uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotides Synthesis and Purification
2.2. NMR Spectroscopy
2.3. CD Spectroscopy
2.4. Cell Cultures and Treatments with the ODNs
2.5. Western Blotting Analysis
2.6. MTT Assay
2.7. RT-qPCR
2.8. Cell Cycle Analysis by Flow Cytometry
2.9. Cell Death Assay
2.10. Statistical Analysis
3. Results
3.1. Structural Insight of the Investigated Sequences
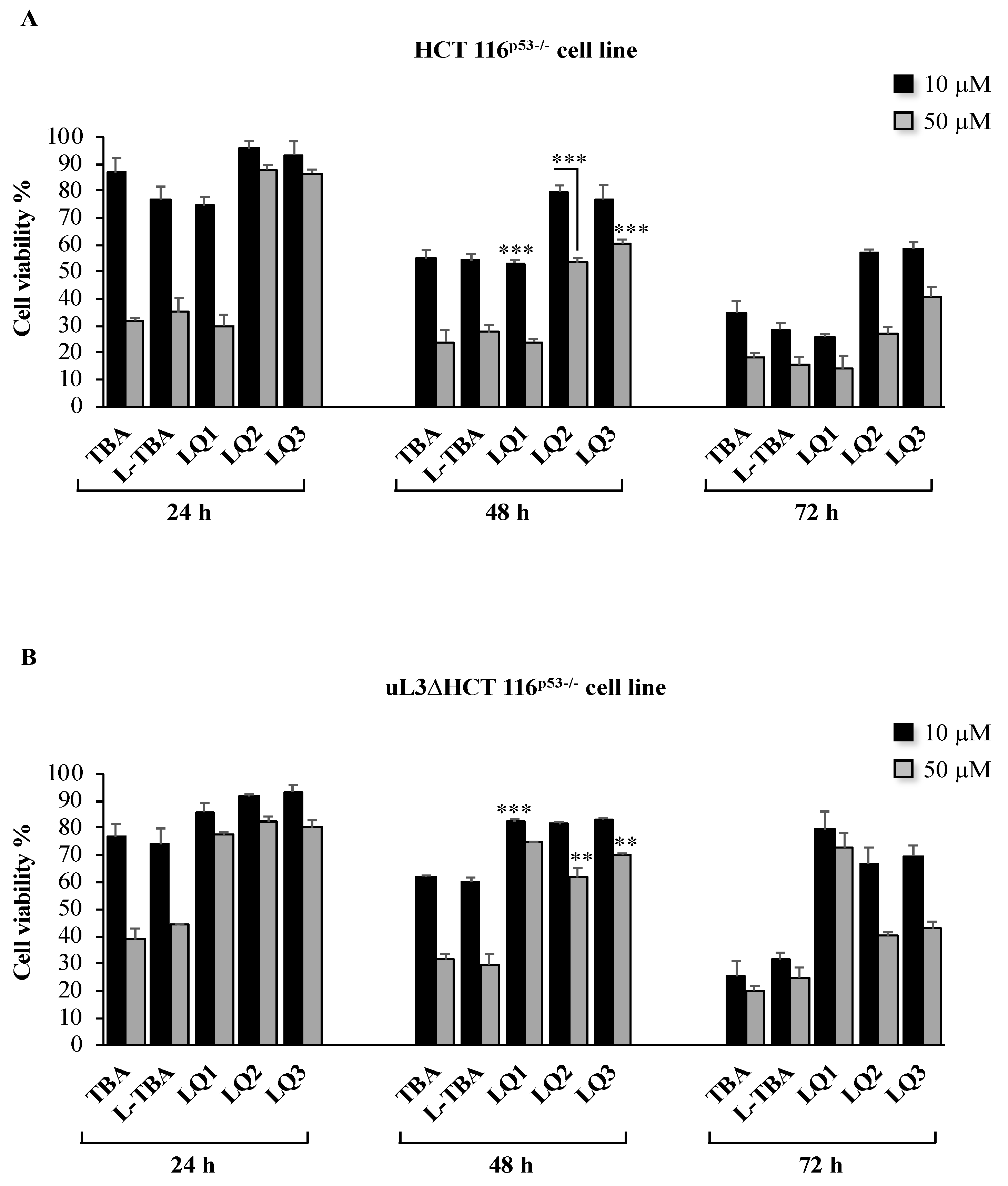
3.2. Cytotoxicity Activity of Investigated ODNs
3.3. LQ1 Causes Nucleolar Stress and Affects rRNA Processing
3.4. Effects of LQ1 on Cell Cycle and Programmed Cell Death
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Varizhuk, A.; Ilyinsky, N.; Smirnov, I.; Pozmogova, G. G4 Aptamers: Trends in Structural Design. Mini Rev. Med. Chem. 2016, 16, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.W.; Nayak, L.V.; Bates, P.J. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010, 38, 1623–1635. [Google Scholar] [CrossRef]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef]
- Zhang, N.; Bing, T.; Liu, X.; Qi, C.; Shen, L.; Wang, L.; Shangguan, D. Cytotoxicity of guanine-based degradation products contributes to the antiproliferative activity of guanine-rich oligonucleotides. Chem. Sci. 2015, 6, 3831–3838. [Google Scholar] [CrossRef]
- Ogloblina, A.M.; Khristich, A.N.; Karpechenko, N.Y.; Semina, S.E.; Belitsky, G.A.; Dolinnaya, N.G.; Yakubovskaya, M.G. Multi-targeted effects of G4-aptamers and their antiproliferative activity against cancer cells. Biochimie 2018, 145, 163–173. [Google Scholar] [CrossRef]
- Dapić, V.; Abdomerović, V.; Marrington, R.; Peberdy, J.; Rodger, A.; Trent, J.O.; Bates, P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 2097–2107. [Google Scholar] [CrossRef]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef]
- Scuotto, M.; Rivieccio, E.; Varone, A.; Corda, D.; Bucci, M.; Vellecco, V.; Cirino, G.; Virgilio, A.; Esposito, V.; Galeone, A.; et al. Site specific replacements of a single loop nucleoside with a dibenzyl linker may switch the activity of TBA from anticoagulant to antiproliferative. Nucleic Acids Res. 2015, 43, 7702–7716. [Google Scholar] [CrossRef]
- Esposito, V.; Russo, A.; Amato, T.; Varra, M.; Vellecco, V.; Bucci, M.; Russo, G.; Virgilio, A.; Galeone, A. Backbone modified TBA analogues endowed with antiproliferative activity. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Russo, A.; Vellecco, V.; Bucci, M.; Russo, G.; Mayol, L.; Virgilio, A.; Galeone, A. Thrombin binding aptamer analogues containing inversion of polarity sites endowed with antiproliferative and anti-motility properties against Calu-6 cells. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2645–2650. