An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of Recombinant AnpIBP and Its Mutants
2.2. Thermal Hysteresis Measurements and Ice Crystal Morphology
2.3. FIPA Analysis
2.4. Crystallization and X-Ray Structure Determination of AnpIBP and Its Mutants
3. Results and Discussion
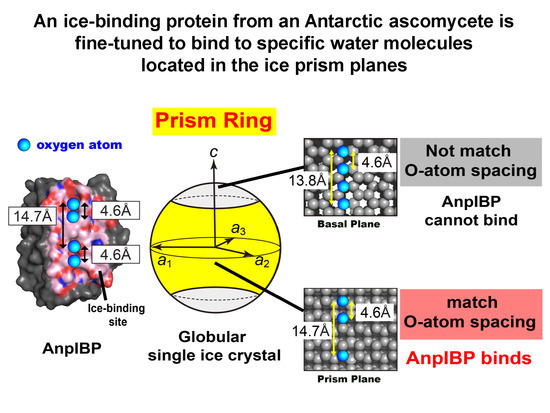
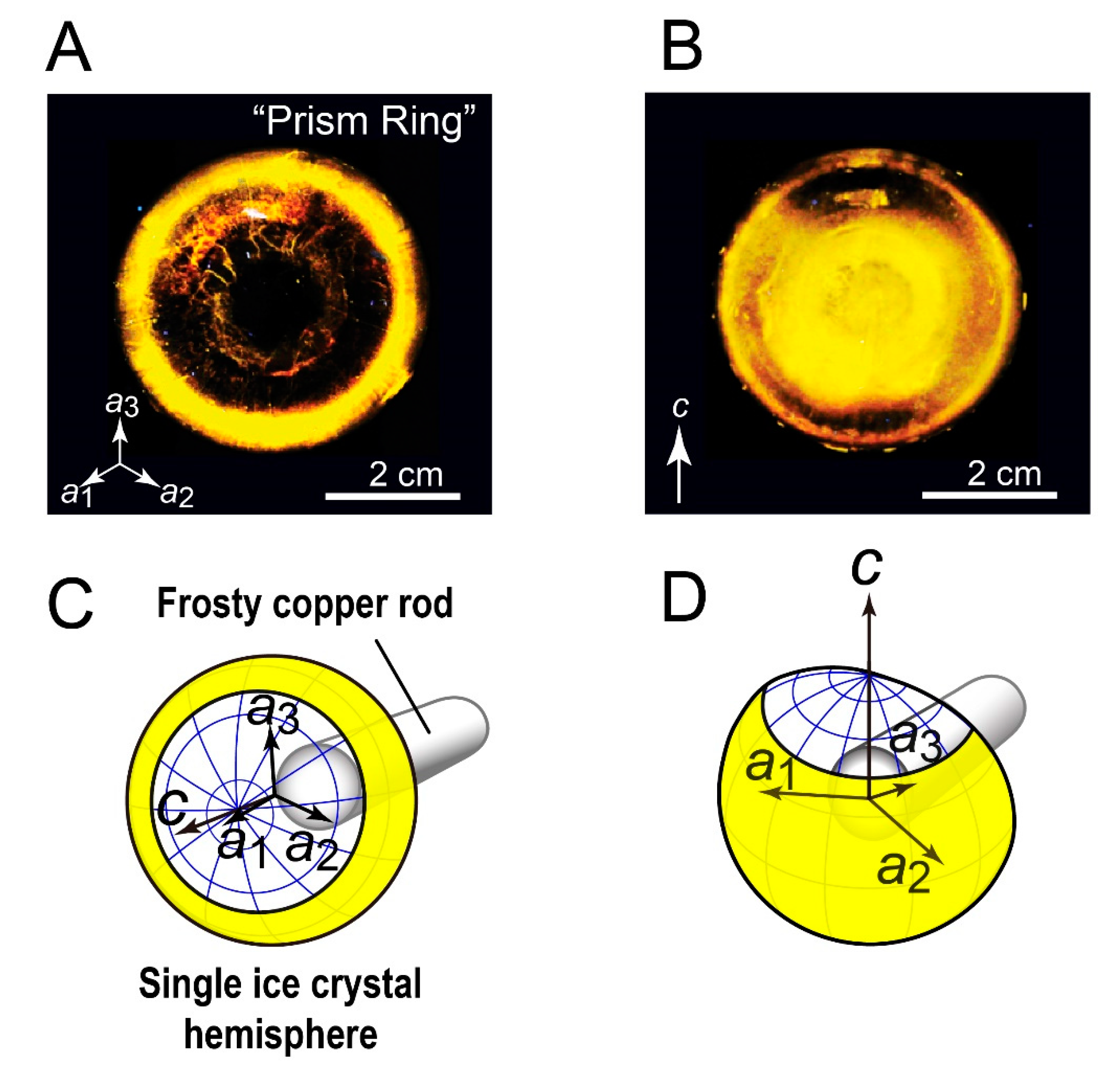
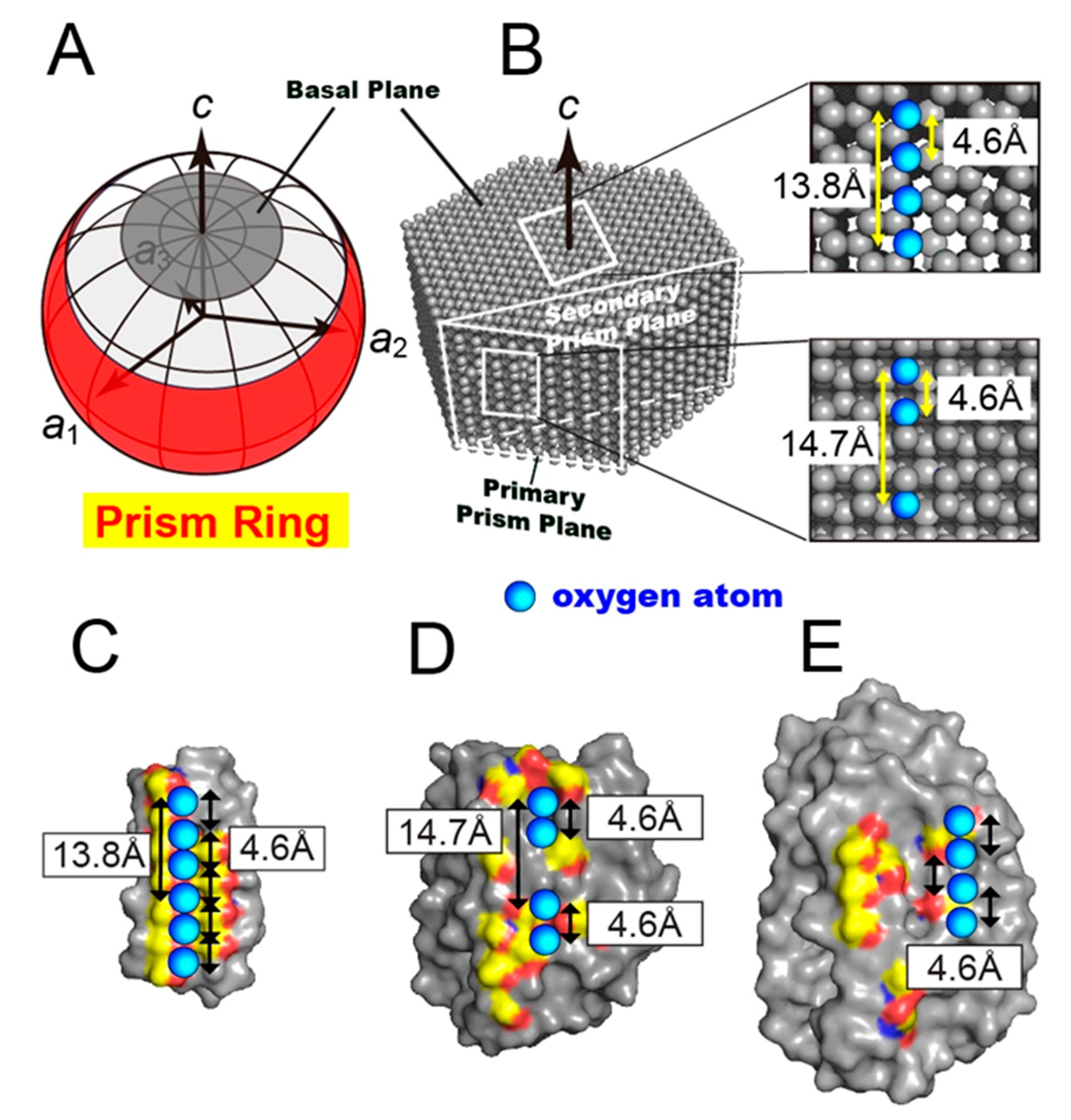
3.1. FIPA of AnpIBP Showed a Novel Prism-Ring Pattern
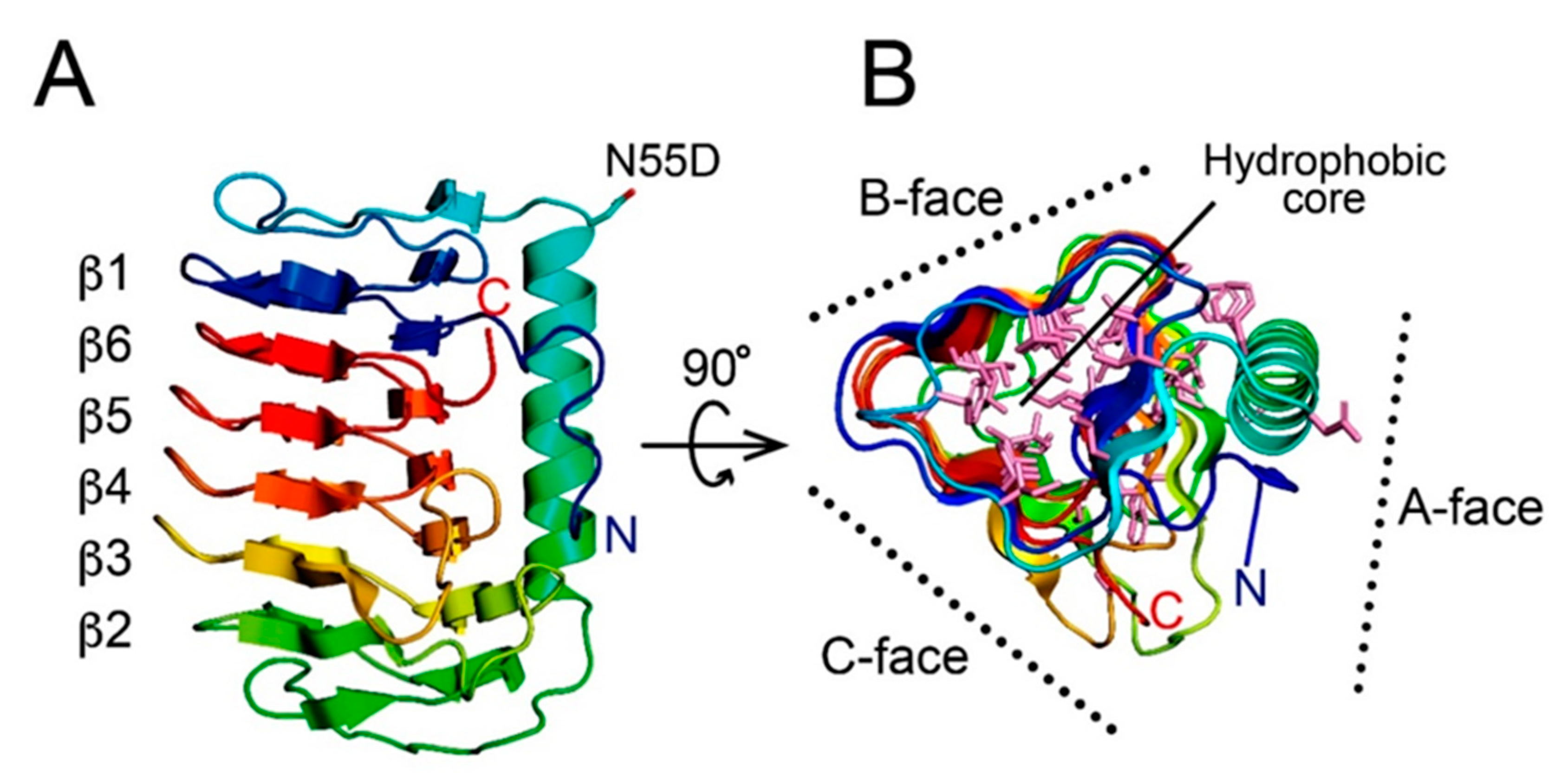
3.2. AnpIBP Folds as an Irregular β-Helical Structure
3.3. The IBS of AnpIBP Located on the B-Face
3.4. Ice-Like Polygonal Water Networks Were Observed on the IBS of AnpIBP
3.5. Ice-Binding Mechanism of AnpIBP
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ball, P. Water as an active constituent in cell biology. Chem. Rev. 2008, 108, 74–108. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.; Onuchic, J.N. Water mediation in protein folding and molecular recognition. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 389–415. [Google Scholar] [CrossRef] [PubMed]
- DeVries, A.L.; Wohlschlag, D.E. Freezing resistance in some Antarctic fishes. Science 1969, 163, 1073–1075. [Google Scholar] [CrossRef]
- Garnham, C.P.; Campbell, R.L.; Davies, P.L. Anchored clathrate waters bind antifreeze proteins to ice. Proc. Natl. Acad. Sci. USA 2011, 108, 7363–7367. [Google Scholar] [CrossRef]
- Davies, P.L. Ice-binding proteins: A remarkable diversity of structures for stopping and starting ice growth. Trends Biochem. Sci. 2014, 39, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Scotter, A.J.; Marshall, C.B.; Graham, L.A.; Gilbert, J.A.; Garnham, C.P.; Davies, P.L. The basis for hyperactivity of antifreeze proteins. Cryobiology 2006, 53, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Graether, S.P.; Sykes, B.D. Cold survival in freeze-intolerant insects: The structure and function of beta-helical antifreeze proteins. Eur. J. Biochem. 2004, 271, 3285–3296. [Google Scholar] [CrossRef]
