Association of microRNA Expression and BRAFV600E Mutation with Recurrence of Thyroid Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Tissue Samples
2.2. RNA and DNA Extraction
2.3. BRAFV600E Mutation Analysis
2.4. qRT-PCR for miRNA Measurement
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
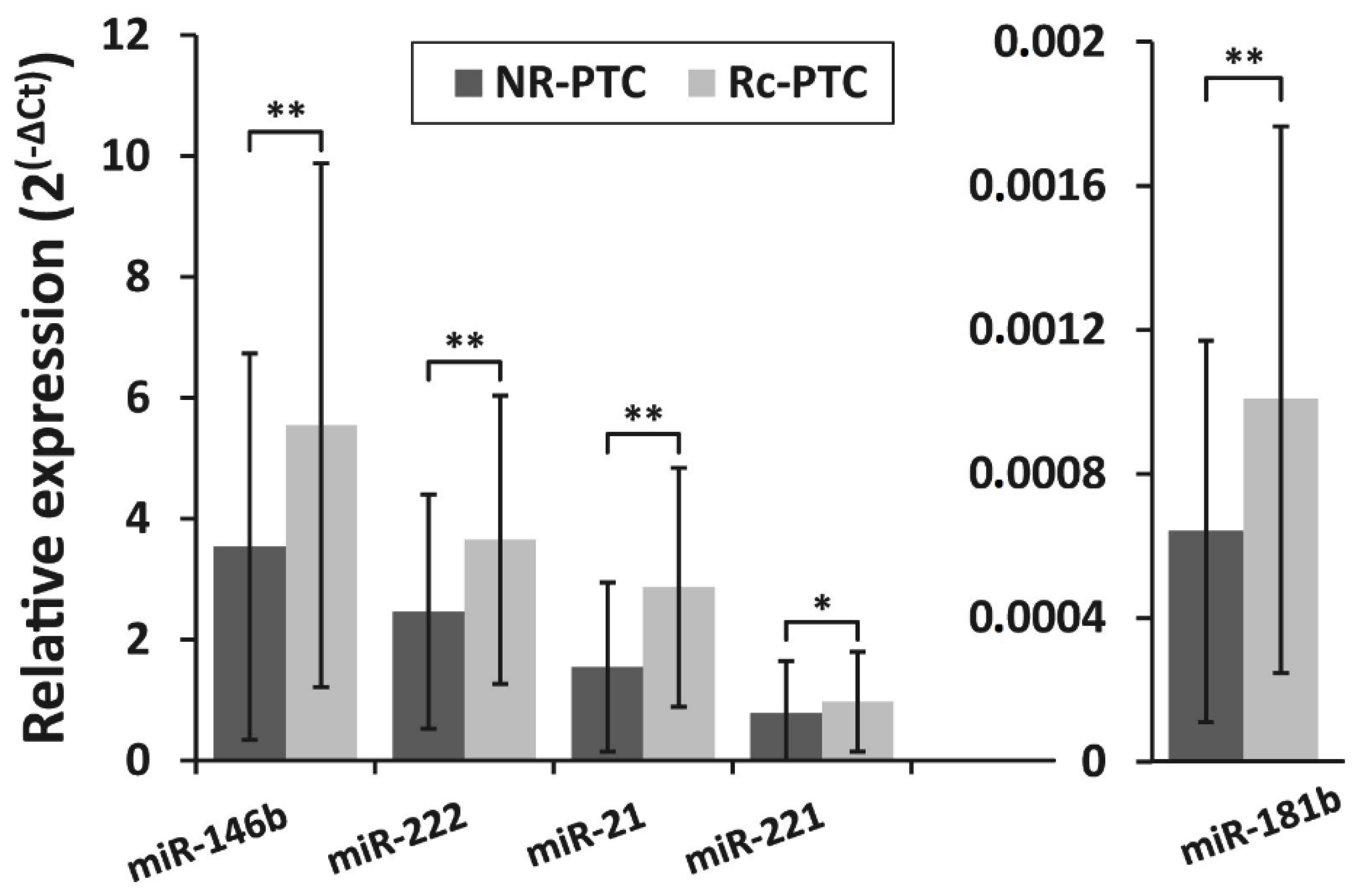
3.2. miRNA Expression Levels in PTC and Their Association With PTC Recurrence
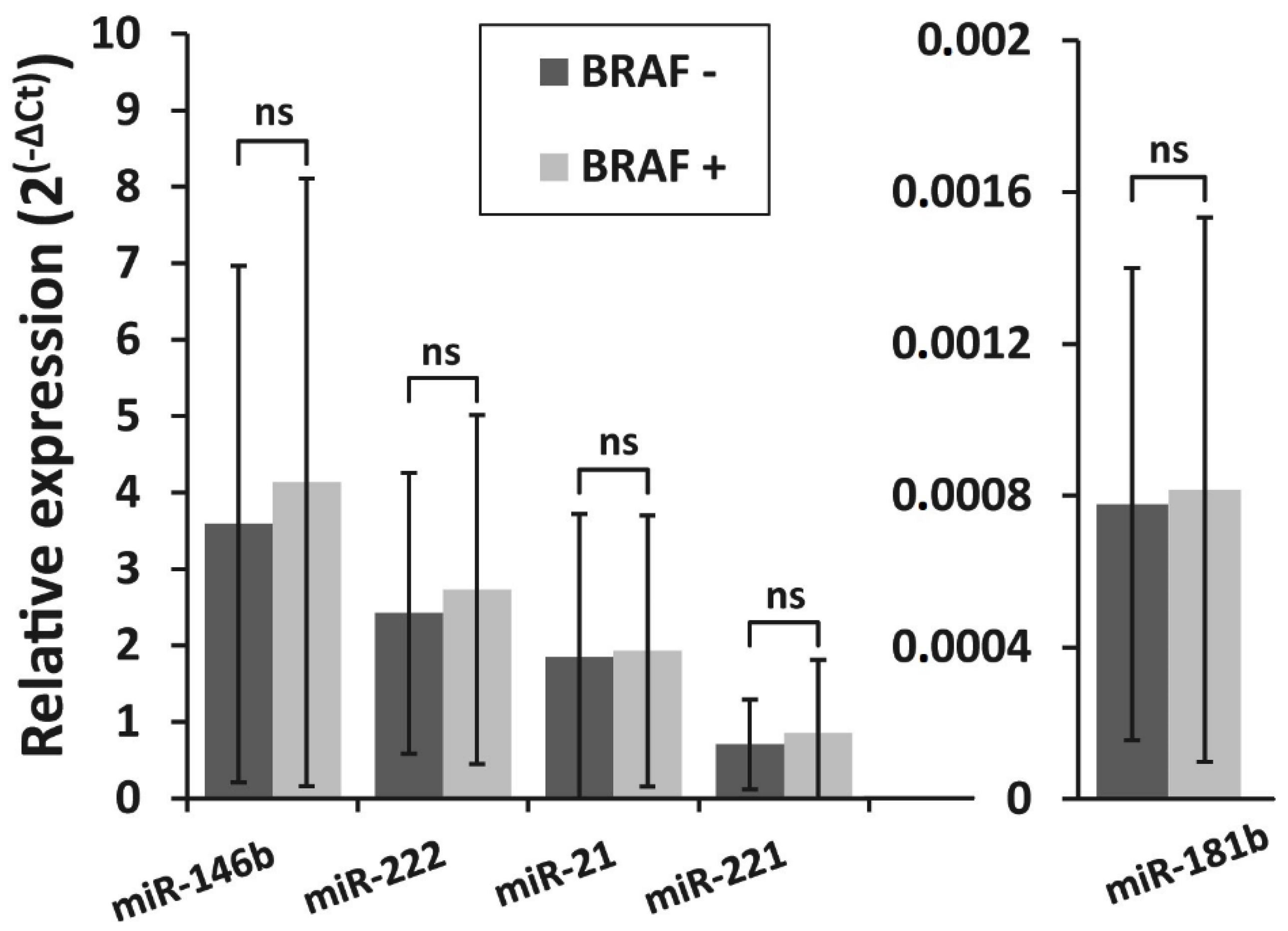
3.3. Detection of BRAFV600E Mutation in PTC Recurrence and Non-Recurrence Groups and Its Association With miRNA Expression
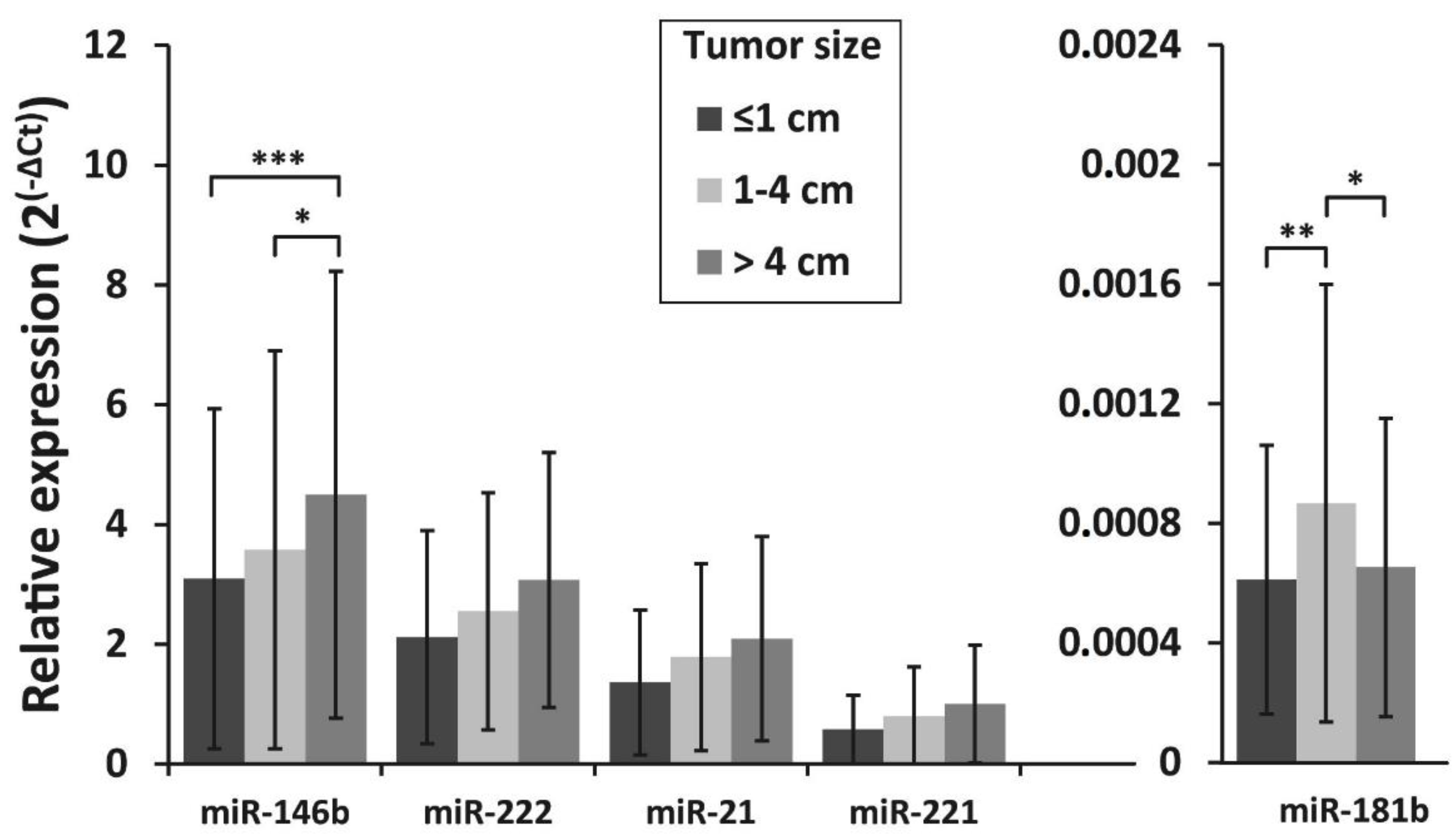
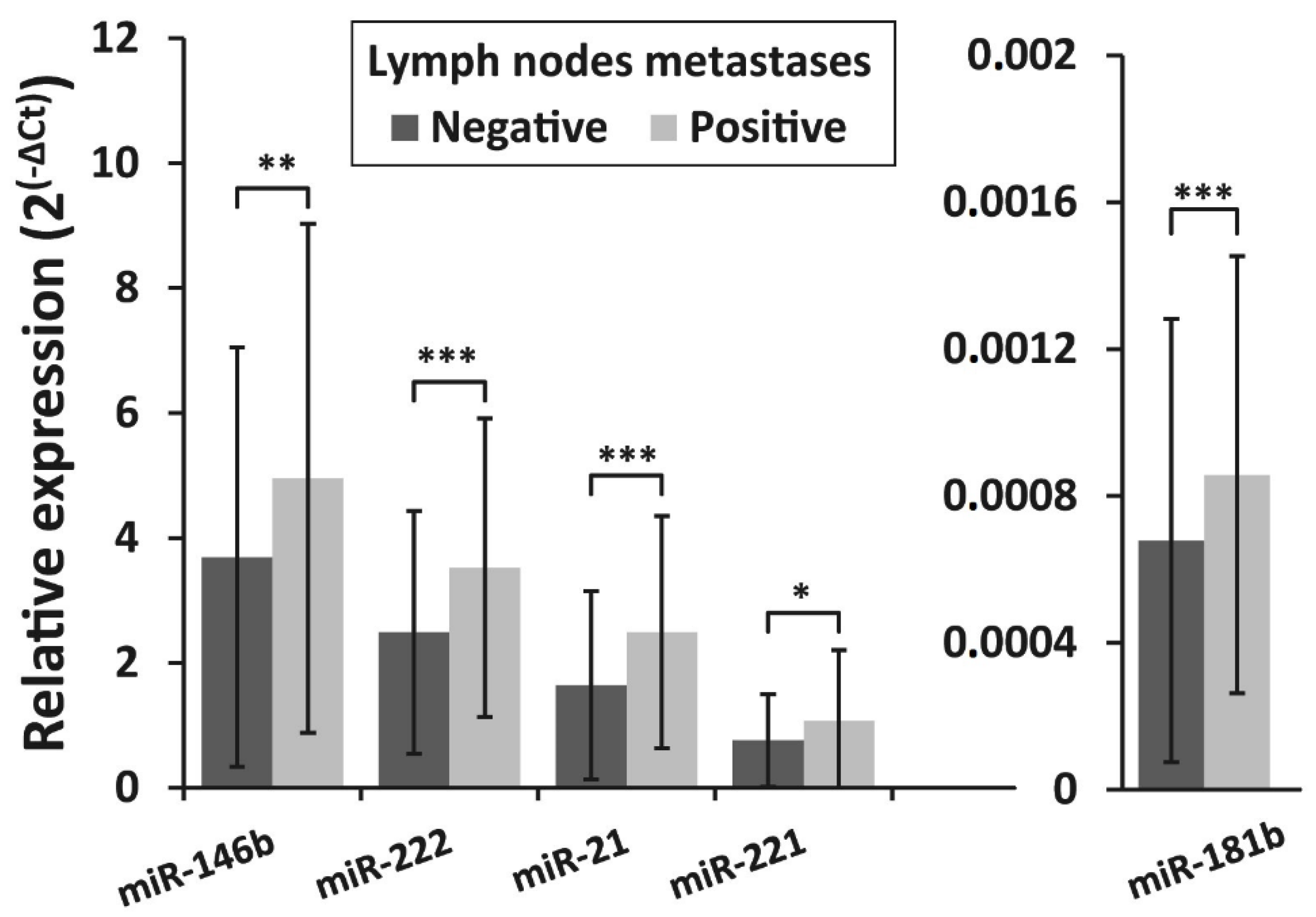
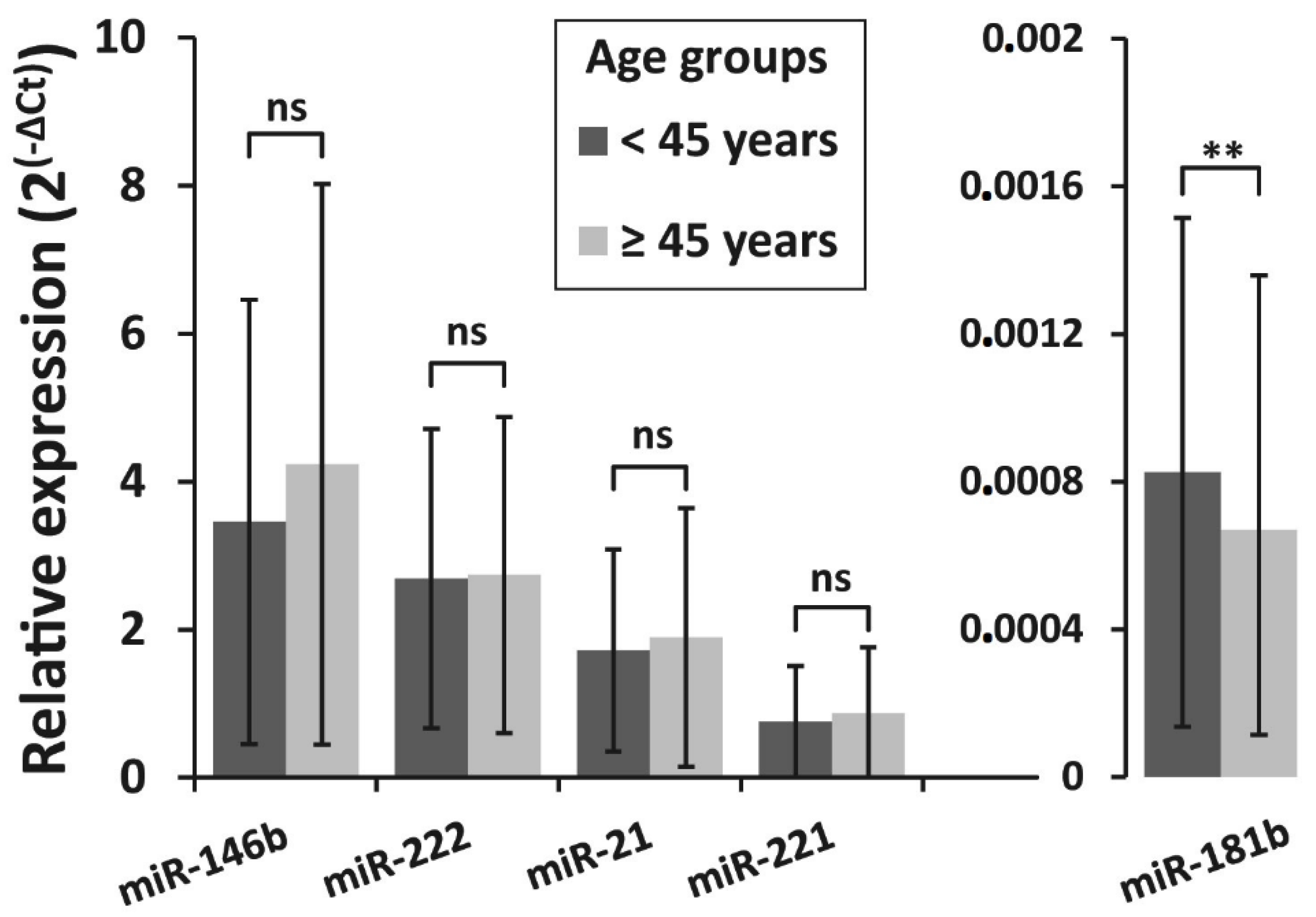
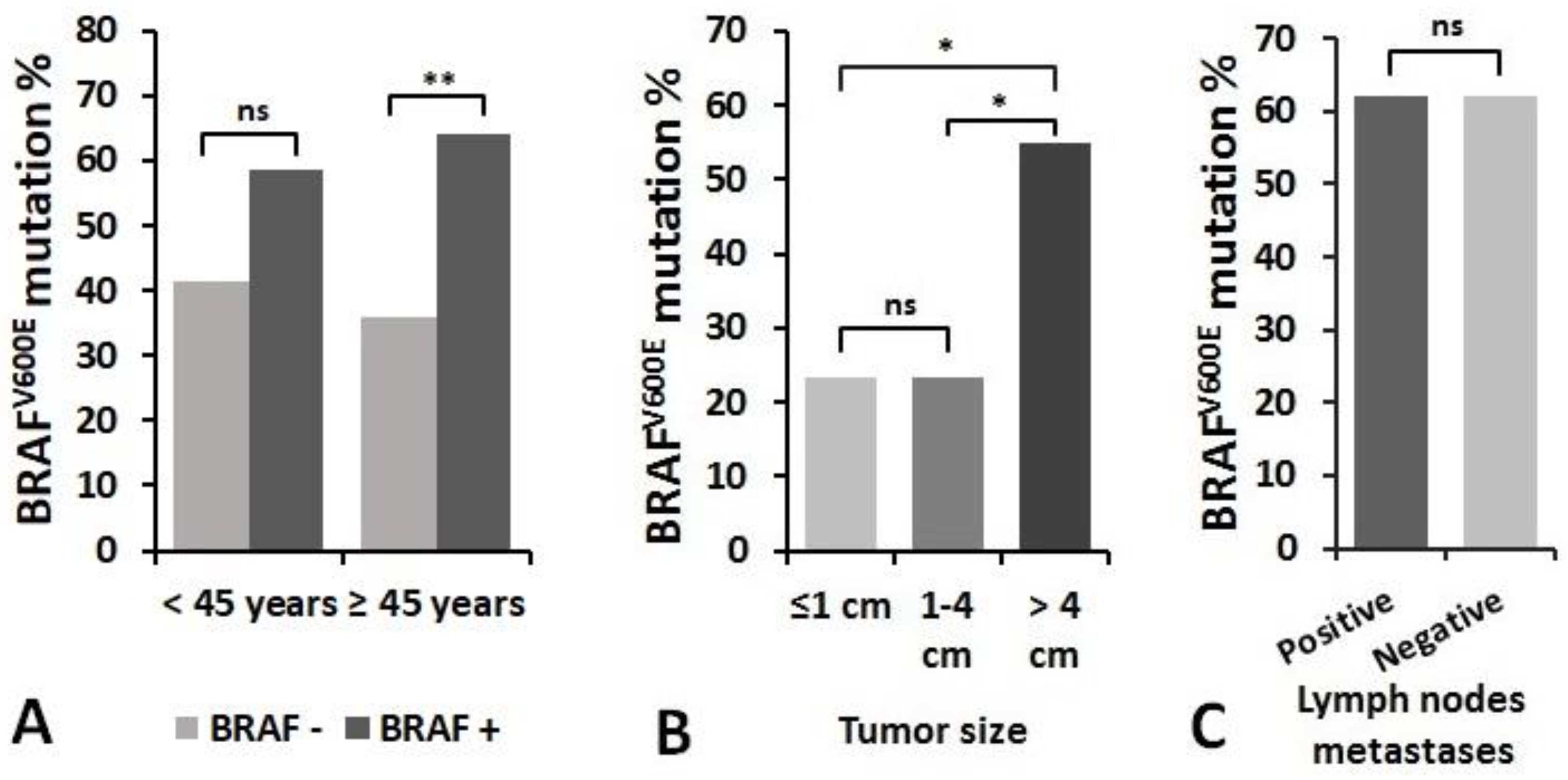
3.4. Association of miRNAs Expression and BRAFV600E Mutation with Clinicopathologic Features of PTC
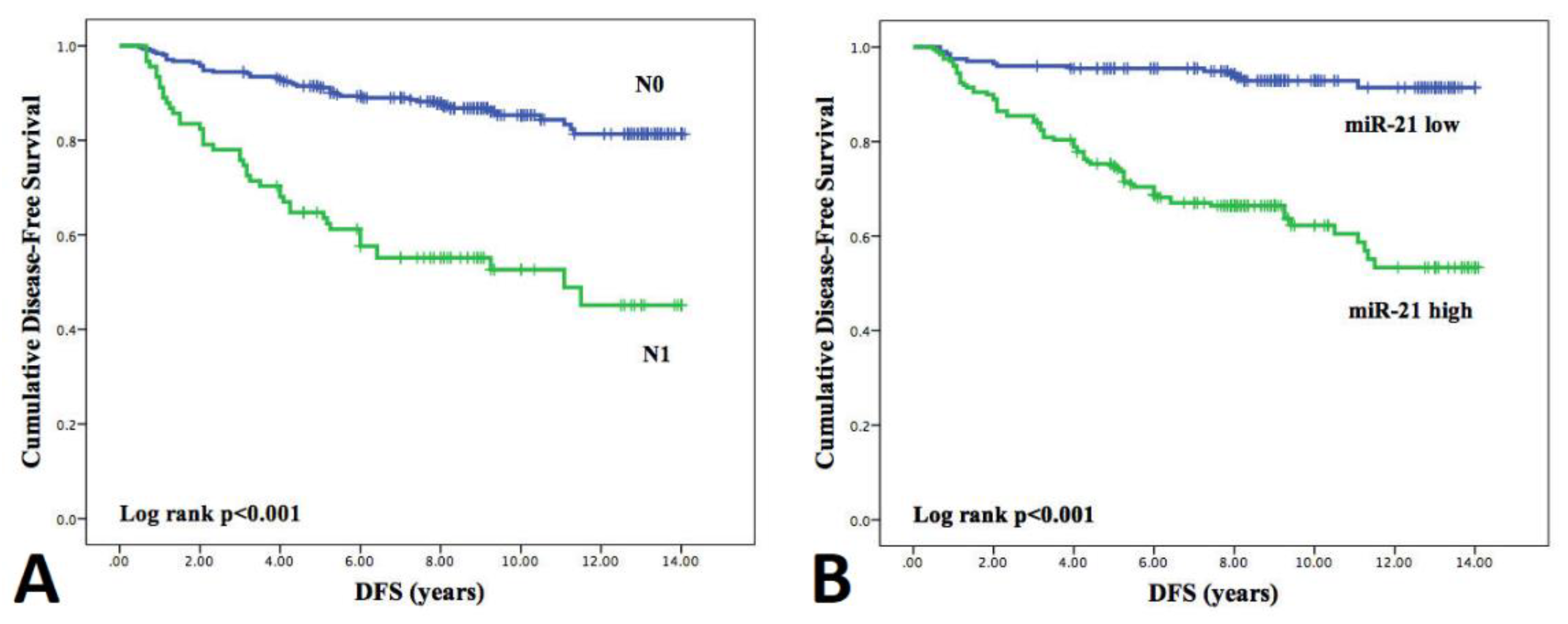
3.5. Influence of Clinicopathological Features and miRNA Expression on Disease-Free Survival (DFS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer Statistics. Ca Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef]
- Han, P.A.; Weng, C.-H.; Khawaja, H.T.; Nagarajan, N.; Schneider, E.B.; Umbricht, C.B.; Witwer, K.W.; Zeiger, M.A. MicroRNA Expression and Association with Clinicopathologic Features in Papillary Thyroid Cancer: A Systematic Review. Thyroid 2015, 25, 1322–1329. [Google Scholar] [CrossRef]
- Eszlinger, M.; Paschke, R. Molecular fine-needle aspiration biopsy diagnosis of thyroid nodules by tumor specific mutations and gene expression patterns. Mol. Cell. Endocrinol. 2010, 322, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C. Cancer: MicroRNA expression provides clues about the aggressiveness of papillary thyroid carcinoma. Nat. Rev. Endocrinol. 2010, 6, 416. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Kitabayashi, N.; Zhou, X.K.; Fahey, T.J.; Scognamiglio, T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod. Pathol. 2008, 21, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.-G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Zhao, J.T.; Clifton-Bligh, R.; Gill, A.; Gundara, J.S.; Ip, J.C.; Glover, A.; Sywak, M.S.; Delbridge, L.W.; Robinson, B.G.; et al. MicroRNA-222 and MicroRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 2013, 119, 4358–4365. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Sheu, S.-Y.; Grabellus, F.; Schwertheim, S.; Worm, K.; Broecker-Preuss, M.; Schmid, U.-P.D.M.K.W. Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br. J. Cancer 2009, 102, 376–382. [Google Scholar] [CrossRef]