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhu, Y.; Wang, C.; Guan, Z.; Zhang, L.; Yang, Z. Alkylation of phosphorothioated thrombin binding aptamers improves the selectivity of inhibition of tumor cell proliferation upon anticoagulation. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1864–1869. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Scuotto, M.; Capuozzo, A.; Santamaria, R.; Varra, M.; Mayol, L.; Virgilio, A.; Galeone, A. A straightforward modification in the thrombin binding aptamer improving the stability, affinity to thrombin and nuclease resistance. Org. Biomol. Chem. 2014, 12, 8840–8843. [Google Scholar] [CrossRef] [PubMed]
- Kotkowiak, W.; Lisowiec-Wachnicka, J.; Grynda, J.; Kierzek, R.; Wengel, J.; Pasternak, A. Thermodynamic, Anticoagulant, and Antiproliferative Properties of Thrombin Binding Aptamer Containing Novel UNA Derivative. Mol. Ther. Nucleic Acids 2018, 10, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Russo, A.; Amato, T.; Vellecco, V.; Bucci, M.; Mayol, L.; Russo, G.; Virgilio, A.; Galeone, A. The “Janus face” of the thrombin binding aptamer: Investigating the anticoagulant and antiproliferative properties through straightforward chemical modifications. Bioorg. Chem. 2018, 76, 202–209. [Google Scholar] [CrossRef]
- Virgilio, A.; Petraccone, L.; Scuotto, M.; Vellecco, V.; Bucci, M.; Mayol, L.; Varra, M.; Esposito, V.; Galeone, A. 5-Hydroxymethyl-2’-deoxyuridine residues in the thrombin binding aptamer: Investigating anticoagulant activity by making a tiny chemical modification. Chembiochem 2014, 15, 2427–2434. [Google Scholar] [CrossRef]
- Turi, Z.; Lacey, M.; Mistrik, M.; Moudry, P. Impaired ribosome biogenesis: Mechanisms and relevance to cancer and aging. Aging 2019, 11, 2512–2540. [Google Scholar] [CrossRef]
- Lessard, F.; Brakier-Gingras, L.; Ferbeyre, G. Ribosomal Proteins Control Tumor Suppressor Pathways in Response to Nucleolar Stress. Bioessays 2019, 41, e1800183. [Google Scholar] [CrossRef]
- Russo, A.; Russo, G.; Cuccurese, M.; Garbi, C.; Pietropaolo, C. The 3′-untranslated region directs ribosomal protein-encoding mRNAs to specific cytoplasmic regions. Biochim. Biophys. Acta 2006, 1763, 833–843. [Google Scholar] [CrossRef]
- Russo, A.; Russo, G. Ribosomal Proteins Control or Bypass p53 during Nucleolar Stress. Int. J. Mol. Sci. 2017, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, P.; Pecoraro, A.; Palma, G.; Russo, G.; Russo, A. Therapeutic Approaches Targeting Nucleolus in Cancer. Cells 2019, 8, 1090. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Carotenuto, P.; Russo, G.; Russo, A. Ribosomal protein uL3 targets E2F1 and Cyclin D1 in cancer cell response to nucleolar stress. Sci. Rep. 2019, 9, 15431. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Carotenuto, P.; Brunella, F.; De Cegli, R.; Russo, G.; Russo, A. Role of uL3 in the Crosstalk between Nucleolar Stress and Autophagy in Colon Cancer Cells. Int. J. Mol. Sci. 2020, 21, 2143. [Google Scholar] [CrossRef]
