Human PCNA Structure, Function and Interactions
Abstract
1. DNA Sliding Clamps
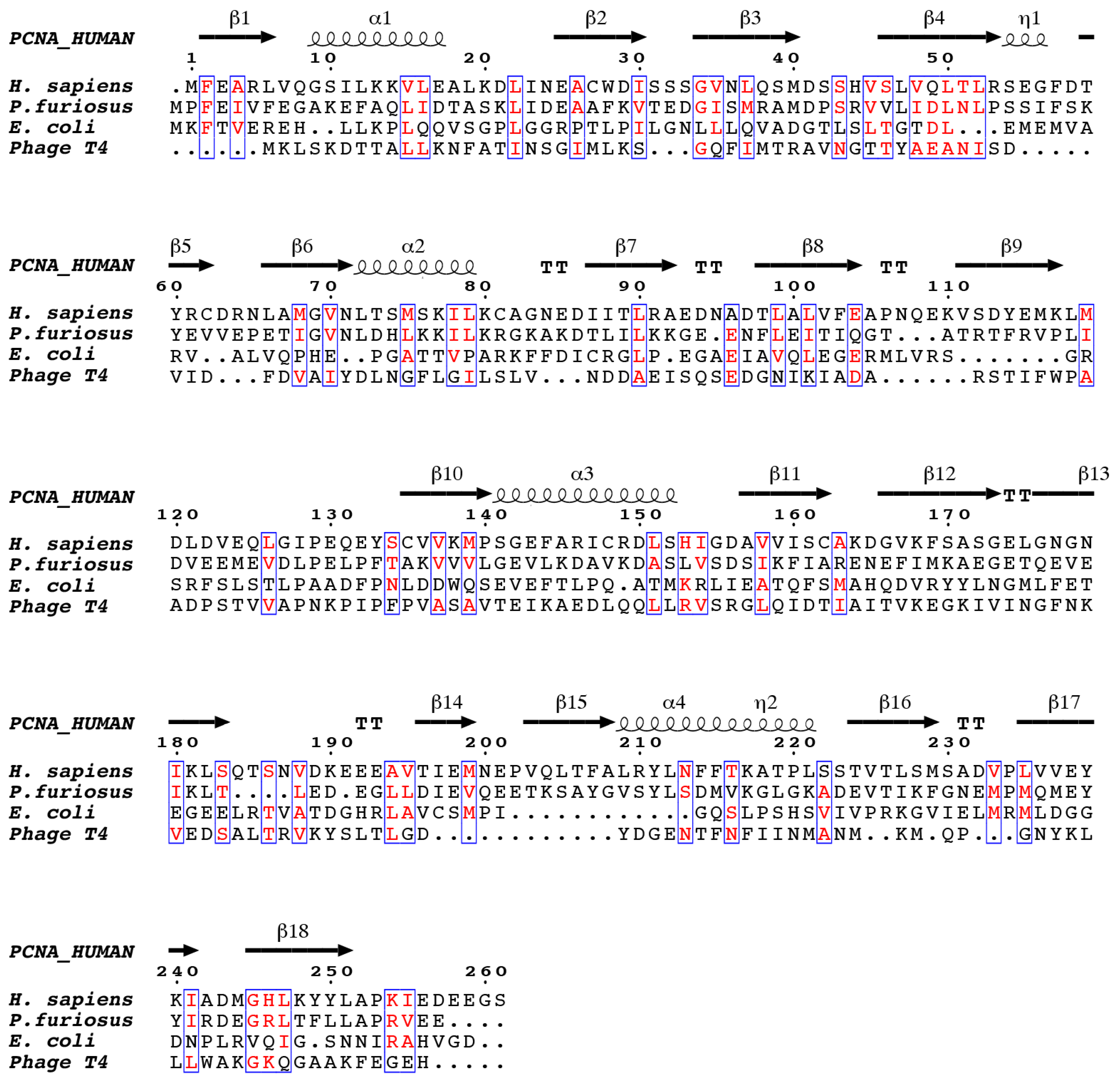
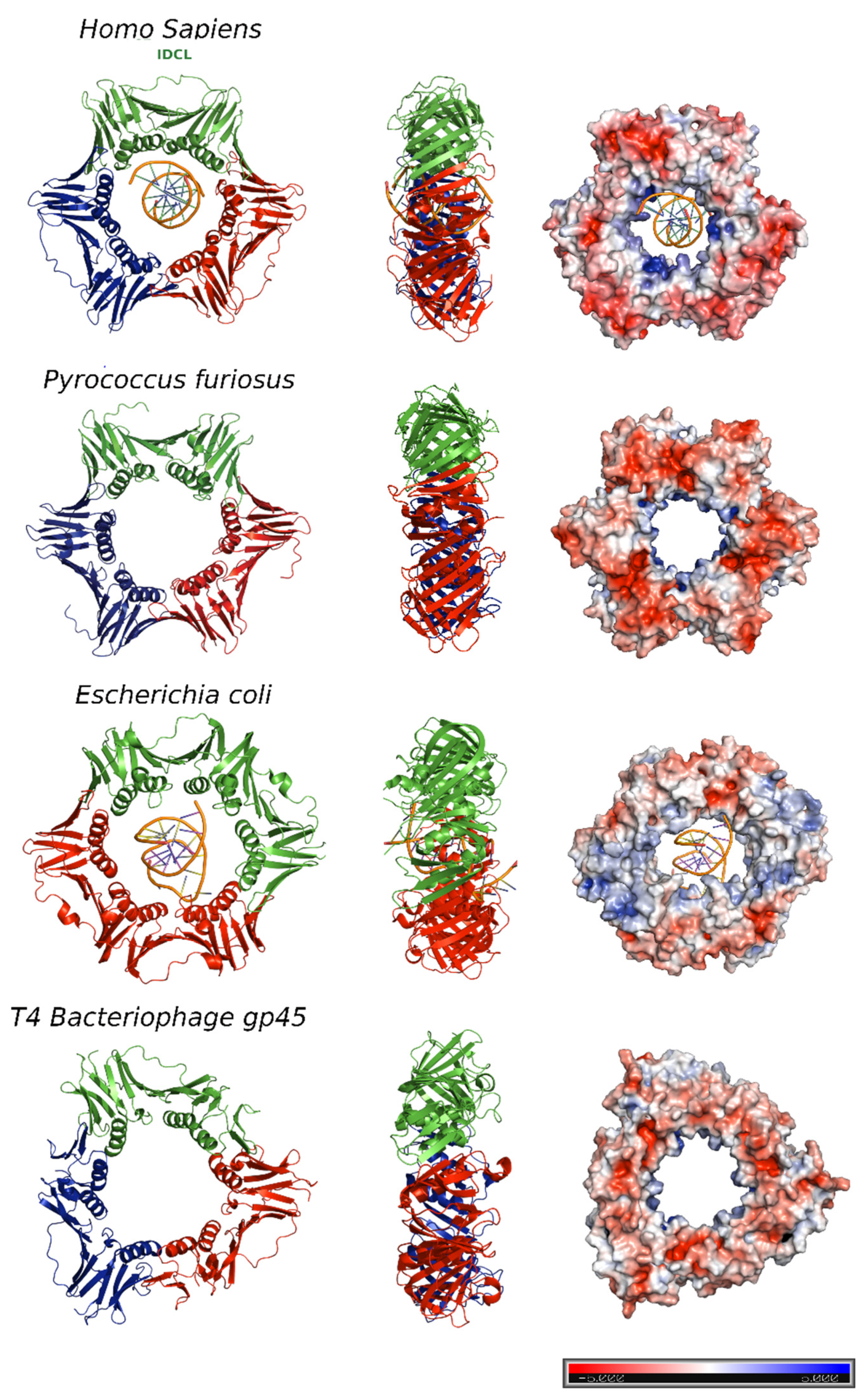
2. Human PCNA Structure
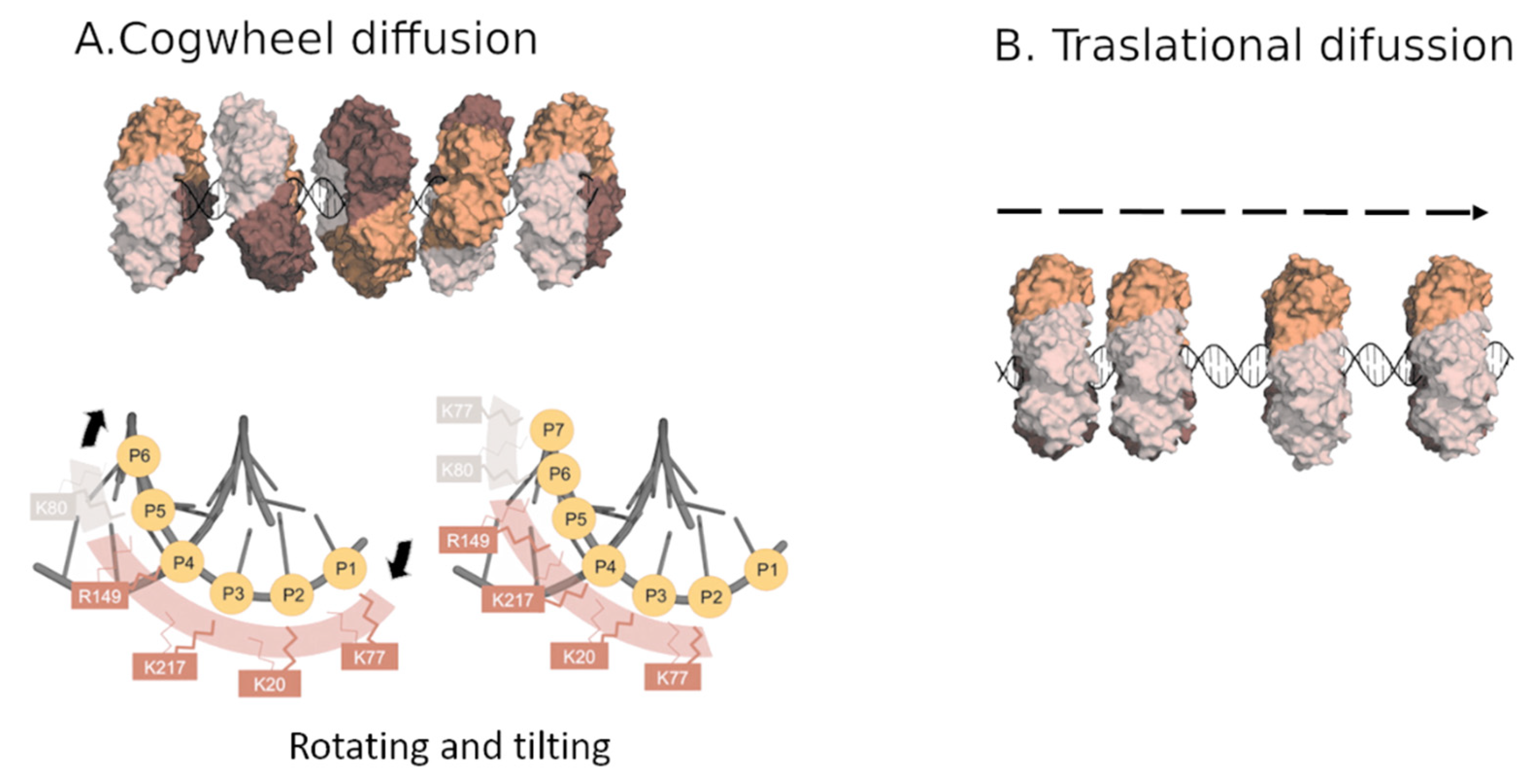
3. PCNA Sliding on DNA
4. PCNA Binding Proteins
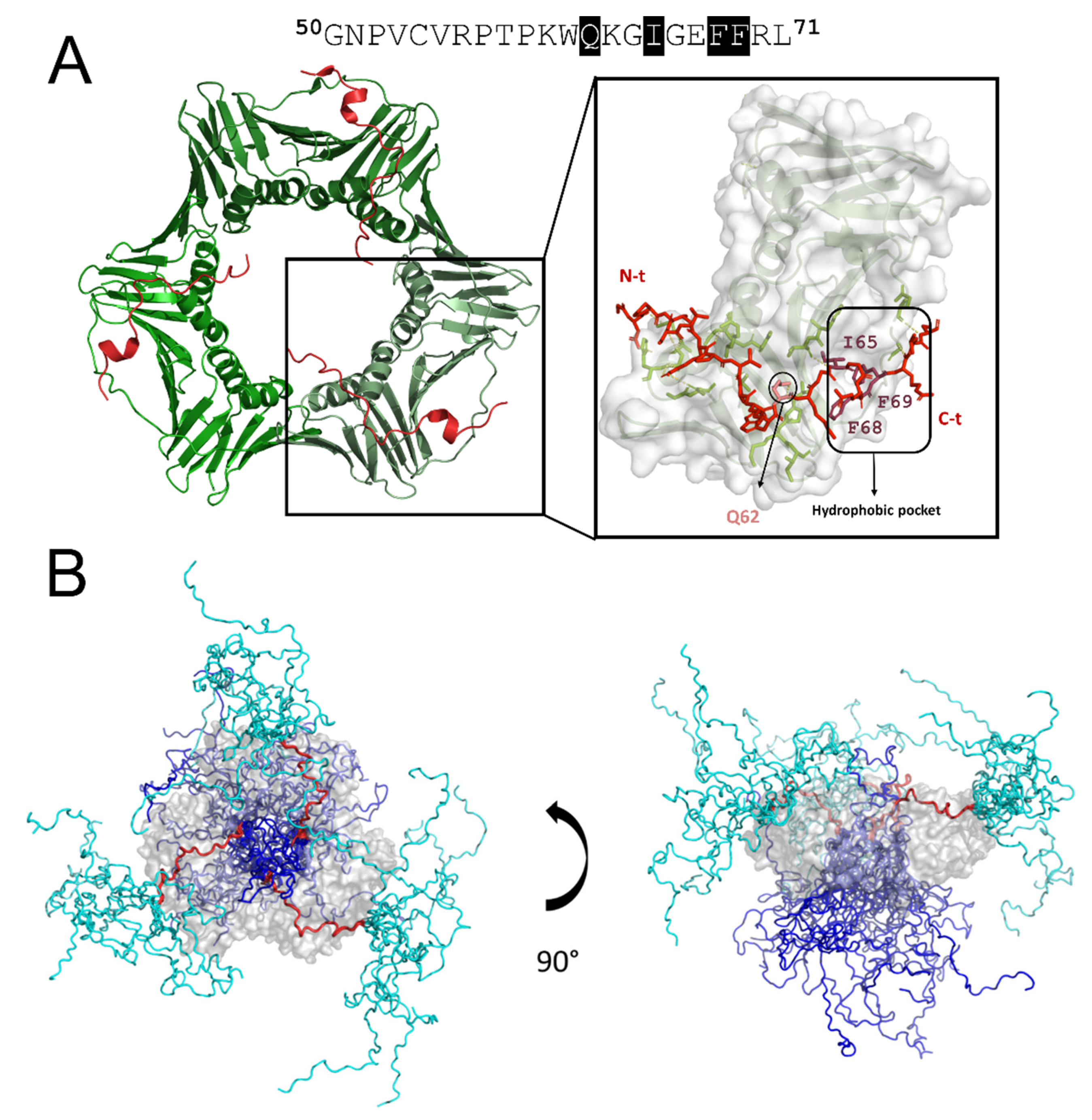
5. The Unique Interaction of PCNA with p15
6. Simultaneous Interactions with the PCNA Ring: The Tool-Belt Model
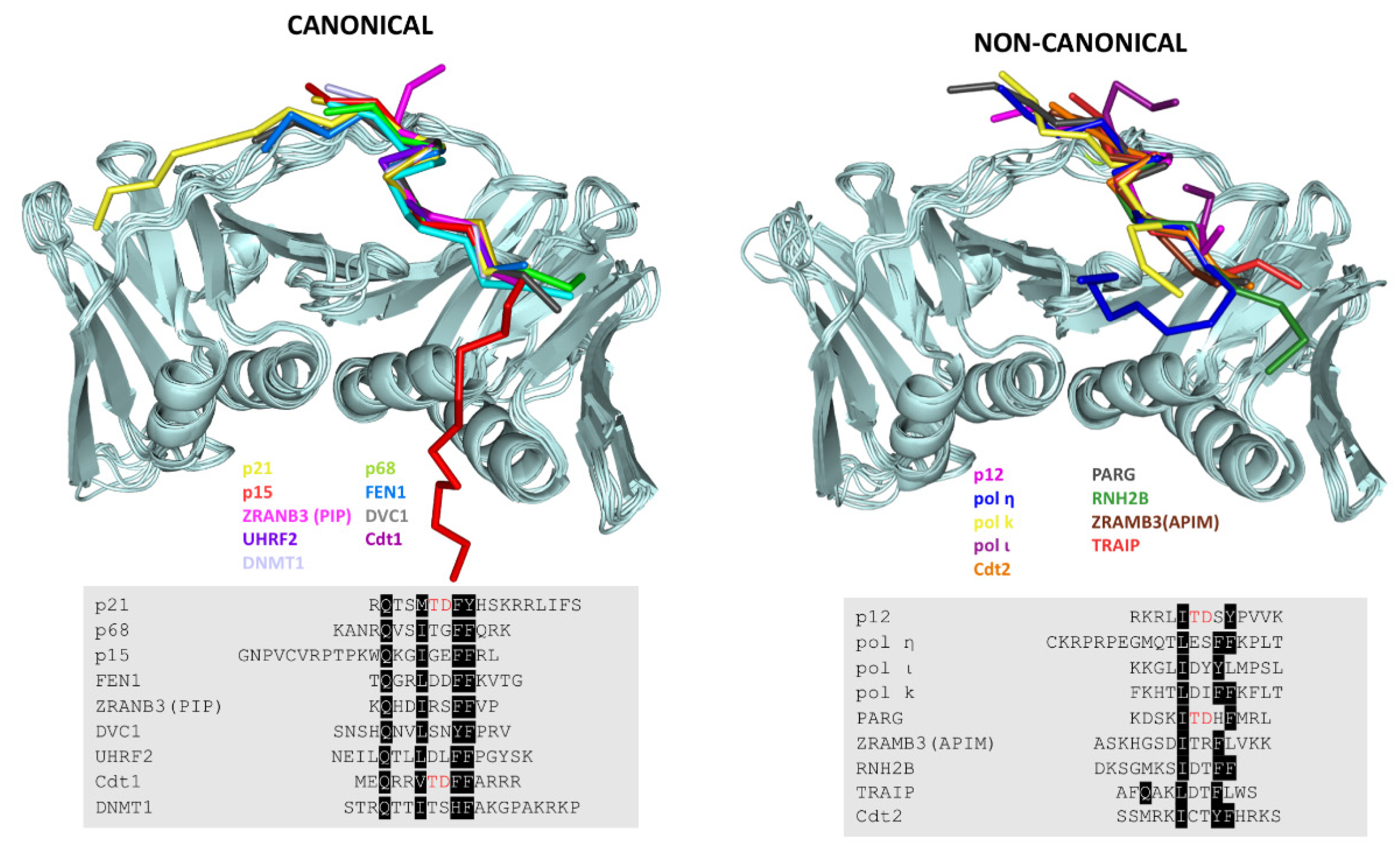
7. Revisiting the PIP Box Definition
8. Post-Translational Modifications of Human PCNA
9. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Kuriyan, J. Sliding Clamps of DNA Polymerases. J. Mol. Biol. 1993, 234, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the Maestro of the Replication Fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Stukenberg, P.T.; Studwell-Vaughan, P.S.; O’Donnell, M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 1991, 266, 11328–11334. [Google Scholar] [PubMed]
- Kelch, B.A.; Makino, D.L.; Donnell, M.O.; Kuriyan, J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jeruzalmi, D.; O’Donnell, M.; Kuriyan, J. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 2002, 12, 217–224. [Google Scholar] [CrossRef]
- Kelch, B.A. Review The Lord of the Rings: Structure and Mechanism of the Sliding Clamp Loader. Biopolymers 2016, 105, 532–546. [Google Scholar] [CrossRef]
- Indiani, C.; Donnell, M.O. The replication clamp-loading machine at work in the three domains of life. Mol. Cell Biol. 2006, 7, 751–761. [Google Scholar] [CrossRef]
- Krishna, T.S.R.; Kong, X.P.; Gary, S.; Burgers, P.M.; Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994, 79, 1233–1243. [Google Scholar] [CrossRef]
- De Biasio, A.; Blanco, F.J. Proliferating cell nuclear antigen structure and interactions: Too many partners for one dancer? Adv. Protein Chem. Struct. Biol. 2013, 91, 1–36. [Google Scholar]
- Kong, X.P.; Onrust, R.; O’Donnell, M.; Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 1992, 69, 425–437. [Google Scholar] [CrossRef]
- Hedglin, M.; Kumar, R.; Benkovic, S.J. Replication Clamps and Clamp Loaders. Cold Spring Harb Perspect Biol. 2013, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Matsumiya, S.; Ishino, Y. Crystal structure of an archaeal DNA sliding clamp: Proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 2001, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Wing, R.; Drew, H.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Crystal structure analysis of a complete turn of B-DNA. Nature 1980, 287, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Kelman, Z.; Donnell, M.O. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995, 23, 3613–3620. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, R.E.; Kim, S.S.; Yurieva, O.; Kuriyan, J.; Kong, X.P.; O’Donnell, M. Structure of a Sliding Clamp on DNA. Cell 2008, 132, 43–54. [Google Scholar] [CrossRef]
