Multiscale Molecular Modeling in G Protein-Coupled Receptor (GPCR)-Ligand Studies
Abstract
1. Introduction
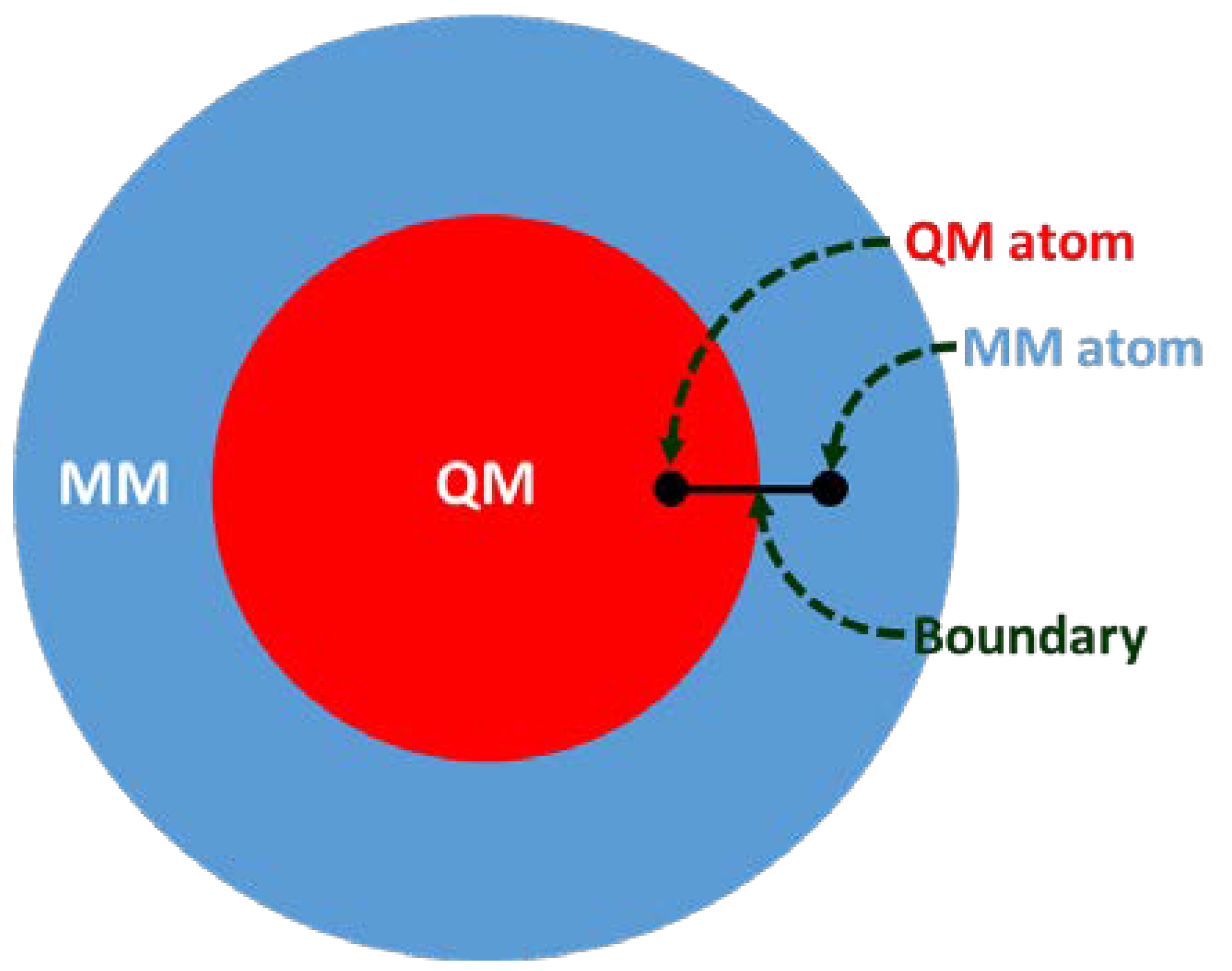
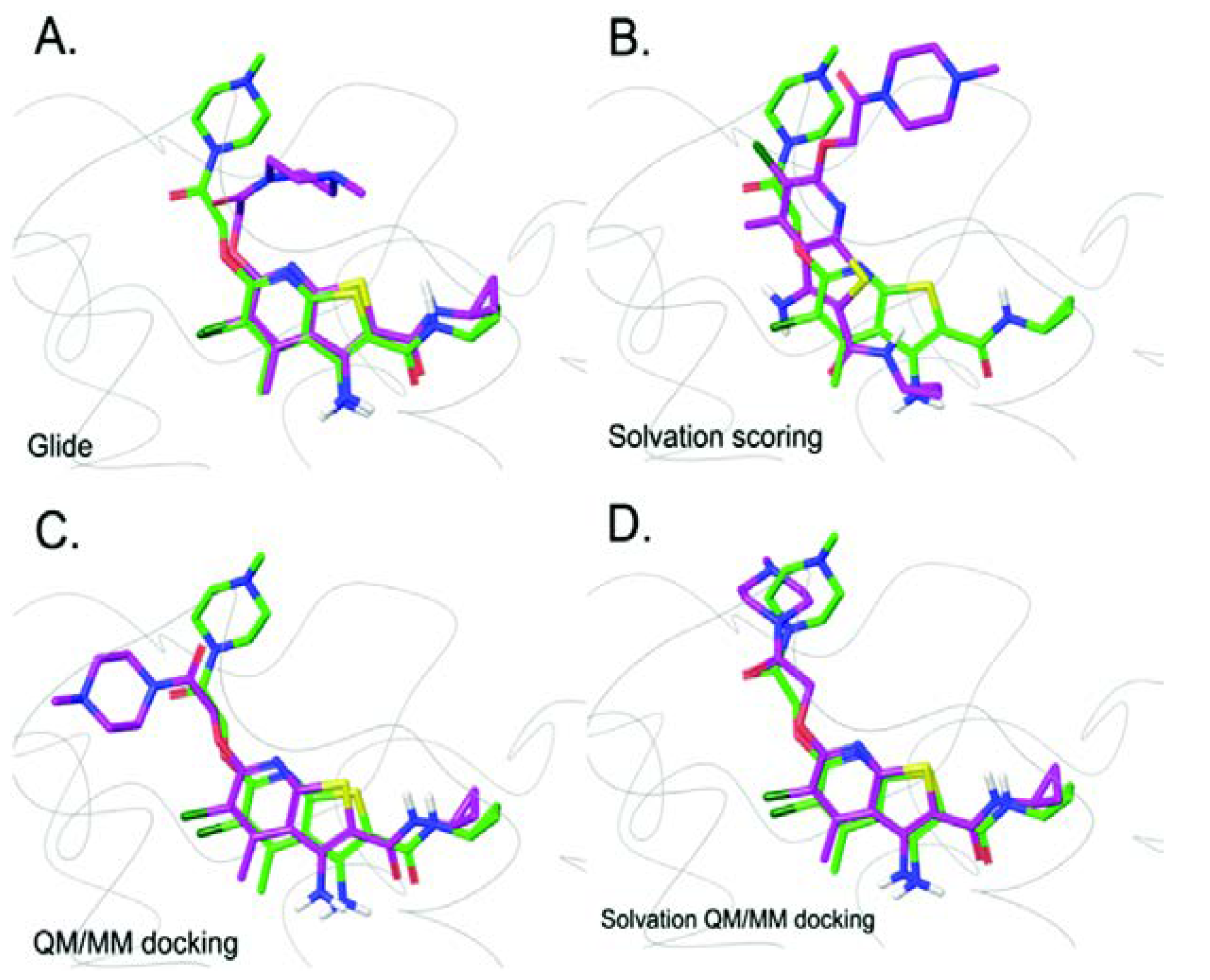
2. Molecular Docking Development using QM/MM Approach
3. Class A Rhodopsin Photoactivity Investigation
4. The QM Approach in GPCR Studies
5. Conclusions and Outlooks
Author Contributions
Funding
Conflicts of Interest
References
- Basith, S.; Cui, M.H.; Macalino, S.J.Y.; Park, J.; Clavio, N.A.B.; Kang, S.; Choi, S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front. Pharmacol. 2018, 9, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.C.S.; Li, Y.; Dahoun, T.; Vogel, H.; Yuan, S.G. New Binding Sites, New Opportunities for GPCR Drug Discovery. Trends Biochem. Sci. 2019, 44, 312–330. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; James, T.; Morao, I.; Bodkin, M.J.; Biggin, P.C. Guiding lead optimization with GPCR structure modeling and molecular dynamics. Curr. Opin. Pharmacol. 2016, 30, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Latorraca, N.R.; Venkatakrishnan, A.J.; Dror, R.O. GPCR Dynamics: Structures in Motion. Chem. Rev. 2017, 117, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.J.; Xu, Y.C. Recent Trends and Applications of Molecular Modeling in GPCR-Ligand Recognition and Structure-Based Drug Design. Int. J. Mol. Sci. 2018, 19, 2105. [Google Scholar] [CrossRef]