- Graether, S.P.; Kuiper, M.J.; Gagné, S.M.; Walker, V.K.; Jia, Z.; Sykes, B.D.; Davies, P.L. Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect. Nature 2000, 406, 325–328. [Google Scholar] [CrossRef]
- Liou, Y.-C.; Tocilj, A.; Davies, P.L.; Jia, Z. Mimicry of ice structure by surface hydroxyls and water of a β-helix antifreeze protein. Nature 2000, 406, 322–324. [Google Scholar] [CrossRef]
- Voets, I.K. From ice-binding proteins to bio-inspired antifreeze materials. Soft Matter 2017, 12, 4808–4823. [Google Scholar] [CrossRef]
- Hobbs, P.V. Ice Physics; Oxford Univ Press: London, UK, 1974; pp. 461–523. [Google Scholar]
- Kallungal, J.P. The Growth of a Single Ice Crystal Parallel to the a-Axis in Subcooled Quiescent and Flowing Water. Ph.D. Thesis, Syracuse University, Syracuse, NY, USA, 1975. [Google Scholar]
- Garnham, C.P.; Natarajan, A.; Middleton, A.J.; Kuiper, M.J.; Braslavsky, I.; Davies, P.L. Compound ice-binding site of an antifreeze protein revealed by mutagenesis and fluorescent tagging. Biochemistry 2010, 49, 9063–9071. [Google Scholar] [CrossRef] [PubMed]
- Basu, K.; Garnham, C.P.; Nishimiya, Y.; Tsuda, S.; Braslavsky, I.; Davies, P. Determining the ice-binding planes of antifreeze proteins by fluorescence-based ice plane affinity. JoVE 2014, 83, e51185. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.T.; Arai, T.; Yamauchi, A.; Miura, A.; Kondo, H.; Ohyama, Y.; Tsuda, S. Ice recrystallization is strongly inhibited when antifreeze proteins bind to multiple ice planes. Sci. Rep. 2019, 9, 2212. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.A.; DeVries, A.L. Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc. Natl. Acad. Sci. USA 1977, 74, 2589–2593. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J. Thermal hysteresis proteins. Int. J. Biochem. Cell Biol. 2001, 33, 105–117. [Google Scholar] [CrossRef]