- McNally, R.; Bowman, G.D.; Goedken, E.R.; O’Donnell, M.; Kuriyan, J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct. Biol. 2010, 10. [Google Scholar] [CrossRef]
- Miyachi, K.; Fritzler, M.J.; Tan, E.M. Autoantibody to a Nuclear Antigen in Proliferating Cells. J. Immunol. 1978, 121, 2228. [Google Scholar]
- Kelman, Z. PCNA: Structure, functions and interactions. Oncogene 1997, 14, 629–640. [Google Scholar] [CrossRef]
- Bravo, R.; Celis, J.E. A search for differential polypeptide synthesis throughout the cell cycle of hela cells. J. Cell Biol. 1980, 84, 795–802. [Google Scholar] [CrossRef]
- Tsurimoto, T.; Stillman, B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 1991, 266, 1950–1960. [Google Scholar]
- Slade, D. Maneuvers on PCNA Rings during DNA Replication and Repair. Genes 2018, 9, 416. [Google Scholar] [CrossRef]
- Gulbis, J.M.; Kelman, Z.; Hurwitz, J.; O’Donnell, M.; Kuriyan, J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 1996, 87, 297–306. [Google Scholar] [CrossRef]
- Kontopidis, G.; Wu, S.; Zheleva, D.I.; Taylor, P.; Mcinnes, C.; Lane, D.P.; Fischer, P.M.; Walkinshaw, M.D. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. 2004, 102, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, R.; Torres, D.; Prieto, J.; Blanco, F.J.; Campos-Olivas, R. Backbone assignment of human proliferating cell nuclear antigen. Biomol. NMR Assign. 2007, 1, 245–247. [Google Scholar] [CrossRef] [PubMed]
- de Biasio, A.; Sánchez, R.; Prieto, J.; Villate, M.; Campos-Olivas, R.; Blanco, F.J. Reduced stability and increased dynamics in the human Proliferating Cell Nuclear Antigen (PCNA) relative to the yeast homolog. PLoS ONE 2011, 6, e16600. [Google Scholar] [CrossRef] [PubMed]
- Naryzhny, S.N.; Zhao, H.; Lee, H. Proliferating cell nuclear antigen (PCNA) may function as a double homotrimer complex in the mammalian cell. J. Biol. Chem. 2005, 280, 13888–13894. [Google Scholar] [CrossRef] [PubMed]
- De March, M.; Merino, N.; Barrera-Vilarmau, S.; Crehuet, R.; Onesti, S.; Blanco, F.J.; De Biasio, A. Structural basis of human PCNA sliding on DNA. Nat. Commun. 2017, 8, 13935. [Google Scholar] [CrossRef]
- De March, M.; De Biasio, A. The dark side of the ring: Role of the DNA sliding surface of PCNA. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 663–673. [Google Scholar] [CrossRef]
- Yao, N.Y.; O’Donnell, M. DNA Replication: How Does a Sliding Clamp Slide? Curr. Biol. 2017, 27, R174–R176. [Google Scholar] [CrossRef]
- Kochaniak, A.B.; Habuchi, S.; Loparo, J.J.; Chang, D.J.; Cimprich, K.A.; Walter, J.C.; van Oijen, A.M. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 2009, 284, 17700–17710. [Google Scholar] [CrossRef]
- Cazzalini, O.; Sommatis, S.; Tillhon, M.; Dutto, I.; Bachi, A.; Rapp, A.; Nardo, T.; Scovassi, A.I.; Necchi, D.; Cardoso, M.C.; et al. CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic Acids Res. 2014, 42, 8433–8448. [Google Scholar] [CrossRef] [PubMed]