- Lee, Y.; Basith, S.; Choi, S. Recent Advances in Structure-Based Drug Design Targeting Class A G Protein-Coupled Receptors Utilizing Crystal Structures and Computational Simulations. J. Med. Chem. 2018, 61, 1–46. [Google Scholar] [CrossRef]
- Lee, Y.; Lazim, R.; Macalino, S.J.Y.; Choi, S. Importance of protein dynamics in the structure-based drug discovery of class A G protein-coupled receptors (GPCRs). Curr. Opin. Struc. Biol. 2019, 55, 147–153. [Google Scholar] [CrossRef]
- Chan, H.C.S.; Wang, J.J.; Palczewski, K.; Filipek, S.; Vogel, H.; Liu, Z.J.; Yuan, S.G. Exploring a new ligand binding site of G proteincoupled receptors. Chem. Sci. 2018, 9, 6480–6489. [Google Scholar] [CrossRef]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef]
- Sloop, K.W.; Emmerson, P.J.; Statnick, M.A.; Willard, F.S. The current state of GPCR-based drug discovery to treat metabolic disease. Brit. J. Pharmacol. 2018, 175, 4060–4071. [Google Scholar] [CrossRef]
- The Nobel Prize in Chemistry 2012. Available online: https://www.nobelprize.org/prizes/chemistry/2012/summary/ (accessed on 10 February 2020).