- Celik, Y.; Graham, L.A.; Mok, Y.F.; Bar, M.; Davies, P.L.; Braslavsky, I. Superheating of ice crystals in antifreeze protein solutions. Proc. Natl. Acad. Sci. USA 2010, 107, 5423–5428. [Google Scholar] [CrossRef]
- Takamichi, M.; Nishimiya, Y.; Miura, A.; Tsuda, S. Effect of annealing time of an ice crystal on the activity of type III antifreeze protein. FEBS J. 2007, 274, 6469–6476. [Google Scholar] [CrossRef]
- Arai, T.; Nishimiya, Y.; Ohyama, Y.; Kondo, H.; Tsuda, S. Calcium-binding generates the semi-clathrate waters on a type II antifreeze protein to adsorb onto an ice crystal surface. Biomolecules 2019, 9, 162. [Google Scholar] [CrossRef]
- Mahatabuddin, S.; Fukami, D.; Arai, T.; Nishimiya, Y.; Shimizu, R.; Shibazaki, C.; Kondo, H.; Adachi, M.; Tsuda, S. Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein. Proc. Natl. Acad. Sci. USA 2018, 115, 5456–5461. [Google Scholar] [CrossRef]
- Sun, T.; Lin, F.-H.; Campbell, R.L.; Allingham, J.S.; Davies, P.L. An antifreeze protein folds with an interior network of more than 400 semi-clathrate waters. Science 2014, 343, 795–798. [Google Scholar] [CrossRef]
- Sharp, K.A. The remarkable hydration of the antifreeze protein Maxi: A computational study. J. Chem. Phys. 2014, 141, 22D510. [Google Scholar] [CrossRef] [PubMed]
- Andorfer, C.A.; Duman, J.G. Isolation and characterization of cDNA clones encoding antifreeze proteins of the pyrochroid beetle Dendroides canadensis. J. Insect Physiol. 2000, 46, 365–372. [Google Scholar] [CrossRef]
