B.; Huh, C.G.; Becker, D.; Geiser, A.; Lyght, M.; Flanders, K.C.; Roberts, A.B.; Sporn, M.B.; Ward, J.M.; Karlsson, S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. USA 1993, 90, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Boivin, G.P.; O’Toole, B.A.; Orsmby, I.E.; Diebold, R.J.; Eis, M.J.; Doetschman, T.; Kier, A.B. Onset and progression of pathological lesions in transforming growth factor-beta 1-deficient mice. Am. J. Pathol. 1995, 146, 276–288. [Google Scholar] [PubMed]
- Annes, J.P.; Munger, J.S.; Rifkin, D.B. Making sense of latent TGFbeta activation. J. Cell Sci. 2003, 116, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Travis, M.A.; Reizis, B.; Melton, A.C.; Masteller, E.; Tang, Q.; Proctor, J.M.; Wang, Y.; Bernstein, X.; Huang, X.; Reichardt, L.F.; et al. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature 2007, 449, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Passerini, L.; Olek, S.; Di Nunzio, S.; Barzaghi, F.; Hambleton, S.; Abinun, M.; Tommasini, A.; Vignola, S.; Cipolli, M.; Amendola, M.; et al. Forkhead box protein 3 (FOXP3) mutations lead to increased TH17 cell numbers and regulatory T-cell instability. J. Allergy Clin. Immunol. 2011, 128, 1376–1379. [Google Scholar] [CrossRef]
- Shi, M.; Zhu, J.; Wang, R.; Chen, X.; Mi, L.; Walz, T.; Springer, T.A. Latent TGF-beta structure and activation. Nature 2011, 474, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.; Gunaltay, S.; McEntee, C.P.; Shuttleworth, E.E.; Smedley, C.; Houston, S.A.; Fenton, T.M.; Levison, S.; Mann, E.R.; Travis, M.A. Human monocytes and macrophages regulate immune tolerance via integrin alphavbeta8-mediated TGFbeta activation. J. Exp. Med. 2018, 215, 2725–2736. [Google Scholar] [CrossRef]
- Li, M.O.; Sanjabi, S.; Flavell, R.A. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity 2006, 25, 455–471. [Google Scholar] [CrossRef]
- Marie, J.C.; Liggitt, D.; Rudensky, A.Y. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity 2006, 25, 441–454. [Google Scholar] [CrossRef]
- Ouyang, W.; Beckett, O.; Ma, Q.; Li, M.O. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 2010, 32, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Li, M.O.; Wan, Y.Y.; Flavell, R.A. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity 2007, 26, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Gutcher, I.; Donkor, M.K.; Ma, Q.; Rudensky, A.Y.; Flavell, R.A.; Li, M.O. Autocrine transforming growth factor-beta1 promotes in vivo Th17 cell differentiation. Immunity 2011, 34, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Konkel, J.E.; Zhang, D.; Zanvit, P.; Chia, C.; Zangarle-Murray, T.; Jin, W.; Wang, S.; Chen, W. Transforming Growth Factor-beta Signaling in Regulatory T Cells Controls T Helper-17 Cells and Tissue-Specific Immune Responses. Immunity 2017, 46, 660–674. [Google Scholar] [CrossRef]
- Zhou, X.; Bailey-Bucktrout, S.L.; Jeker, L.T.; Penaranda, C.; Martinez-Llordella, M.; Ashby, M.; Nakayama, M.; Rosenthal, W.; Bluestone, J.A. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 2009, 10, 1000–1007. [Google Scholar] [CrossRef]
- Bovenschen, H.J.; van de Kerkhof, P.C.; van Erp, P.E.; Woestenenk, R.; Joosten, I.; Koenen, H.J. Foxp3+ regulatory T cells of psoriasis patients easily differentiate into IL-17A-producing cells and are found in lesional skin. J. Investig. Dermatol. 2011, 131, 1853–1860. [Google Scholar] [CrossRef]
- Hovhannisyan, Z.; Treatman, J.; Littman, D.R.; Mayer, L. Characterization of interleukin-17-producing regulatory T cells in inflamed intestinal mucosa from patients with inflammatory bowel diseases. Gastroenterology 2011, 140, 957–965. [Google Scholar] [CrossRef]
- Bailey-Bucktrout, S.L.; Martinez-Llordella, M.; Zhou, X.; Anthony, B.; Rosenthal, W.; Luche, H.; Fehling, H.J.; Bluestone, J.A. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity 2013, 39, 949–962. [Google Scholar] [CrossRef]
- Tsuji, M.; Komatsu, N.; Kawamoto, S.; Suzuki, K.; Kanagawa, O.; Honjo, T.; Hori, S.; Fagarasan, S. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science 2009, 323, 1488–1492. [Google Scholar] [CrossRef]