- Dai, L.; Wang, Y.; Chen, L.; Zheng, J.; Li, J.; Wu, X. MiR-221, a potential prognostic biomarker for recurrence in papillary thyroid cancer. World J. Surg. Oncol. 2017, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Yip, L.; Kelly, L.; Shuai, Y.; Armstrong, M.J.; Nikiforov, Y.E.; Carty, S.E.; Nikiforova, M.N.; Linwah, Y.; Yongli, S. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann. Surg. Oncol. 2011, 18, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-K.; Chen, R.-F.; Chou, F.-F.; Chang, H.-W.; Chen, Y.-J.; Lee, Y.-F.; Yang, K.D.; Cheng, J.-T.; Huang, H.-Y.; Liu, R.-T. miR-146b is Highly Expressed in Adult Papillary Thyroid Carcinomas with High Risk Features Including Extrathyroidal Invasion and the BRAFV600EMutation. Thyroid 2010, 20, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.-T.; Lee, C.-H. BRAF Mutation in Papillary Thyroid Carcinoma: Pathogenic Role and Clinical Implications. J. Chin. Med Assoc. 2010, 73, 113–128. [Google Scholar] [CrossRef]
- Tufano, R.P.; Teixeira, G.; Bishop, J.; Carson, K.A.; Xing, M. BRAF Mutation in Papillary Thyroid Cancer and Its Value in Tailoring Initial Treatment. Medicine 2012, 91, 274–286. [Google Scholar] [CrossRef]
- Lassalle, S.; Hofman, V.; Ilie, M.; Butori, C.; Bozec, A.; Santini, J.; Vielh, P.; Hofman, P. Clinical impact of the detection of BRAF mutations in thyroid pathology: Potential usefulness as diagnostic, prognostic and theragnostic applications. Curr. Med. Chem. 2010, 17, 1839–1850. [Google Scholar] [CrossRef]
- Lee, J.-H.; Lee, E.-S.; Kim, Y.-S. Clinicopathologic significance ofBRAF V600E mutation in papillary carcinomas of the thyroid. Cancer 2007, 110, 38–46. [Google Scholar] [CrossRef]
- Xing, M. BRAF Mutation in Papillary Thyroid Cancer: Pathogenic Role, Molecular Bases, and Clinical Implications. Endocr. Rev. 2007, 28, 742–762. [Google Scholar] [CrossRef]
- Kim, S.-J.; Lee, K.E.; Myong, J.P.; Park, J.-H.; Jeon, Y.K.; Min, H.S.; Park, S.Y.; Jung, K.C.; Koo, D.H.; Youn, Y.-K. BRAFV600E Mutation is Associated with Tumor Aggressiveness in Papillary Thyroid Cancer. World J. Surg. 2011, 36, 310–317. [Google Scholar] [CrossRef]