- Arai, T.; Fukami, D.; Hoshino, T.; Kondo, H.; Tsuda, S. Ice-binding proteins from the fungus Antarctomyces psychrotrophicus possibly originate from two different bacteria through horizontal gene transfer. FEBS J. 2018, 286, 946–962. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Hanada, Y.; Sugimoto, H.; Hoshino, T.; Garnham, C.P.; Davies, P.L.; Tsuda, S. Ice-binding site of snow mold fungus antifreeze protein deviates from structural regularity and high conservation. Proc. Natl. Acad. Sci. USA 2012, 109, 9360–9365. [Google Scholar] [CrossRef] [PubMed]
- Bayer-Giraldi, M.; Weikusat, I.; Besir, H.; Dieckmann, G. Characterization of an antifreeze protein from the polar diatom Fragilariopsis cylindrus and its relevance in sea ice. Cryobiology 2011, 63, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Kiko, R. Acquisition of freeze protection in a sea-ice crustacean through horizontal gene transfer? Polar Biol. 2010, 33, 543–556. [Google Scholar] [CrossRef]
- Vance, T.D.R.; Bayer-Giraldi, M.; Davies, P.L.; Mangiagalli, M. Ice-binding proteins and the ‘domain of unknown function’ 3494 family. FEBS J. 2019, 286, 855–873. [Google Scholar] [CrossRef]
- Hanada, Y.; Nishimiya, Y.; Miura, A.; Tsuda, S.; Kondo, H. Hyperactive antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. has a compound ice-binding site without repetitive sequence. FEBS J. 2014, 281, 3576–3590. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, A.K.; Do, H.; Park, K.S.; Moh, S.H.; Chi, Y.M. Structural basis for antifreeze activity of ice-binding protein from arctic yeast. J. Biol. Chem. 2012, 287, 11460–11468. [Google Scholar] [CrossRef]
- McPherson, A. Current approaches to macromolecular crystallization. Eur. J. Biochem. 1990, 189, 1–23. [Google Scholar] [CrossRef]
- Igarashi, N.; Ikuta, K.; Miyoshi, T.; Matsugaki, N.; Yamada, Y.; Yousef, M.S.; Wakatsuki, S. X-ray beam stabilization at BL-17A, the protein microcrystallography beamline of the Photon Factory. J. Synchrotron Rad. 2008, 15, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993, 26, 795–800. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Brunger, A.T. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992, 355, 472–475. [Google Scholar] [CrossRef]