- Oldenhove, G.; Bouladoux, N.; Wohlfert, E.A.; Hall, J.A.; Chou, D.; Dos Santos, L.; O’Brien, S.; Blank, R.; Lamb, E.; Natarajan, S.; et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 2009, 31, 772–786. [Google Scholar] [CrossRef] [PubMed]
- McClymont, S.A.; Putnam, A.L.; Lee, M.R.; Esensten, J.H.; Liu, W.; Hulme, M.A.; Hoffmuller, U.; Baron, U.; Olek, S.; Bluestone, J.A.; et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J. Immunol. 2011, 186, 3918–3926. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Kitani, A.; Fuss, I.; Strober, W. Cutting edge: Regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J. Immunol. 2007, 178, 6725–6729. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.O.; Nurieva, R.; Martinez, G.J.; Kang, H.S.; Chung, Y.; Pappu, B.P.; Shah, B.; Chang, S.H.; Schluns, K.S.; Watowich, S.S.; et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 2008, 29, 44–56. [Google Scholar] [CrossRef]
- Sharma, M.D.; Hou, D.Y.; Baban, B.; Koni, P.A.; He, Y.; Chandler, P.R.; Blazar, B.R.; Mellor, A.L.; Munn, D.H. Reprogrammed foxp3(+) regulatory T cells provide essential help to support cross-presentation and CD8(+) T cell priming in naive mice. Immunity 2010, 33, 942–954. [Google Scholar] [CrossRef]
- Da Silva Martins, M.; Piccirillo, C.A. Functional stability of Foxp3+ regulatory T cells. Trends Mol. Med. 2012, 18, 454–462. [Google Scholar] [CrossRef]
- Dominguez-Villar, M.; Hafler, D.A. Regulatory T cells in autoimmune disease. Nat. Immunol. 2018, 19, 665–673. [Google Scholar] [CrossRef]
- Chaudhry, A.; Rudra, D.; Treuting, P.; Samstein, R.M.; Liang, Y.; Kas, A.; Rudensky, A.Y. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 2009, 326, 986–991. [Google Scholar] [CrossRef]
- Koch, M.A.; Tucker-Heard, G.; Perdue, N.R.; Killebrew, J.R.; Urdahl, K.B.; Campbell, D.J. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009, 10, 595–602. [Google Scholar] [CrossRef]
- Zheng, Y.; Chaudhry, A.; Kas, A.; de Roos, P.; Kim, J.M.; Chu, T.T.; Corcoran, L.; Treuting, P.; Klein, U.; Rudensky, A.Y. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 2009, 458, 351–356. [Google Scholar] [CrossRef]
- Chung, Y.; Tanaka, S.; Chu, F.; Nurieva, R.I.; Martinez, G.J.; Rawal, S.; Wang, Y.H.; Lim, H.; Reynolds, J.M.; Zhou, X.H.; et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 2011, 17, 983–988. [Google Scholar] [CrossRef]
- Linterman, M.A.; Pierson, W.; Lee, S.K.; Kallies, A.; Kawamoto, S.; Rayner, T.F.; Srivastava, M.; Divekar, D.P.; Beaton, L.; Hogan, J.J.; et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 2011, 17, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Lu, H.; Ichiyama, K.; Chen, X.; Zhang, Y.B.; Mistry, N.A.; Tanaka, K.; Lee, Y.H.; Nurieva, R.; Zhang, L.; et al. Generation of RORgammat(+) Antigen-Specific T Regulatory 17 Cells from Foxp3(+) Precursors in Autoimmunity. Cell Rep. 2017, 21, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.G.; Mendoza, A.; Hemmers, S.; Moltedo, B.; Niec, R.E.; Schizas, M.; Hoyos, B.E.; Putintseva, E.V.; Chaudhry, A.; Dikiy, S.; et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 2017, 546, 421–425. [Google Scholar] [CrossRef]
- Kryczek, I.; Wu, K.; Zhao, E.; Wei, S.; Vatan, L.; Szeliga, W.; Huang, E.; Greenson, J.; Chang, A.; Rolinski, J.; et al. IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J. Immunol. 2011, 186, 4388–4395. [Google Scholar] [CrossRef]
- Liu, T.; Song, C.H.; Liu, A.M.; Xie, C.; Zhao, F.; Chen, X.; Cheng, L.; Yang, P.C. Forkhead box P3+ T cells express interleukin-17 in nasal mucosa of patients with both allergic rhinitis and polyposis. Clin. Exp. Immunol. 2011, 163, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Hansmann, L.; Schmidl, C.; Kett, J.; Steger, L.; Andreesen, R.; Hoffmann, P.; Rehli, M.; Edinger, M. Dominant Th2 differentiation of human regulatory T cells upon loss of FOXP3 expression. J. Immunol. 2012, 188, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, H.M.; Benoist, C.; Mathis, D. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J. Exp. Med. 2004, 200, 1221–1230. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, P.; Li, J.; Kulkarni, A.B.; Perruche, S.; Chen, W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 2008, 9, 632–640. [Google Scholar] [CrossRef]