- Amaro, R.E.; Mulholland, A.J. Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat. Rev. Chem. 2018, 2. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Chavali, S.; Masuho, I.; Jahn, L.J.; Martemyanov, K.A.; Gloriam, D.E.; Babu, M.M. Pharmacogenomics of GPCR Drug Targets. Cell 2018, 172, 41. [Google Scholar] [CrossRef] [PubMed]
- Bondar, A.N.; Lemieux, M.J. Reactions at Biomembrane Interfaces. Chem. Rev. 2019, 119, 6162–6183. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2018, 47, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Vreven, T.; Byun, K.S.; Komaromi, I.; Dapprich, S.; Montgomery, J.A.; Morokuma, K.; Frisch, M.J. Combining Quantum Mechanics Methods with Molecular Mechanics Methods in ONIOM. J. Chem. Theory Comput. 2006, 2, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Senn, H.M.; Thiel, W. QM/MM Methods for Biomolecular Systems. Angew. Chem. Int. Edit. 2009, 48, 1198–1229. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.W.; Sameera, W.M.C.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.F.; Liu, F.Y.; et al. The ONIOM Method and Its Applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef]
- The Nobel Prize in Chemistry 2013. Available online: https://www.nobelprize.org/prizes/chemistry/2013/summary/ (accessed on 10 February 2020).