- Mahatabuddin, S.; Hanada, Y.; Nishimiya, Y.; Miura, A.; Kondo, H.; Davies, P.L.; Tsuda, S. Concentration-dependent oligomerization of an alpha-helical antifreeze polypeptide makes it hyperactive. Sci. Rep. 2017, 7, 42501. [Google Scholar] [CrossRef]
- Tsuda, S.; Yamauchi, A.; Moffiz Uddin Khan, N.M.; Arai, T.; Mahatabuddin, S.; Miura, A.; Kondo, H. Fish-derived antifreeze proteins and antifreeze glycoprotein exhibit a different ice-binding property with increasing concentration. Biomolecules 2020, 10, 423. [Google Scholar] [CrossRef]
- Cheng, J.; Hanada, Y.; Miura, A.; Tsuda, S.; Kondo, H. Hydrophobic ice-binding sites confer hyperactivity of an antifreeze protein from a snow mold fungus. Biochem. J. 2016, 473, 4011–4026. [Google Scholar] [CrossRef]
- Vance, T.D.R.; Graham, L.A.; Davies, P.L. An ice-binding and tandem beta-sandwich domain-containing protein in Shewanella frigidimarina is a potential new type of ice adhesin. FEBS J. 2018, 285, 1511–1527. [Google Scholar] [CrossRef] [PubMed]
- Do, H.; Kim, S.J.; Kim, H.J.; Lee, J.H. Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Mangiagalli, M.; Sarusi, G.; Kaleda, A.; Dolev, M.B.; Nardone, V.; Vena, V.F.; Lotti, M.; Nardini, M. Structure of a bacterial ice binding protein with two face of interaction with ice. FEBS J. 2018, 285, 1635–1666. [Google Scholar] [CrossRef]
- Wang, C.; Pakhomova, S.; Newcomer, M.E.; Christner, B.C.; Luo, B.H. Structural basis of antifreeze activity of a bacterial multi-domain antifreeze protein. PLoS ONE 2017, 12, e0187169. [Google Scholar] [CrossRef] [PubMed]
- Leinala, E.K.; Davies, P.L.; Jia, Z. Crystal structure of beta-helical antifreeze protein points to a general ice binding model. Structure 2002, 10, 619–627. [Google Scholar] [CrossRef]