- De March, M.; Barrera-vilarmau, S.; Crespan, E.; Mentegari, E.; Romano-Moreno, M.; Maga, G.; Merino, N.; Gonzalez-Magaña, A.; Crehuet, R.; Onesti, S.; et al. p15 PAF binding to PCNA modulates the DNA sliding surface. Nucleic Acids Res. 2018, 1–13. [Google Scholar]
- Billon, P.; Côté, J. Novel mechanism of PCNA control through acetylation of its sliding surface. Mol. Cell. Oncol. 2017, 4, 2–4. [Google Scholar] [CrossRef]
- Warbrick, E. The puzzle of PCNA’s many partners. Bioessays 2000, 22, 997–1006. [Google Scholar] [CrossRef]
- Maga, G.; Hübscher, U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003, 116, 3051–3060. [Google Scholar] [CrossRef] [PubMed]
- Click, T.H.; Ganguly, D.; Chen, J. Intrinsically disordered proteins in a physics-based world. Int. J. Mol. Sci. 2010, 11, 5292–5309. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Cordeiro, T.N.; Chen, P.; De Biasio, A.; Sibille, N. Disentangling polydispersity in the PCNA−p15 PAF complex, a disordered, transient and multivalent macromolecular assembly. Nucleic Acids Res. 2017, 45, 1501–1515. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradović, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Unfolded Proteins and Protein Folding Studied by NMR. Chem. Rev. 2004, 104, 3607–3622. [Google Scholar] [CrossRef]
- Prestel, A.; Wichmann, N.; Martins, J.M.; Marabini, R.; Kassem, N.; Broendum, S.S.; Otterlei, M.; Nielsen, O.; Willemoës, M.; Ploug, M.; et al. The PCNA interaction motifs revisited: Thinking outside the PIP-box. Cell. Mol. Life Sci. 2019, 76, 4923–4943. [Google Scholar] [CrossRef]
- Havens, C.G.; Walter, J.C. Docking of a specialized PIP box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 2009, 23, 1–7. [Google Scholar] [CrossRef]
- Liang, Z.; Diamond, M.; Smith, J.A.; Schnell, M.; Daniel, R. Proliferating cell nuclear antigen is required for loading of the SMCX/KMD5C histone demethylase onto chromatin. Epigenetics Chromatin 2011, 4, 18. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, P.; Liu, L.; Lee, M.Y.W.T. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 2001, 40, 4512–4520. [Google Scholar] [CrossRef]
- De Biasio, A.; Campos-Olivas, R.; Sánchez, R.; López-Alonso, J.P.; Pantoja-Uceda, D.; Merino, N.; Villate, M.; Martin-Garcia, J.M.; Castillo, F.; Luque, I.; et al. Proliferating cell nuclear antigen (PCNA) interactions in solution studied by NMR. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Bruning, J.B.; Shamoo, Y. Structural and Thermodynamic Analysis of Human PCNA with Peptides Derived from DNA Polymerase-δ p66 Subunit and Flap Endonuclease-1. Structure 2004, 12, 2209–2219. [Google Scholar] [CrossRef]