- Chakraborty, A.; Narkar, A.; Mukhopadhyaya, R.; Kane, S.; D’Cruz, A.; Rajan, M.G.R. BRAF V600E Mutation in Papillary Thyroid Carcinoma: Significant Association with Node Metastases and Extra Thyroidal Invasion. Endocr. Pathol. 2011, 23, 83–93. [Google Scholar] [CrossRef]
- Nakayama, H.; Yoshida, A.; Nakamura, Y.; Hayashi, H.; Miyagi, Y.; Wada, N.; Rino, Y.; Masuda, M.; Imada, T. Clinical significance of BRAF (V600E) mutation and Ki-67 labeling index in papillary thyroid carcinomas. Anticancer. Res. 2007, 27, 3645–3649. [Google Scholar] [PubMed]
- Pelttari, H.; Schalin-Jantti, C.; Arola, J.; Löyttyniemi, E.; Knuutila, S.; Valimaki, M.J. BRAF V600E mutation does not predict recurrence after long-term follow-up in TNM stage I or II papillary thyroid carcinoma patients. APMIS 2011, 120, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Cahill, S.; Smyth, P.; Denning, K.; Flavin, R.; Li, J.; Potratz, A.; Guenther, S.M.; Henfrey, R.; O’Leary, J.J.; Sheils, O. Effect of BRAFV600E mutation on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol. Cancer 2007, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.-Y.; Schwertheim, S.; Handke, S.; Worm, K.; Grabellus, F.; Schmid, U.-P.D.M.K.W. Lack of Correlation between BRAF V600E Mutational Status and the Expression Profile of a Distinct Set of miRNAs in Papillary Thyroid Carcinoma. Horm. Metab. Res. 2009, 41, 482–487. [Google Scholar] [CrossRef]
- Cheng, S.; Serra, S.; Mercado, M.; Ezzat, S.; Asa, S.L. A High-Throughput Proteomic Approach Provides Distinct Signatures for Thyroid Cancer Behavior. Clin. Cancer Res. 2011, 17, 2385–2394. [Google Scholar] [CrossRef]
- Khoo, M.L.C.; Freeman, J.L.; Witterick, I.J.; Irish, J.C.; Rotstein, L.E.; Gullane, P.J.; Asa, S.L. Underexpression of p27/Kip in thyroid papillary microcarcinomas with gross metastatic disease. Arch. Otolaryngol. Head Neck Surg. 2002, 128, 253–257. [Google Scholar] [CrossRef]
- Vickery, A.L.; Carcangiu, M.L.; Johannessen, J.V.; Sobrinho-Simoes, M. Papillary carcinoma. Semin. Diagn. Pathol. 1985, 2, 2004 57. [Google Scholar]
- Sondermann, A.; Andreghetto, F.M.; Moulatlet, A.C.B.; Victor, E.D.S.; De Castro, M.G.; Nunes, F.D.; Brandão, L.G.; Severino, P. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin. Exp. Metastasis 2015, 32, 521–530. [Google Scholar] [CrossRef]