- Russo, A.; Siciliano, G.; Catillo, M.; Giangrande, C.; Amoresano, A.; Pucci, P.; Pietropaolo, C.; Russo, G. hnRNP H1 and intronic G runs in the splicing control of the human rpL3 gene. Biochim. Biophys. Acta 2010, 1799, 419–428. [Google Scholar] [CrossRef]
- Russo, A.; Maiolino, S.; Pagliara, V.; Ungaro, F.; Tatangelo, F.; Leone, A.; Scalia, G.; Budillon, A.; Quaglia, F.; Russo, G. Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles. Oncotarget 2016, 7, 79670–79687. [Google Scholar] [CrossRef]
- Russo, A.; Saide, A.; Smaldone, S.; Faraonio, R.; Russo, G. Role of uL3 in Multidrug Resistance in p53-Mutated Lung Cancer Cells. Int. J. Mol. Sci. 2017, 18, 547. [Google Scholar] [CrossRef]
- Virgilio, A.; Varra, M.; Scuotto, M.; Capuozzo, A.; Irace, C.; Mayol, L.; Esposito, V.; Galeone, A. Expanding the potential of G-quadruplex structures: Formation of a heterochiral TBA analogue. Chembiochem 2014, 15, 652–655. [Google Scholar] [CrossRef]
- Pagliara, V.; Saide, A.; Mitidieri, E.; D’Emmanuele di Villa Bianca, R.; Sorrentino, R.; Russo, G.; Russo, A. 5-FU targets rpL3 to induce mitochondrial apoptosis via cystathionine-β-synthase in colon cancer cells lacking p53. Oncotarget 2016, 7, 50333–50348. [Google Scholar] [CrossRef]
- Miniaci, M.C.; Irace, C.; Capuozzo, A.; Piccolo, M.; Di Pascale, A.; Russo, A.; Lippiello, P.; Lepre, F.; Russo, G.; Santamaria, R. Cysteine Prevents the Reduction in Keratin Synthesis Induced by Iron Deficiency in Human Keratinocytes. J Cell Biochem 2016, 117, 402–412. [Google Scholar] [CrossRef]
- De Filippis, D.; Russo, A.; De Stefano, D.; Cipriano, M.; Esposito, D.; Grassia, G.; Carnuccio, R.; Russo, G.; Iuvone, T. Palmitoylethanolamide inhibits rMCP-5 expression by regulating MITF activation in rat chronic granulomatous inflammation. Eur. J. Pharmacol. 2014, 725, 64–69. [Google Scholar] [CrossRef] [PubMed]
- D’Emmanuele di Villa Bianca, R.; Mitidieri, E.; Fusco, F.; Russo, A.; Pagliara, V.; Tramontano, T.; Donnarumma, E.; Mirone, V.; Cirino, G.; Russo, G.; et al. Urothelium muscarinic activation phosphorylates CBS(Ser227) via cGMP/PKG pathway causing human bladder relaxation through H2S production. Sci Rep 2016, 6, 31491. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- De Filippis, D.; Russo, A.; D’Amico, A.; Esposito, G.; Pietropaolo, C.; Concetta, P.; Cinelli, M.; Russo, G.; Iuvone, T. Cannabinoids reduce granuloma-associated angiogenesis in rats by controlling transcription and expression of mast cell protease-5. Br. J. Pharmacol. 2008, 154, 1672–1679. [Google Scholar] [CrossRef]