- Yu, P.; Huang, B.; Shen, M.; Lau, C.; Chan, E.; Michel, J.; Xiong, Y.; Payan, D.G.; Luo, Y. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene 2001, 20, 484–489. [Google Scholar] [CrossRef]
- Xie, C.; Yao, M.; Dong, Q. Proliferating cell unclear antigen-associated factor (PAF15): A novel oncogene. Int. J. Biochem. Cell Biol. 2014, 50, 127–131. [Google Scholar] [CrossRef]
- Simpson, F.; Lammerts Van Bueren, K.; Butterfield, N.; Bennetts, J.S.; Bowles, J.; Adolphe, C.; Simms, L.A.; Young, J.; Walsh, M.D.; Leggett, B.; et al. The PCNA-associated factor KIAA0101/p15PAF binds the potential tumor suppressor product p33ING1b. Exp. Cell Res. 2006, 312, 73–85. [Google Scholar] [CrossRef]
- Mailand, N.; Gibbs-Seymour, I.; Bekker-Jensen, S. Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 2013, 14, 269–282. [Google Scholar] [CrossRef]
- Emanuele, M.J.; Ciccia, A.; Elia, A.E.H.; Elledge, S.J. Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc. Natl. Acad. Sci. USA 2011, 108, 9845–9850. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M.; Takehara, A.; Matsuda, K.; Eguchi, H.; Ohigashi, H.; Ishikawa, O.; Shinomura, Y.; Imai, K.; Nakamura, Y.; Nakagawa, H. Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res. 2007, 67, 2568–2576. [Google Scholar] [CrossRef] [PubMed]
- De Biasio, A.; Ibañez De Opakua, A.; Cordeiro, T.N.; Villate, M.; Merino, N.; Sibille, N.; Lelli, M.; Diercks, T.; Bernado, P.; Blanco, F.J. P15PAF Is an intrinsically disordered protein with nonrandom structural preferences at sites of interaction with other proteins. Biophys. J. 2014, 106, 865–874. [Google Scholar] [CrossRef]
- De Biasio, A.; de Opakua, A.I.; Mortuza, G.B.; Molina, R.; Cordeiro, T.N.; Castillo, F.; Villate, M.; Merino, N.; Delgado, S.; Gil-Cartón, D.; et al. Structure of p15(PAF)-PCNA complex and implications for clamp sliding during DNA replication and repair. Nat. Commun. 2015, 6, 6439. [Google Scholar] [CrossRef]
- González-Magaña, A.; De Opakua, A.I.; Merino, N.; Monteiro, H.; Diercks, T.; Murciano-Calles, J.; Luque, I.; Bernadó, P.; Cordeiro, T.N.; De Biasio, A.; et al. Double Monoubiquitination Modifies the Molecular Recognition Properties of p15PAF Promoting Binding to the Reader Module of Dnmt1. ACS Chem. Biol. 2019, 14, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- Stodola, J.L.; Burgers, P.M. Resolving individual steps of Okazaku Fragment maturation at a msec time-scale. Nat. Struct. Mol. Biol. 2016, 23, 402–408. [Google Scholar] [CrossRef]
- Dovrat, D.; Stodola, J.L.; Burgers, P.M.J.; Aharoni, A. Sequential switching of binding partners on PCNA during in vitro Okazaki fragment maturation. Proc. Natl. Acad. Sci. USA 2014, 111, 14118–14123. [Google Scholar] [CrossRef]
- Sakurai, S.; Kitano, K.; Yamaguchi, H.; Hamada, K.; Okada, K.; Fukuda, K.; Uchida, M.; Ohtsuka, E.; Morioka, H.; Hakoshima, T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005, 24, 683–693. [Google Scholar] [CrossRef]