- Yang, Z.; Yuan, Z.; Fan, Y.; Deng, X.; Zheng, Q. Integrated analyses of microRNA and mRNA expression profiles in aggressive papillary thyroid carcinoma. Mol. Med. Rep. 2013, 8, 1353–1358. [Google Scholar] [CrossRef]
- Acibucu, F.; Dökmetaş, H.; Tutar, Y.; Elagoz, S.; Kilicli, F.; Acıbucu, F.; Kılıclı, F. Correlations between the Expression Levels of Micro-RNA146b, 221, 222 and p27Kip1 protein mRNA and the Clinicopathologic Parameters in Papillary Thyroid Cancers. Exp. Clin. Endocrinol. Diabetes 2014, 122, 137–143. [Google Scholar] [CrossRef]
- Meng, W.; McElroy, J.P.; Volinia, S.; Palatini, J.; Warner, S.; Ayers, L.W.; Palanichamy, K.; Chakravarti, A.; Lautenschlaeger, T. Comparison of MicroRNA Deep Sequencing of Matched Formalin-Fixed Paraffin-Embedded and Fresh Frozen Cancer Tissues. PLoS ONE 2013, 8, e64393. [Google Scholar] [CrossRef]
- Agretti, P.; Ferrarini, E.; Rago, T.; Candelieri, A.; De Marco, G.; Dimida, A.; Niccolai, F.; Molinaro, A.; Di Coscio, G.; Pinchera, A.; et al. MicroRNA expression profile helps to distinguish benign nodules from papillary thyroid carcinomas starting from cells of fine-needle aspiration. Eur. J. Endocrinol. 2012, 167, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Mančíková, V.; Castelblanco, E.; Piñeiro-Yáñez, E.; Perales-Patón, J.; A De Cubas, A.; Inglada, L.; Matias-Guiu, X.; Capel, I.; Bella, M.; Lerma, E.; et al. MicroRNA deep-sequencing reveals master regulators of follicular and papillary thyroid tumors. Mod. Pathol. 2015, 28, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Tetzlaff, M.T.; Liu, A.; Xu, X.; Master, S.R.; Baldwin, N.A.; Tobias, J.W.; Livolsi, V.A.; Baloch, Z. Differential Expression of miRNAs in Papillary Thyroid Carcinoma Compared to Multinodular Goiter Using Formalin Fixed Paraffin Embedded Tissues. Endocr. Pathol. 2007, 18, 163–173. [Google Scholar] [CrossRef]
- Sykorova, V.; Dvořáková, Š.; Ryska, A.; Vcelak, J.; Vaclavikova, E.; Laco, J.; Kodetova, D.; Kodet, R.; Cibula, A.; Dušková, J.; et al. BRAFV600E mutation in the pathogenesis of a large series of papillary thyroid carcinoma in Czech Republic. J. Endocrinol. Investig. 2010, 33, 318–324. [Google Scholar] [CrossRef]
- Kebebew, E.; Weng, J.; Bauer, J.; Ranvier, G.; Clark, O.H.; Duh, Q.-Y.; Shibru, D.; Bastian, B.C.; Griffin, A. The Prevalence and Prognostic Value of BRAF Mutation in Thyroid Cancer. Ann. Surg. 2007, 246, 466–471. [Google Scholar] [CrossRef]