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Sherman, B.W. Use of the Glide extra precision methodology for docking and scoring. Abstr. Pap. Am. Chem. S. 2006, 232, 42. [Google Scholar]
- Bitencourt-Ferreira, G.; de Azevedo, W.F. Docking with SwissDock. In Docking Screens for Drug Discovery; de Azevedo, W.F., Jr., Ed.; Springer: New York, NY, USA, 2019; pp. 189–202. [Google Scholar]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Dtsch. Chem. Ges. 1894, 27, 2985–2993. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Nabuurs, S.B.; Wagener, M.; De Vlieg, J. A flexible approach to induced fit docking. J. Med. Chem. 2007, 50, 6507–6518. [Google Scholar] [CrossRef]
- Amaro, R.E.; Baudry, J.; Chodera, J.; Demir, Ö.; McCammon, J.A.; Miao, Y.; Smith, J.C. Ensemble Docking in Drug Discovery. Biophys. J. 2018, 114, 2271–2278. [Google Scholar] [CrossRef]
- Sekharan, S.; Ertem, M.Z.; Zhuang, H.Y.; Block, E.; Matsunami, H.; Zhang, R.N.; Wei, J.N.; Pan, Y.; Batista, V.S. QM/MM Model of the Mouse Olfactory Receptor MOR244-3 Validated by Site-Directed Mutagenesis Experiments. Biophys. J. 2014, 107, 5–8. [Google Scholar] [CrossRef]
- Ahmed, L.; Zhang, Y.T.; Block, E.; Buehl, M.; Corr, M.J.; Cormanich, R.A.; Gundala, S.; Matsunami, H.; O’Hagan, D.; Ozbil, M.; et al. Molecular mechanism of activation of human musk receptors OR5AN1 and OR1A1 by (R)-muscone and diverse other musk-smelling compounds. Proc. Natl. Acad. Sci. USA 2018, 115, 3950–3958. [Google Scholar] [CrossRef]
- Zanatta, G.; Nunes, G.; Bezerra, E.M.; da Costa, R.F.; Martins, A.; Caetano, E.W.S.; Freire, V.N.; Gottfried, C. Antipsychotic Haloperidol Binding to the Human Dopamine D3 Receptor: Beyond Docking Through QM/MM Refinement Toward the Design of Improved Schizophrenia Medicines. Acs Chem. Neurosci. 2014, 5, 1041–1054. [Google Scholar] [CrossRef]
- Chien, E.Y.T.; Liu, W.; Zhao, Q.A.; Katritch, V.; Han, G.W.; Hanson, M.A.; Shi, L.; Newman, A.H.; Javitch, J.A.; Cherezov, V.; et al. Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist. Science 2010, 330, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Newcomer, J.W. Second-generation (atypical) antipsychotics and metabolic effects - A comprehensive literature review. Cns Drugs 2005, 19, 1–93. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.E.; Guallar, V.; Berne, B.J.; Friesner, R. Importance of accurate charges in molecular docking: Quantum mechanical/molecular mechanical (QM/MM) approach. J. Comput. Chem. 2005, 26, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.E.; Rinaldo, D. Extension of QM/MM Docking and its Applications to Metalloproteins. J. Comput. Chem. 2009, 30, 2609–2616. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Hah, J.M.; Cho, A.E. Correlation between Performance of QM/MM Docking and Simple Classification of Binding Sites. J. Chem. Inf. Model 2009, 49, 2382–2387. [Google Scholar] [CrossRef]
- Burger, S.K.; Thompson, D.C.; Ayers, P.W. Quantum Mechanics/Molecular Mechanics Strategies for Docking Pose Refinement: Distinguishing between Binders and Decoys in Cytochrome c Peroxidase. J. Chem. Inf. Model 2011, 51, 93–101. [Google Scholar] [CrossRef]
- Chaskar, P.; Zoete, V.; Rohrig, U.F. On-the-Fly QM/MM Docking with Attracting Cavities. J. Chem. Inf. Model 2017, 57, 73–84. [Google Scholar] [CrossRef]
- Beuming, T.; Sherman, W. Current Assessment of Docking into GPCR Crystal Structures and Homology Models: Successes, Challenges, and Guidelines. J. Chem. Inf. Model. 2012, 52, 3263–3277. [Google Scholar] [CrossRef]
- Yu, J.H.; Mannes, P.; Jung, Y.H.; Ciancetta, A.; Bitant, A.; Lieberman, D.I.; Khaznadar, S.; Auchampach, J.A.; Gao, Z.G.; Jacobson, K.A. Structure activity relationship of 2-arylalkynyl-adenine derivatives as human A(3) adenosine receptor antagonists. Medchemcomm 2018, 9, 1920–1932. [Google Scholar] [CrossRef]