- Kitz, A.; de Marcken, M.; Gautron, A.S.; Mitrovic, M.; Hafler, D.A.; Dominguez-Villar, M. AKT isoforms modulate Th1-like Treg generation and function in human autoimmune disease. EMBO Rep. 2016, 17, 1169–1183. [Google Scholar] [CrossRef]
- Dominguez-Villar, M.; Baecher-Allan, C.M.; Hafler, D.A. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat. Med. 2011, 17, 673–675. [Google Scholar] [CrossRef] [PubMed]
- Di Giovangiulio, M.; Rizzo, A.; Franze, E.; Caprioli, F.; Facciotti, F.; Onali, S.; Favale, A.; Stolfi, C.; Fehling, H.J.; Monteleone, G.; et al. Tbet Expression in Regulatory T Cells Is Required to Initiate Th1-Mediated Colitis. Front. Immunol. 2019, 10, 2158. [Google Scholar] [CrossRef] [PubMed]
- Korn, T.; Reddy, J.; Gao, W.; Bettelli, E.; Awasthi, A.; Petersen, T.R.; Backstrom, B.T.; Sobel, R.A.; Wucherpfennig, K.W.; Strom, T.B.; et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat. Med. 2007, 13, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.G.; Mathis, D.; Benoist, C. Singular role for T-BET+CXCR3+ regulatory T cells in protection from autoimmune diabetes. Proc. Natl. Acad. Sci. USA 2016, 113, 14103–14108. [Google Scholar] [CrossRef]
- Toker, A.; Engelbert, D.; Garg, G.; Polansky, J.K.; Floess, S.; Miyao, T.; Baron, U.; Duber, S.; Geffers, R.; Giehr, P.; et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 2013, 190, 3180–3188. [Google Scholar] [CrossRef]
- Hua, J.; Inomata, T.; Chen, Y.; Foulsham, W.; Stevenson, W.; Shiang, T.; Bluestone, J.A.; Dana, R. Pathological conversion of regulatory T cells is associated with loss of allotolerance. Sci. Rep. 2018, 8, 7059. [Google Scholar] [CrossRef]
- Wang, S.; Gao, X.; Shen, G.; Wang, W.; Li, J.; Zhao, J.; Wei, Y.Q.; Edwards, C.K. Interleukin-10 deficiency impairs regulatory T cell-derived neuropilin-1 functions and promotes Th1 and Th17 immunity. Sci. Rep. 2016, 6, 24249. [Google Scholar] [CrossRef]
- Amarnath, S.; Mangus, C.W.; Wang, J.C.; Wei, F.; He, A.; Kapoor, V.; Foley, J.E.; Massey, P.R.; Felizardo, T.C.; Riley, J.L.; et al. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci. Transl. Med. 2011, 3, 111ra120. [Google Scholar] [CrossRef]
- Vocanson, M.; Rozieres, A.; Hennino, A.; Poyet, G.; Gaillard, V.; Renaudineau, S.; Achachi, A.; Benetiere, J.; Kaiserlian, D.; Dubois, B.; et al. Inducible costimulator (ICOS) is a marker for highly suppressive antigen-specific T cells sharing features of TH17/TH1 and regulatory T cells. J. Allergy Clin. Immunol. 2010, 126, 280–289.e7. [Google Scholar] [CrossRef]
- Zhu, J.; Jankovic, D.; Oler, A.J.; Wei, G.; Sharma, S.; Hu, G.; Guo, L.; Yagi, R.; Yamane, H.; Punkosdy, G.; et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 2012, 37, 660–673. [Google Scholar] [CrossRef]
- Koch, M.A.; Thomas, K.R.; Perdue, N.R.; Smigiel, K.S.; Srivastava, S.; Campbell, D.J. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity 2012, 37, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, G.X.; Wysocka, M.; Wu, C.Y.; Trinchieri, G.; Rostami, A. The suppressive effect of TGF-beta on IL-12-mediated immune modulation specific to a peptide Ac1-11 of myelin basic protein (MBP): A mechanism involved in inhibition of both IL-12 receptor beta1 and beta2. J. Neuroimmunol. 2000, 108, 53–63. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, G.; Na, H.; Kuen, D.-S.; Kim, B.-S.; Chung, Y. Autocrine TGF-β1 Maintains the Stability of Foxp3+ Regulatory T Cells via IL-12Rβ2 Downregulation. Biomolecules 2020, 10, 819. https://doi.org/10.3390/biom10060819
Choi G, Na H, Kuen D-S, Kim B-S, Chung Y. Autocrine TGF-β1 Maintains the Stability of Foxp3+ Regulatory T Cells via IL-12Rβ2 Downregulation. Biomolecules. 2020; 10(6):819. https://doi.org/10.3390/biom10060819
Chicago/Turabian StyleChoi, Garam, Hyeongjin Na, Da-Sol Kuen, Byung-Seok Kim, and Yeonseok Chung. 2020. "Autocrine TGF-β1 Maintains the Stability of Foxp3+ Regulatory T Cells via IL-12Rβ2 Downregulation" Biomolecules 10, no. 6: 819. https://doi.org/10.3390/biom10060819
APA StyleChoi, G., Na, H., Kuen, D.-S., Kim, B.-S., & Chung, Y. (2020). Autocrine TGF-β1 Maintains the Stability of Foxp3+ Regulatory T Cells via IL-12Rβ2 Downregulation. Biomolecules, 10(6), 819. https://doi.org/10.3390/biom10060819