- James, A.; Wang, Y.; Raje, H.; Rosby, R.; DiMario, P. Nucleolar stress with and without p53. Nucleus 2014, 5, 402–426. [Google Scholar] [CrossRef]
- Deryło, K.; Michalec-Wawiórka, B.; Krokowski, D.; Wawiórka, L.; Hatzoglou, M.; Tchórzewski, M. The uL10 protein, a component of the ribosomal P-stalk, is released from the ribosome in nucleolar stress. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 34–47. [Google Scholar] [CrossRef]
- Aubert, M.; O’Donohue, M.F.; Lebaron, S.; Gleizes, P.E. Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases. Biomolecules 2018, 8, 123. [Google Scholar] [CrossRef]
- O’Donohue, M.F.; Choesmel, V.; Faubladier, M.; Fichant, G.; Gleizes, P.E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010, 190, 853–866. [Google Scholar] [CrossRef]
- Kousholt, A.N.; Menzel, T.; Sørensen, C.S. Pathways for genome integrity in G2 phase of the cell cycle. Biomolecules 2012, 2, 579–607. [Google Scholar] [CrossRef]
- Willis, S.; Day, C.L.; Hinds, M.G.; Huang, D.C. The Bcl-2-regulated apoptotic pathway. J. Cell Sci. 2003, 116, 4053–4056. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Mangan, H.; Gailín, M.; McStay, B. Integrating the genomic architecture of human nucleolar organizer regions with the biophysical properties of nucleoli. FEBS J. 2017, 284, 3977–3985. [Google Scholar] [CrossRef] [PubMed]
- Núñez Villacís, L.; Wong, M.S.; Ferguson, L.L.; Hein, N.; George, A.J.; Hannan, K.M. New Roles for the Nucleolus in Health and Disease. Bioessays 2018, 40, e1700233. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef]
- Huang, Y.; Sramkoski, R.M.; Jacobberger, J.W. The kinetics of G2 and M transitions regulated by B cyclins. PLoS ONE 2013, 8, e80861. [Google Scholar] [CrossRef] [PubMed]
- Charrier-Savournin, F.B.; Château, M.T.; Gire, V.; Sedivy, J.; Piette, J.; Dulic, V. p21-Mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol. Biol. Cell 2004, 15, 3965–3976. [Google Scholar] [CrossRef]
- Shamloo, B.; Usluer, S. p21 in Cancer Research. Cancers 2019, 11, 1178. [Google Scholar] [CrossRef]



| Name | Sequence [a] | Tm (°C) |
|---|---|---|
| TBA | 5′-GGTTGGTGTGGTTGG-3′ | 50 [b] |
| L-TBA | 5′-ggttggtgtggttgg-3′ | 50 [b] |
| LQ1 | 5′-ggTTggtgtggTTgg-3′ | 52 [b] |
| LQ2 | 5′-ggTtgggtgtggTtgg-3′ | 49 |
| LQ3 | 5′-ggtTgggtgtggtTgg-3′ | 49 |
| Gene | Sequence |
|---|---|
| CDK1 | Forward: 5′ – CATGGCTACCACTTGACCTGT – 3′ Reverse: 5′ – AAGCCGGGATCTACCATACC – 3′ |
| CycA | Forward: 5′ – TTCATTTAGCACTCTACACAGTCACGG – 3′ Reverse: 5′ – TTGAGGTAGGTCTGGTGAAGGTCC – 3′ |
| CycB | Forward: 5′ – CAGTCAGACCAAAATACCTACTGGGT – 3′ Reverse: 5′ – ACACCAACCAGCTGCAGCATCTTCTT – 3′ |