- Kim, M.; Cho, A.E. Incorporating QM and solvation into docking for applications to GPCR targets. Phys. Chem. Chem. Phys. 2016, 18, 28281–28289. [Google Scholar] [CrossRef]
- Gascon, J.A.; Batista, V.S. QM/MM study of energy storage and molecular rearrangements due to the primary event in vision. Biophys. J. 2004, 87, 2931–2941. [Google Scholar] [CrossRef] [PubMed]
- Send, R.; Sundholm, D. Stairway to the Conical Intersection: A Computational Study of the Retinal Isomerization. J. Phys. Chem. A 2007, 111, 8766–8773. [Google Scholar] [CrossRef] [PubMed]
- Send, R.; Sundholm, D. Coupled-cluster studies of the lowest excited states of the 11-cis-retinal chromophore. Phys. Chem. Chem. Phys. 2007, 9, 2862–2867. [Google Scholar] [CrossRef] [PubMed]
- Schick, G.A.; Cooper, T.M.; Holloway, R.A.; Murray, L.P.; Birge, R.R. Energy storage in the primary photochemical events of rhodopsin and isorhodopsin. Biochemistry 1987, 26, 2556–2562. [Google Scholar] [CrossRef]
- Cooper, A. Energetics of rhodopsin and isorhodopsin. FEBS Lett. 1979, 100, 382–384. [Google Scholar] [CrossRef]
- Cooper, A. Energy uptake in the first step of visual excitation. Nature 1979, 282, 531–533. [Google Scholar] [CrossRef]
- Altun, A.; Yokoyama, S.; Morokuma, K. Mechanism of Spectral Tuning Going from Retinal in Vacuo to Bovine Rhodopsin and its Mutants: Multireference ab Initio Quantum Mechanics/Molecular Mechanics Studies. J. Phys. Chem. B 2008, 112, 16883–16890. [Google Scholar] [CrossRef]
- Altun, A.; Yokoyama, S.; Morokuma, K. Spectral tuning in visual pigments: An ONIOM(QM: MM) study on bovine rhodopsin and its mutants. J. Phys. Chem. B 2008, 112, 6814–6827. [Google Scholar] [CrossRef]
- Sekharan, S.; Altun, A.; Morokuma, K. Photochemistry of Visual Pigment in a G(q) Protein-Coupled Receptor (GPCR)-Insights from Structural and Spectral Tuning Studies on Squid Rhodopsin. Chem. Eur. J. 2010, 16, 1744–1749. [Google Scholar] [CrossRef]
- Sekharan, S.; Morokuma, K. Drawing the Retinal Out of Its Comfort Zone: An ONIOM(QM/MM) Study of Mutant Squid Rhodopsin. J. Phys. Chem. Lett. 2010, 1, 668–672. [Google Scholar] [CrossRef][Green Version]
- Hernandez-Rodriguez, E.W.; Sanchez-Garcia, E.; Crespo-Otero, R.; Montero-Alejo, A.L.; Montero, L.A.; Thiel, W. Understanding Rhodopsin Mutations Linked to the Retinitis pigmentosa Disease: A QM/MM and DFT/MRCI Study. J. Phys. Chem. B 2012, 116, 1060–1076. [Google Scholar] [CrossRef] [PubMed]
- Rakoczy, E.P.; Kiel, C.; McKeone, R.; Stricher, F.; Serrano, L. Analysis of Disease-Linked Rhodopsin Mutations Based on Structure, Function, and Protein Stability Calculations. J. Mol. Biol. 2011, 405, 584–606. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, A.; Hwa, J. Rhodopsin and retinitis pigmentosa: Shedding light on structure and function. Receptors Channels 2002, 8, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.C.; Nanbu, S.; Ishida, T. QM/MM Trajectory Surface Hopping Approach to Photoisomerization of Rhodopsin and Isorhodopsin: The Origin of Faster and More Efficient Isomerization for Rhodopsin. J. Phys. Chem. B 2012, 116, 8009–8023. [Google Scholar] [CrossRef]
- Mattle, D.; Kuhn, B.; Aebi, J.; Bedoucha, M.; Kekilli, D.; Grozinger, N.; Alker, A.; Rudolph, M.G.; Schmid, G.; Schertler, G.F.X.; et al. Ligand Channel in Pharmacologically stabilized Rhodopsin. Proc. Natl. Acad. Sci. USA 2018, 115, 3640–3645. [Google Scholar] [CrossRef]
- Pedraza-Gonzalez, L.; De Vico, L.; Marin, M.D.; Fanelli, F.; Olivucci, M. a-ARM: Automatic Rhodopsin Modeling with Chromophore Cavity Generation, Ionization State Selection, and External Counterion Placement. J. Chem. Theory Comput. 2019, 15, 3134–3152. [Google Scholar] [CrossRef]