- Lancey, C.; Tehseen, M.; Raducanu, V.S.; Rashid, F.; Merino, N.; Ragan, T.J.; Savva, C.G.; Zaher, M.S.; Shirbini, A.; Blanco, F.J.; et al. Structure of the processive human Pol δ holoenzyme. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Hishiki, A.; Hashimoto, H.; Hanafusa, T.; Kamei, K.; Ohashi, E.; Shimizu, T.; Ohmori, H.; Sato, M. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 2009, 284, 10552–10560. [Google Scholar] [CrossRef]
- Gonzalez-Magaña, A.; De Opakua, A.I.; Romano-Moreno, M.; Murciano-Calles, J.; Merino, N.; Luque, I.; Rojas, A.L.; Onesti, S.; Blanco, F.J.; De Biasio, A. The p12 subunit of human polymerase uses an atypical PIP box for molecular recognition of proliferating cell nuclear antigen (PCNA). J. Biol. Chem. 2019, 294, 3947–3956. [Google Scholar] [CrossRef] [PubMed]
- Boehm, E.M.; Washington, M.T. R.I.P. to the PIP: PCNA-binding motif no longer considered specific: PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. BioEssays 2016, 38, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Leung, W.; Baxley, R.M.; Moldovan, G.L.; Bielinsky, A.K. Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Kashiwaba, S.-i.; Kanao, R.; Masuda, Y.; Kusumoto-Matsuo, R.; Hanaoka, F.; Masutani, C. USP7 Is a Suppressor of PCNA Ubiquitination and Oxidative-Stress-Induced Mutagenesis in Human Cells. Cell Rep. 2015, 13, 2072–2080. [Google Scholar] [CrossRef] [PubMed]
- Hedglin, M.; Pandey, B.; Benkovic, S.J. Characterization of human translesion DNA synthesis across a UV-induced DNA lesion. Elife 2016, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Vujanovic, M.; Krietsch, J.; Raso, M.C.; Terraneo, N.; Zellweger, R.; Schmid, J.A.; Taglialatela, A.; Huang, J.W.; Holland, C.L.; Zwicky, K.; et al. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol. Cell 2017, 67, 882–890.e5. [Google Scholar] [CrossRef]
- Hibbert, R.G.; Sixma, T.K. Intrinsic flexibility of ubiquitin on proliferating cell nuclear antigen (PCNA) in translesion synthesis. J. Biol. Chem. 2012, 287, 39216–39223. [Google Scholar] [CrossRef]
- Gali, H.; Juhasz, S.; Morocz, M.; Hajdu, I.; Fatyol, K.; Szukacsov, V.; Burkovics, P.; Haracska, L. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012, 40, 6049–6059. [Google Scholar] [CrossRef]
- Elia, A.E.H.; Boardman, A.P.; Wang, D.C.; Huttlin, E.L.; Everley, R.A.; Dephoure, N.; Zhou, C.; Koren, I.; Gygi, S.P.; Elledge, S.J. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cell. 2015, 59, 867–881. [Google Scholar] [CrossRef]
- Park, J.M.; Yang, S.W.; Yu, K.R.; Ka, S.H.; Lee, S.W.; Seol, J.H.; Jeon, Y.J.; Chung, C.H. Modification of PCNA by ISG15 Plays a Crucial Role in Termination of Error-Prone Translesion DNA Synthesis. Mol. Cell 2014, 54, 626–638. [Google Scholar] [CrossRef]