- Zhou, Y.-L.; Liu, C.; Dai, X.-X.; Zhang, X.-H.; Wang, O.-C. Overexpression of miR-221 is associated with aggressive clinicopathologic characteristics and the BRAF mutation in papillary thyroid carcinomas. Med Oncol. 2012, 29, 3360–3366. [Google Scholar] [CrossRef]
- Rosenbaum, E.; Hosler, G.; Zahurak, M.; Cohen, Y.; Sidransky, D.; Westra, W.H. Mutational activation of BRAF is not a major event in sporadic childhood papillary thyroid carcinoma. Mod. Pathol. 2004, 18, 898–902. [Google Scholar] [CrossRef]
- Pelizzo, M.R.; Boschin, I.M.; Barollo, S.; Pennelli, G.; Toniato, A.; Zambonin, L.; Vianello, F.; Piotto, A.; Ide, E.C.; Pagetta, C.; et al. BRAF analysis by fine needle aspiration biopsy of thyroid nodules improves preoperative identification of papillary thyroid carcinoma and represents a prognostic factor. A mono-institutional experience. Clin. Chem. Lab. Med. 2011, 49, 325–329. [Google Scholar] [CrossRef]
- Basolo, F.; Torregrossa, L.; Giannini, R.; Miccoli, M.; Lupi, C.; Sensi, E.; Berti, P.; Elisei, R.; Vitti, P.; Baggiani, A.; et al. Correlation between the BRAF V600E Mutation and Tumor Invasiveness in Papillary Thyroid Carcinomas Smaller than 20 Millimeters: Analysis of 1060 Cases. J. Clin. Endocrinol. Metab. 2010, 95, 4197–4205. [Google Scholar] [CrossRef]
- Frasca, F.; Nucera, C.; Pellegriti, G.; Gangemi, P.; Attard, M.; Stella, M.; Loda, M.; Vella, V.; Giordano, C.; Trimarchi, F.; et al. BRAF(V600E) mutation and the biology of papillary thyroid cancer. Endocrine-Related Cancer 2008, 15, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Ji, M.; Bao, R.; Yu, H.; Wang, Y.; Hou, P.; Zhang, Y.; Shan, Z.; Teng, W.; Xing, M. Association of High Iodine Intake with the T1799ABRAFMutation in Papillary Thyroid Cancer. J. Clin. Endocrinol. Metab. 2009, 94, 1612–1617. [Google Scholar] [CrossRef]
- Ito, Y.; Yoshida, H.; Maruo, R.; Morita, S.; Takano, T.; Hirokawa, M.; Yabuta, T.; Fukushima, M.; Inoue, H.; Tomoda, C.; et al. BRAF mutation in papillary thyroid carcinoma in a Japanese population: Its lack of correlation with high-risk clinicopathological features and disease-free survival of patients. Endocr. J. 2008, 56, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Goutas, N.; Vlachodimitropoulos, D.; Bouka, M.; Lazaris, A.C.; Nasioulas, G.; Gazouli, M. BRAF and K-RAS mutation in a Greek papillary and medullary thyroid carcinoma cohort. Anticancer. Res. 2008, 28, 305–308. [Google Scholar] [PubMed]