- Fedorov, D.G.; Kitaura, K. Extending the power of quantum chemistry to large systems with the fragment molecular orbital method. J. Phys. Chem. A 2007, 111, 6904–6914. [Google Scholar] [CrossRef]
- Kitaura, K.; Ikeo, E.; Asada, T.; Nakano, T.; Uebayasi, M. Fragment molecular orbital method: An approximate computational method for large molecules. Chem. Phys. Lett. 1999, 313, 701–706. [Google Scholar] [CrossRef]
- Fedorov, D.G.; Nagata, T.; Kitaura, K. Exploring chemistry with the fragment molecular orbital method. Phys. Chem. Chem. Phys. 2012, 14, 7562–7577. [Google Scholar] [CrossRef]
- Hayashi, S.; Taikhorshid, E.; Schulten, K. Photochemical Reaction Dynamics of the Primary Event of Vision Studied by Means of a Hybrid Molecular Simulation. Biophys. J. 2009, 96, 403–416. [Google Scholar] [CrossRef]
- Heifetz, A.; Aldeghi, M.; Chudyk, E.I.; Fedorov, D.G.; Bodkin, M.J.; Biggin, P.C. Using the fragment molecular orbital method to investigate agonist-orexin-2 receptor interactions. Biochem. Soc. T. 2016, 44, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; Chudyk, E.I.; Gleave, L.; Aldeghi, M.; Cherezov, V.; Fedorov, D.G.; Biggin, P.C.; Bodkin, M.J. The Fragment Molecular Orbital Method Reveals New Insight into the Chemical Nature of GPCR-Ligand Interactions. J. Chem. Inf. Model 2016, 56, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; James, T.; Southey, M.; Morao, I.; Aldeghi, M.; Sarrat, L.; Fedorov, D.G.; Bodkin, M.J.; Townsend-Nicholson, A. Characterising GPCR-ligand interactions using a fragment molecular orbital-based approach. Curr. Opin. Struc. Biol. 2019, 55, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; Trani, G.; Aldeghi, M.; MacKinnon, C.H.; McEwan, P.A.; Brookfield, F.A.; Chudyk, E.I.; Bodkin, M.; Pei, Z.H.; Burch, J.D.; et al. Fragment Molecular Orbital Method Applied to Lead Optimization of Novel Interleukin-2 Inducible T-Cell Kinase (ITK) Inhibitors. J. Med. Chem. 2016, 59, 4352–4363. [Google Scholar] [CrossRef]
- Morao, I.; Fedorov, D.G.; Robinson, R.; Southey, M.; Townsend-Nicholson, A.; Bodkin, M.J.; Heifetz, A. Rapid and Accurate Assessment of GPCR-Ligand Interactions Using the Fragment Molecular Orbital-Based Density-Functional Tight-Binding Method. J. Comput. Chem. 2017, 38, 1987–1990. [Google Scholar] [CrossRef]
- Tokiwa, T.; Nakano, S.; Yamamoto, Y.; Ishikawa, T.; Ito, S.; Sladek, V.; Fukuzawa, K.; Mochizuki, Y.; Tokiwa, H.; Misaizu, F.; et al. Development of an Analysis Toolkit, AnalysisFMO, to Visualize Interaction Energies Generated by Fragment Molecular Orbital Calculations. J. Chem. Inf. Model 2019, 59, 25–30. [Google Scholar] [CrossRef]
- Watanabe, C.; Watanabe, H.; Okiyama, Y.; Takaya, D.; Fukuzawa, K.; Tanaka, S.; Honma, T. Development of an automated fragment molecular orbital (FMO) calculation protocol toward construction of quantum mechanical calculation database for large biomolecules. Chem-Bio Inform. J. 2019, 19, 5–18. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakliang, P.; Lazim, R.; Chang, H.; Choi, S. Multiscale Molecular Modeling in G Protein-Coupled Receptor (GPCR)-Ligand Studies. Biomolecules 2020, 10, 631. https://doi.org/10.3390/biom10040631
Nakliang P, Lazim R, Chang H, Choi S. Multiscale Molecular Modeling in G Protein-Coupled Receptor (GPCR)-Ligand Studies. Biomolecules. 2020; 10(4):631. https://doi.org/10.3390/biom10040631
Chicago/Turabian StyleNakliang, Pratanphorn, Raudah Lazim, Hyerim Chang, and Sun Choi. 2020. "Multiscale Molecular Modeling in G Protein-Coupled Receptor (GPCR)-Ligand Studies" Biomolecules 10, no. 4: 631. https://doi.org/10.3390/biom10040631
APA StyleNakliang, P., Lazim, R., Chang, H., & Choi, S. (2020). Multiscale Molecular Modeling in G Protein-Coupled Receptor (GPCR)-Ligand Studies. Biomolecules, 10(4), 631. https://doi.org/10.3390/biom10040631