- Guan, J.; Yu, S.; Zheng, X. NEDDylation antagonizes ubiquitination of proliferating cell nuclear antigen and regulates the recruitment of polymerase η in response to oxidative DNA damage. Protein Cell 2018, 9, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Takawa, M.; Cho, H.S.; Hayami, S.; Toyokawa, G.; Kogure, M.; Yamane, Y.; Iwai, Y.; Maejima, K.; Ueda, K.; Masuda, A.; et al. Histone lysine methyltransferase setd8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012, 72, 3217–3227. [Google Scholar] [CrossRef] [PubMed]
- Peng, A.; Xu, X.; Wang, C.; Yang, J.; Wang, S.; Dai, J.; Ye, L. EZH2 promotes DNA replication by stabilizing interaction of POLδ and PCNA via methylation-mediated PCNA trimerization. Epigenetics Chromatin 2018, 11. [Google Scholar]
- Yu, Y.; Cai, J.P.; Tu, B.; Wu, L.; Zhao, Y.; Liu, X.; Li, L.; McNutt, M.A.; Feng, J.; He, Q.; et al. Proliferating cell nuclear antigen is protected from degradation by forming a complex with MutT homolog2. J. Biol. Chem. 2009, 284, 19310–19320. [Google Scholar] [CrossRef]
- Wang, S.-C.; Nakajima, Y.; Yu, Y.-L.; Xia, W.; Chen, C.-T.; Yang, C.-C.; McIntush, E.W.; Li, L.-Y.; Hawke, D.H.; Kobayashi, R.; et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006, 8, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Ortega, J.; Gu, L.; Chang, Z.; Li, G.M. Phosphorylation of proliferating cell nuclear antigen promotes cancer progression by activating the ATM/Akt/GSK3β/Snail signaling pathway. J. Biol. Chem. 2019, 294, 7037–7045. [Google Scholar] [CrossRef]
- Ortega, J.; Li, J.Y.; Lee, S.; Tong, D.; Gu, L.; Li, G.M. Phosphorylation of PCNA by EGFR inhibits mismatch repair and promotes misincorporation during DNA synthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 5667–5672. [Google Scholar] [CrossRef]
- Lo, Y.-H.; Ho, P.-C.; Chen, M.-S.; Hugo, E.; Ben-Jonathan, N.; Wang, S.-C. Phosphorylation at tyrosine 114 of Proliferating Cell Nuclear Antigen (PCNA) is required for adipogenesis in response to high fat diet. Biochem. Biophys Res. Commun. 2013, 23, 1–7. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Ding, M.; Yu, Y. Site-specific characterization of the Asp-and Glu-ADP-ribosylated proteome. Nat. Methods 2013, 10, 981–984. [Google Scholar] [CrossRef]
- Punchihewa, C.; Inoue, A.; Hishiki, A.; Fujikawa, Y.; Connelly, M.; Evison, B.; Shao, Y.; Heath, R.; Kuraoka, I.; Rodrigues, P.; et al. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J. Biol. Chem. 2012, 287, 14289–14300. [Google Scholar] [CrossRef]
- Horsfall, A.J.; Abell, A.D.; Bruning, J. Targeting PCNA with peptide mimetics for therapeutic purposes. ChemBioChem 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lo, Y.-H.; E.Waltz, S.; Gray, J.K.; Hung, M.-C.; Wang, S.-C. Targeting Tyrosine Phosphorylation of PCNA Inhibits Prostate Cancer Growth. Mol. Cancer Ther. 2011, 10, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Sebesta, M.; Cooper, C.D.O.; Ariza, A.; Carnie, C.J.; Ahel, D. Structural insights into the function of ZRANB3 in replication stress response. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, M.; Jiang, T. Crystal structure of human PCNA in complex with the PIP box of DVC1. Biochem. Biophys. Res. Commun. 2016, 474, 264–270. [Google Scholar] [CrossRef]