- Jeffrey, G.A. An Introduction to Hydrogen Bonding; Oxford University: Oxford, NY, USA, 1997. [Google Scholar]
- Luzar, A.; Chandler, D. Effect of environment on hydrogen bond dynamics in liquid water. Phys. Rev. Lett. 1996, 76, 928–931. [Google Scholar] [CrossRef]
- Mantz, Y.A.; Geiger, F.M.; Molina, L.T.; Molina, M.J.; Trout, B.L. First-principles molecular-dynamics study of surface disordering of the (0001) face of hexagonal ice. J. Chem. Phys. 2000, 113, 10733–10743. [Google Scholar] [CrossRef]
- Hayward, J.A.; Haymet, A.D.J. The ice/water interface: Molecular dynamics simulations of the basal, prism, {2021}, and {2110} Interfaces of Ice Ih. J. Chem. Phys. 2001, 114, 3713–3726. [Google Scholar] [CrossRef]
- Gallagher, K.R.; Sharp, K.A. Analysis of thermal hysteresis protein hydration using the random network model. Biophys. Chem. 2003, 105, 195–209. [Google Scholar] [CrossRef]
- Yang, C.; Sharp, K.A. The mechanism of the type III antifreeze protein action: A computational study. Biophys. Chem. 2004, 109, 137–148. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamauchi, A.; Arai, T.; Kondo, H.; Sasaki, Y.C.; Tsuda, S. An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes. Biomolecules 2020, 10, 759. https://doi.org/10.3390/biom10050759
Yamauchi A, Arai T, Kondo H, Sasaki YC, Tsuda S. An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes. Biomolecules. 2020; 10(5):759. https://doi.org/10.3390/biom10050759
Chicago/Turabian StyleYamauchi, Akari, Tatsuya Arai, Hidemasa Kondo, Yuji C. Sasaki, and Sakae Tsuda. 2020. "An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes" Biomolecules 10, no. 5: 759. https://doi.org/10.3390/biom10050759
APA StyleYamauchi, A., Arai, T., Kondo, H., Sasaki, Y. C., & Tsuda, S. (2020). An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes. Biomolecules, 10(5), 759. https://doi.org/10.3390/biom10050759