| Bax | Forward: 5′ – CCCGAGAGGTCTTTTCCGAG – 3′ Reverse: 5′ – CCAGCCCATGATGGTTCTGAT – 3′ |
| B23/NPM | Forward: 5′ – AGAAAAAGCGCCAGTGAAGA – 3′ Reverse: 5′ – TGGTGTT GATGATTGGTTTTGA – 3′ |
| β-actin | Forward: 5′ – CCAACCGCGAGAAGATGA – 3′ Reverse: 5′ – CCAGAGGCGTACAGGGATAG – 3′ |
| Bcl-2 | Forward: 5′ – ATGTGTGTGGAGAGCGTCAACC – 3′ Reverse: 5′ – GCATCCCAGCCTCCGTTATC – 3′ |
| p21 | Forward: 5′ – CCTCAAATCGTCCAGCGACCTT – 3′ Reverse: 5′ – CATTGTGGGAGGAGCTGTGAAA – 3′ |
| uL3 | Forward: 5′ – CAAAGGCTACAAAGGGGT – 3′ Reverse: 5′ – CTCAGTGCGGTGATGGTAG – 3′ |
| uL5 | Forward: 5′ – GGGATCCAGGAACACATCGA – 3′ Reverse: 5′ – AGAAGTCCAGGCCGTAGATACCA – 3′ |
| uL11 | Forward: 5′ – AGTCGTATACCTGAGGTGCACCGGA – 3′ Reverse: 5′ – GCCATCAACATTACAGCCCACTGAC – 3′ |
| uL18 | Forward: 5′ – TGGAACCGTCCCAAAATGTC – 3′ Reverse: 5′ – GAGGAAGCTTGCCTTCTTTTGAG – 3′ |
| uS12 | Forward: 5′ – CGAGACCAGAAGTGGCATGA – 3′ Reverse: 5′ – GCATGAGAAGCACCTCCAAAAG – 3′ |
| 47S | Forward: 5′ – GCTGACACGCTGTCCTCTG – 3′ Reverse: 5′ – ACGCGCGAGAGAACAGCAG – 3′ |
| 45S | Forward: 5′ – GCCTTCTCTAGCGATCTGAGAG – 3′ Reverse: 5′ – CCATAACGGAGGCAGAGACA – 3′ |
| 36S | Forward: 5′ – GCGGAGGTTTAAAGACCC – 3′ Reverse: 5′ – CCAGACGAGACAGCAAAC – 3′ |
| 32S | Forward: 5′ – GTCAGCGGAGGAGAAGAA – 3′ Reverse: 5′ – CTCGATCAGAAGGACTTGG – 3′ |
| 30S | Forward: 5′ – CCTCTGACGCGGCAGACAGC – 3′ Reverse: 5′ – CTCCAGGAGCACCGCAAGGG – 3′ |
| 18S precursors | Forward: 5′ – GTTCAAAGCAGGCCCGAGCC – 3′ Reverse: 5′ – AGCGGCGCAATACGAATGCC – 3′ |
| 28S | Forward: 5′ – CAGGGGAATCCGACTGTTTA – 3′ Reverse: 5′ – ATGACGAGGCATTTGGCTAC – 3′ |
| 18S | Forward: 5′ – AAACGGCTACCACATCCAAG – 3′ Reverse: 5′ – CCTCCAATGGATCCTCGTTA – 3′ |
| 5.8S | Forward: 5′ – CTCTTAGCGGTGGATCACTC – 3′ Reverse: 5′ – GACGCTCAGACAGGCGTAG – 3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecoraro, A.; Virgilio, A.; Esposito, V.; Galeone, A.; Russo, G.; Russo, A. uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells. Biomolecules 2020, 10, 583. https://doi.org/10.3390/biom10040583
Pecoraro A, Virgilio A, Esposito V, Galeone A, Russo G, Russo A. uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells. Biomolecules. 2020; 10(4):583. https://doi.org/10.3390/biom10040583
Chicago/Turabian StylePecoraro, Annalisa, Antonella Virgilio, Veronica Esposito, Aldo Galeone, Giulia Russo, and Annapina Russo. 2020. "uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells" Biomolecules 10, no. 4: 583. https://doi.org/10.3390/biom10040583
APA StylePecoraro, A., Virgilio, A., Esposito, V., Galeone, A., Russo, G., & Russo, A. (2020). uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells. Biomolecules, 10(4), 583. https://doi.org/10.3390/biom10040583