- Jimenji, T.; Matsumura, R.; Kori, S.; Arita, K. Structure of PCNA in complex with DNMT1 PIP box reveals the basis for the molecular mechanism of the interaction. Biochem. Biophys. Res. Commun. 2019, 516, 578–583. [Google Scholar] [CrossRef]
- Chen, W.; Wu, M.; Hang, T.; Wang, C.; Zhang, X.; Zang, J. Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA. Biochem. Biophys. Res. Commun. 2017, 494, 575–580. [Google Scholar] [CrossRef]
- Hoffmann, S.; Smedegaard, S.; Nakamura, K.; Mortuza, G.B.; Räschle, M.; de Opakua, A.I.; Oka, Y.; Feng, Y.; Blanco, F.J.; Mann, M.; et al. TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress. J. Cell Biol. 2016, 212, 63–75. [Google Scholar] [CrossRef]
- Hara, K.; Uchida, M.; Tagata, R.; Yokoyama, H.; Ishikawa, Y.; Hishiki, A.; Hashimoto, H. Structure of proliferating cell nuclear antigen (PCNA) bound to an APIM peptide reveals the universality of PCNA interaction. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 214–221. [Google Scholar] [CrossRef]
- Kaufmann, T.; Grishkovskaya, I.; Polyansky, A.A.; Kostrhon, S.; Kukolj, E.; Olek, K.M.; Herbert, S.; Beltzung, E.; Mechtler, K.; Peterbauer, T.; et al. A novel non-canonical PIP-box mediates PARG interaction with PCNA. Nucleic Acids Res. 2017, 45, 9741–9759. [Google Scholar] [CrossRef]
- Hayashi, A.; Giakoumakis, N.N.; Heidebrecht, T.; Ishii, T.; Panagopoulos, A.; Caillat, C.; Takahara, M.; Hibbert, R.G.; Suenaga, N.; Stadnik-spiewak, M.; et al. Direct binding of Cdt2 to PCNA is important for targeting the CRL4 Cdt2 E3 ligase activity to Cdt1. Life Sci. Alliance 2018, 1, 1–17. [Google Scholar] [CrossRef]
- Bubeck, D.; Reijns, M.A.M.; Graham, S.C.; Astell, K.R.; Jones, E.Y.; Jackson, A.P. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011, 39, 3652–3666. [Google Scholar] [CrossRef] [PubMed]
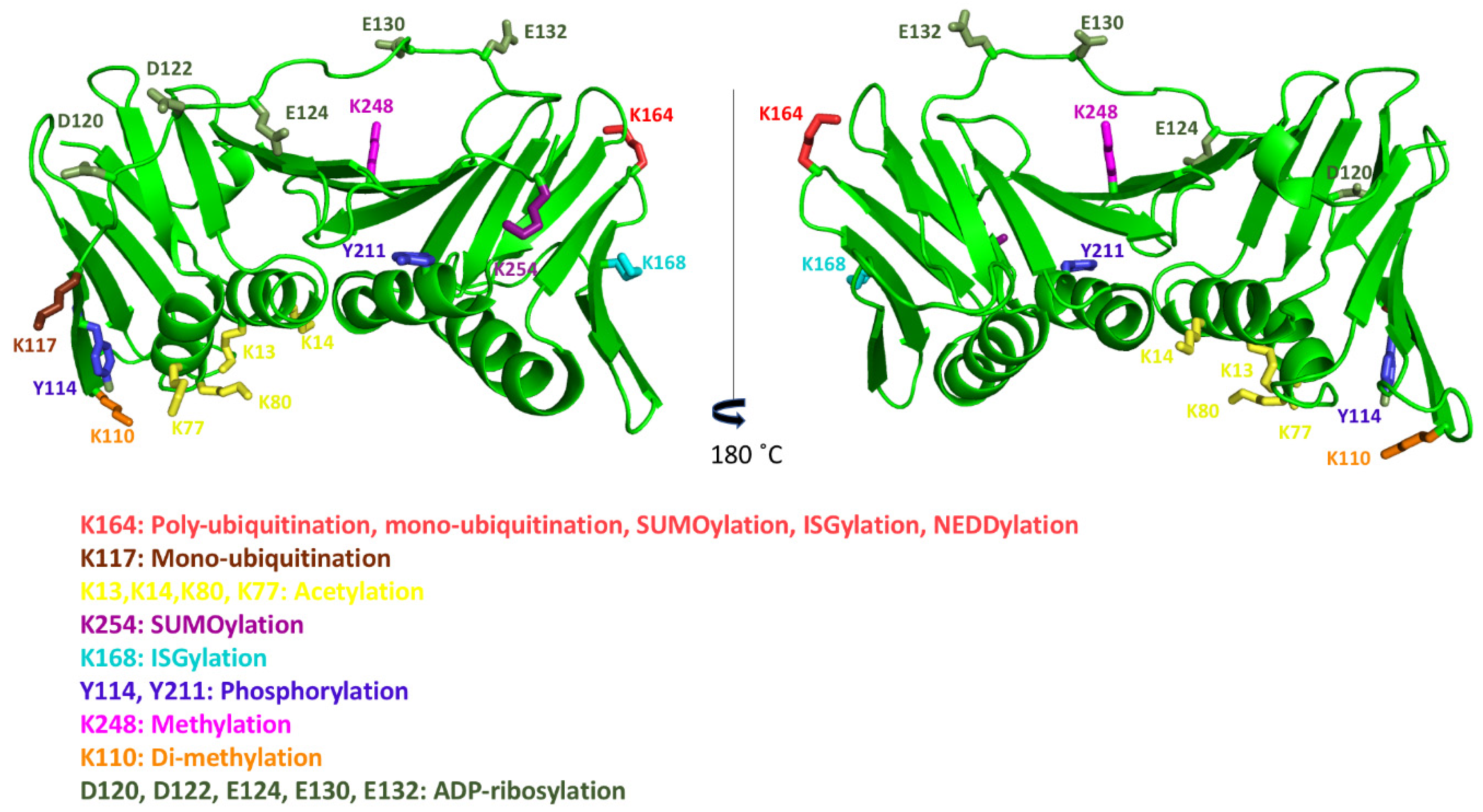
| Modification | Target Residue | Enzyme | Function | Ref. |
|---|---|---|---|---|
| Monoubiquitination | K164 | Rad18, RNF8 CRL4Cdt2 | Promotes DNA synthesis at damaged sites | [65] |
| K117 | Unknown | Unknown | [69] | |
| Polyubiquitination | K164 | Rad5 HLTF/SHPRH | Promotes TS | [66] |
| Acetylation | K13,K14, K77, K80 | CPB/p300 | Promotes PCNA degradation after NER | [31] |
| SUMOylation | K164 | UBC9(E2) | Inhibits DSBs formation | [68] |
| K254 | Unknown | Unknown | [72] | |
| ISGylation | K164, K168 | EFP | Terminates TLS | [70] |
| NEDDylation | K164 * | Rad18 | Regulates Pol η recruitment in DDR pathway | [71] |
| Phosphorylation | Y114 * | Unknown | Promotes adipogenesis in response to fatty diet | [76] |