- Oler, G.; Cerutti, J. High prevalence ofBRAFmutation in a Brazilian cohort of patients with sporadic papillary thyroid carcinomas. Cancer 2009, 115, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Ye, L.; Zhang, X.; Shen, L.; Han, R.; Wei, Q.; Fei, X.; Chen, X.; Wang, W.; Wang, S.; et al. BRAF mutation correlates with recurrent papillary thyroid carcinoma in Chinese patients. Curr. Oncol. 2014, 21, 740–e747. [Google Scholar] [CrossRef]
- Barbaro, D.; Incensati, R.M.; Materazzi, G.; Boni, G.; Grosso, M.; Panicucci, E.; Lapi, P.; Pasquini, C.; Miccoli, P. The BRAF V600E mutation in papillary thyroid cancer with positive or suspected pre-surgical cytological finding is not associated with advanced stages or worse prognosis. Endocrine 2013, 45, 462–468. [Google Scholar] [CrossRef]
- Kim, T.H.; Park, Y.J.; Lim, J.A.; Ahn, H.Y.; Lee, E.-K.; Lee, E.-K.; Kim, K.W.; Hahn, S.K.; Youn, Y.-K.; Kim, K.H.; et al. The association of the BRAFV600E mutation with prognostic factors and poor clinical outcome in papillary thyroid cancer. Cancer 2011, 118, 1764–1773. [Google Scholar] [CrossRef]
- Czarniecka, A.; Kowal, M.; Rusinek, D.; Krajewska, J.; Jarząb, M.; Stobiecka, E.; Chmielik, E.; Zembala-Nozyńska, E.; Poltorak, S.; Sacher, A.; et al. The Risk of Relapse in Papillary Thyroid Cancer (PTC) in the Context of BRAFV600E Mutation Status and Other Prognostic Factors. PLoS ONE 2015, 10, e0132821. [Google Scholar] [CrossRef]
- Nair, C.G.; Babu, M.; Biswas, L.; Jacob, P.; Ramachandran, R.; Revathy, A.K.; Nair, K. Lack of Association of B-type Raf Kinase V600E Mutation with High-risk Tumor Features and Adverse Outcome in Conventional and Follicular Variants of Papillary Thyroid Carcinoma. Indian J. Endocrinol. Metab. 2017, 21, 329–333. [Google Scholar] [CrossRef]
- Yarchoan, M.; Livolsi, V.A.; Brose, M.S. BRAF Mutation and Thyroid Cancer Recurrence. J. Clin. Oncol. 2015, 33, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Selcuklu, S.D.; Donoghue, M.T.; Spillane, C. miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 2009, 37, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Geraldo, M.V.; Yamashita, A.S.; Kimura, E.T. MicroRNA miR-146b-5p regulates signal transduction of TGF-β by repressing SMAD4 in thyroid cancer. Oncogene 2011, 31, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-K.; Chi, S.-Y.; Huang, C.-H.; Chou, F.-F.; Huang, C.-C.; Liu, R.-T.; Kang, H.-Y. IRAK1, a Target of miR-146b, Reduces Cell Aggressiveness of Human Papillary Thyroid Carcinoma. J. Clin. Endocrinol. Metab. 2016, 101, 4357–4366. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ouyang, Y.; Che, J.; Li, X.; Zhao, Y.; Yang, K.; Zhao, X.; Chen, Y.; Fan, C.; Yuan, W.-E. Potential Value of miR-221/222 as Diagnostic, Prognostic, and Therapeutic Biomarkers for Diseases. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shi, W.; Wu, C.; Ju, J.; Jiang, J. miR-181b as a key regulator of the oncogenic process and its clinical implications in cancer (Review). Biomed. Rep. 2013, 2, 7–11. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | All Patients (n = 400) | Non-Recurrence Group (n = 313) | Recurrence Group (n = 87) | p |
|---|---|---|---|---|
| Sex, n (%) Male Female | 53 (13.3) 347 (86.7) | 33 (10.6) 280 (89.4) | 20 (23.0) 67 (77.0) | 0.003 |
| Age at initial surgery (years) Median (min-max) <45 years, n (%) ≥45 years, n (%) | 50 (18-83) 131 (32.8) 269 (67.2) | 53 (18-83) 91 (28.8) 222 (71.2) | 47 (18-75) 41 (47.1) 46 (52.9) | <0.001 0.001 |
| Tumor size *, n (%) pT1a pT1b pT2 pT3 pT4 | 109 (27.3) 81 (20.3) 23 (5.8) 172 (43.0) 15 (3.8) | 97 (30.8) 64 (20.5) 18 (5.8) 128 (41.0) 6 (1.9) | 12 (13.8) 17 (19.5) 5 (5.7) 44 (50.6) 9 (10.3) | <0.001 <0.001 0.007 <0.001 0.439 |
| Lymph node metastases at initial surgery, n (%) | 91 (22.8) | 48 (15.4) | 43 (49.4) | <0.001 |
| Variable | OR (95% CI) | p-Value |
|---|---|---|
| Tumor size (T1 vs. T2–4) | 1.17 (0.56–2.44) | 0.680 |
| Lymph nodes (pos vs. neg) | 3.66 (1.59–8.45) | 0.002 |
| BRAFV600E (pos vs. neg) | 1.01 (0.45–2.11) | 0.994 |
| miR- 146b | 1.09 (0.93–1.27) | 0.279 |
| miR-222 | 1.21 (0.87–1.68) | 0.248 |
| miR-21 | 1.50 (1.12–2.00) | 0.006 |
| miR-221 | 0.86 (0.48–1.54) | 0.862 |
| miR-181b | 1.08 (0.81–1.43) | 0.618 |
| Univariate Analysis DFS | ||
| HR (95% CI) | p-Value | |
| Gender (female vs. male) | 0.47 (0.29-0.78) | 0.003 |
| Age (<45 years vs. ≥45 years) | 1.77 (1.16-2.7) | 0.008 |
| Tumor size (T1 vs. T2-4) | 0.52 (0.33-0.83) | 0.004 |