| Y211 | EGR | Protects PCNA from degradation and inhibits MMR | [75] [77] | |
| Methylation | K248 | SETD8 | Promotes maturation of Okazaki fragments | [72] |
| Di-methylation | K110 * | EZH2 | Promotes DNA synthesis by Pol δ | [73] |
| Protein | Activity | Sequence | T(°C) | Kd (µM) | Method * | PDB Code | Ref |
|---|---|---|---|---|---|---|---|
| CANONICAL | |||||||
| p21 | CDK1 inhibitor | 139GRKRRQTSMTDFYHSKRRLIFS160 | 30 | 0.080 | ITC, XR, NMR | 1AXC | [22] |
| p68 | Polymeraseδ subunit | 451GKANRQVSITGFFQRK466 | 30 | 16 | ITC, XR, NMR | 1U76 | [46] |
| FEN1 | Endonuclease | 331SRQGSTQGRLDDFFKVTG350 | 30 | 59.9 | ITC, NMR | 1U7B | [46] |
| p15PAF | Replication/repair | 50GNPVCVRPTPKWQKGIGEFFRLSPKDSE77 | 25 | 5.56 | ITC, XR, NMR | 4D2G 6GWS | [54] [32] |
| ZRANB3 | Helicase/Endonuclease | 511FTHFEKEKQHDIRSFFVPQPKK532 | 25 | 4.8 | ITC,XR | 5MLO | [83] |
| DVC1 | Adaptor protein | 321SNSHQNVLSNYFPRV336 | 25 | 15.55 | ITC, XR | 5IY4 | [84] |
| DNMT1 | Methyltransferase | 161STRQTTITSHFAKGPAKRKP180 | 25 | 1 | ITC, XR | 6K3A | [85] |
| UHRF2 | E3 ubiquitin ligase | 784NEILQTLLDLFFPGYSK800 | 20 | 25.7 | ITC, XR | 5ICO | [86] |
| Cdt1 | Replication factor | 1MEQRRVTDFFARRR14 | ND | 7.2 | FP, XR | 6QCG | [87] |
| RecQ5 | Helicase | 952KTSPGRSVKEEAQNLIRHFFHGRARCESE980 | 35 | 210 | NMR | - | [61] |
| NON-CANONICAL | |||||||
| pol ı | TLS polymerase | 419CAKKGLIDYYLMPSLST435 | 25 | 0.39 | SPR,XR | 2ZVM | [60] |
| polƞ | TLS polymerase | 694CKRPRREGMQTLESFFKPLTH713 | 25 | 0.4 | SPR,XR | 2ZVK | [60] |
| Pol ĸ | TLS polymerase | 856CIKPNNPKHTLDIFFK870 | 25 | ND | SPR,XR | 2ZVL | [60] |
| ZRANB3 | Helicase/endonuclease | 1058QVRRQSLASKHGSDITRFLVKK1079 | 25 | 9.24 | ITC, XR | 5MLW 5YD8 | [83] [88] |
| PARG | Glycosylase | 402QHGKKDSKITDHFMRLPKA420 | 25 | 3.3 | ITC, XR | 5MAV | [89] |
| p12 | Polymeraseδ subunit | 1MGRKRLITDSYPVVKRREG19 | 25 | 38 | ITC,XR, NMR | 6HVO | [61] |
| Cdt2 | E3 ubiquitin ligase | 704SSMRKICTYFHRKS717 | ND | 0.057 | FP, XR | 6QC0 | [90] |
| TRAIP | E3 ubiquitin ligase | 447KQRVRVKTVPSLFQAKLDTFLWS469 | 25 | 30.7 | ITC, XR | 4ZTD | [87] |
| RNH2B | RNase | 290DKSGMKSIDTFFGVKNKKKIGKV312 | - | - | XR | 3P87 | [91] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Magaña, A.; Blanco, F.J. Human PCNA Structure, Function and Interactions. Biomolecules 2020, 10, 570. https://doi.org/10.3390/biom10040570
González-Magaña A, Blanco FJ. Human PCNA Structure, Function and Interactions. Biomolecules. 2020; 10(4):570. https://doi.org/10.3390/biom10040570
Chicago/Turabian StyleGonzález-Magaña, Amaia, and Francisco J. Blanco. 2020. "Human PCNA Structure, Function and Interactions" Biomolecules 10, no. 4: 570. https://doi.org/10.3390/biom10040570
APA StyleGonzález-Magaña, A., & Blanco, F. J. (2020). Human PCNA Structure, Function and Interactions. Biomolecules, 10(4), 570. https://doi.org/10.3390/biom10040570