| Lymph nodes (pos vs. neg) | 4.24 (2.78-6.47) | <0.001 |
| BRAFV600E (pos vs. neg) | 1.01 (0.62-1.64) | 0.964 |
| miR-146b (high vs. low) | 2.62 (1.65-4.15) | <0.001 |
| miR-222 (high vs. low) | 2.39 (1.52-3.77) | <0.001 |
| miR-21 (high vs. low) | 6.45 (3.63-11.44) | <0.001 |
| miR-221 (high vs. low) | 1.82 (1.76-2.79) | 0.007 |
| miR-181b (high vs. low) | 3.15 (1.93-5.15) | <0.001 |
| Multivariate Analysis DFS | ||
| HR (95% CI) | p-Value | |
| Gender (female vs. male) | 0.85 (0.46-1.56) | 0.603 |
| Age (<45 years vs. ≥45 years) | 1.37 (0.83-2.26) | 0.214 |
| Tumor size (T1 vs. T2-4) | 1.19 (0.7-2.04) | 0.523 |
| Lymph nodes (pos vs. neg) | 1.94 (1.12-3.36) | 0.017 |
| BRAFV600E (pos vs. neg) | 0.8 (0.47-1.36) | 0.410 |
| miR-146b (high vs. low) | 1.49 (0.77-2.89) | 0.233 |
| miR-222 (high vs. low) | 1.03 (0.51-2.08) | 0.94 |
| miR-21 (high vs. low) | 5.84 (2.79-12.28) | <0.001 |
| miR-221 (high vs. low) | 1.51 (0.89-2.57) | 0.131 |
| miR-181b (high vs. low) | 1.32 (0.75-2.33) | 0.340 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pamedytyte, D.; Simanaviciene, V.; Dauksiene, D.; Leipute, E.; Zvirbliene, A.; Sarauskas, V.; Dauksa, A.; Verkauskiene, R.; Zilaitiene, B. Association of microRNA Expression and BRAFV600E Mutation with Recurrence of Thyroid Cancer. Biomolecules 2020, 10, 625. https://doi.org/10.3390/biom10040625
Pamedytyte D, Simanaviciene V, Dauksiene D, Leipute E, Zvirbliene A, Sarauskas V, Dauksa A, Verkauskiene R, Zilaitiene B. Association of microRNA Expression and BRAFV600E Mutation with Recurrence of Thyroid Cancer. Biomolecules. 2020; 10(4):625. https://doi.org/10.3390/biom10040625
Chicago/Turabian StylePamedytyte, Daina, Vaida Simanaviciene, Dalia Dauksiene, Enrika Leipute, Aurelija Zvirbliene, Valdas Sarauskas, Albertas Dauksa, Rasa Verkauskiene, and Birute Zilaitiene. 2020. "Association of microRNA Expression and BRAFV600E Mutation with Recurrence of Thyroid Cancer" Biomolecules 10, no. 4: 625. https://doi.org/10.3390/biom10040625
APA StylePamedytyte, D., Simanaviciene, V., Dauksiene, D., Leipute, E., Zvirbliene, A., Sarauskas, V., Dauksa, A., Verkauskiene, R., & Zilaitiene, B. (2020). Association of microRNA Expression and BRAFV600E Mutation with Recurrence of Thyroid Cancer. Biomolecules, 10(4), 625. https://doi.org/10.3390/biom10040625





