BioShell 3.0: Library for Processing Structural Biology Data
Abstract
1. Introduction
2. Methods
2.1. Command Line Utilities
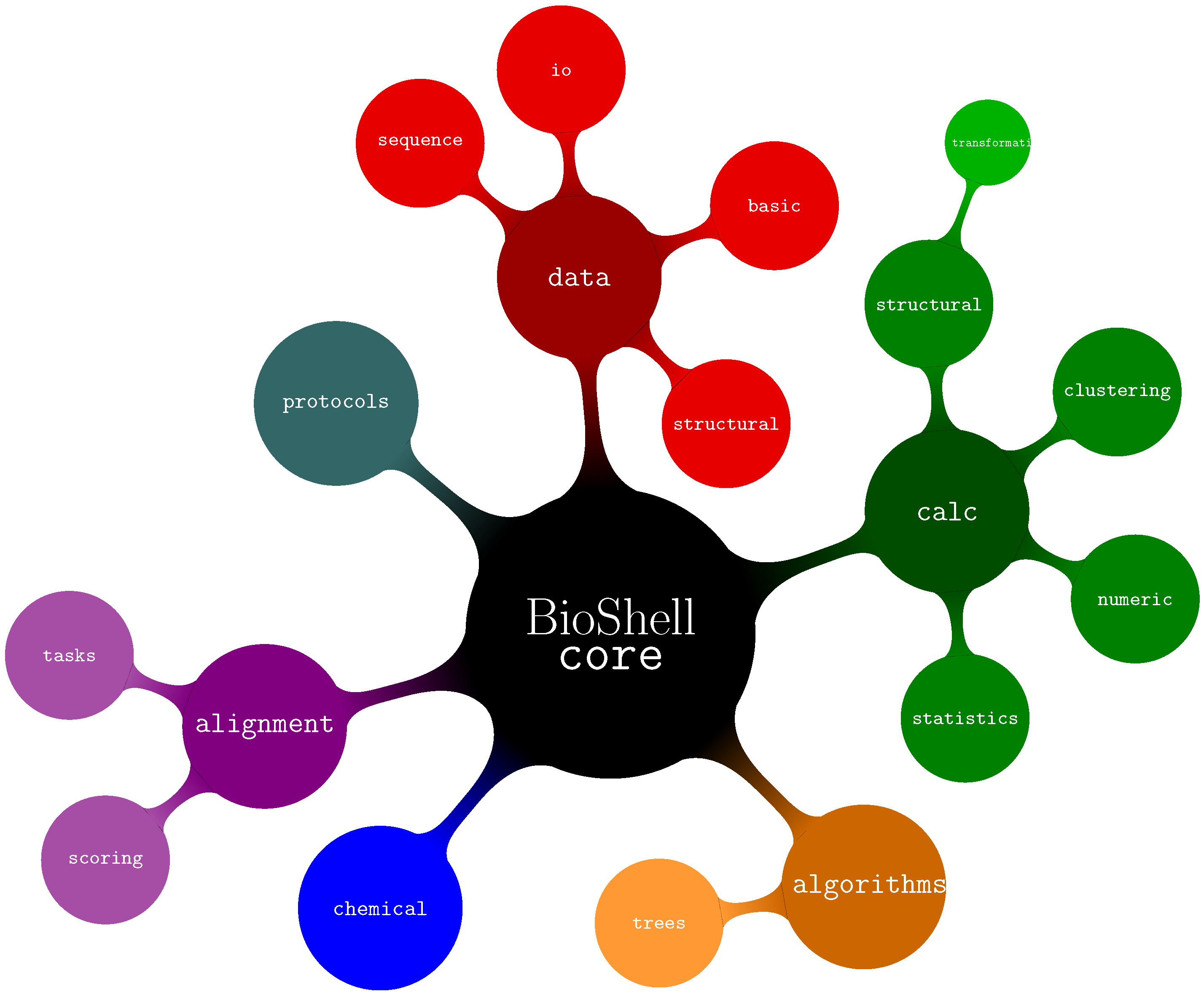
2.2. C++ Software Library
- algorithms—several algorithms used by BioShell such as Union Find, routines to work on trees and graphs.
- alignments—classes related to storing, assessing, and computing alignments between sequences as well as protein structures.
- calc—calculations on biomacromolecular structures (core::calc::structural), data clustering (core::calc::clustering) and generic numerical and statistical routines.
- chemical—classes representing biochemical concepts such as atoms and amino acids
- data—I/O routines core::data::io, data representation of sequences. core::data::sequence and structures core::data::structural, generic data types such as 3D vectors and specialized matrices core::data::basic.
- protocols—classes optimised to perform specific, computationally demanding tasks such as pairwise crmsd calculations. The actual computations are performed by modules from other namespaces (primarily from core::calc::structural). It might be easier for a user to directly employ the latter for small-scale computations. For large scale projects, however, the protocols submodule provides mechanisms for distributing jobs between threads, filtering results and other post- and pre-processing operations.
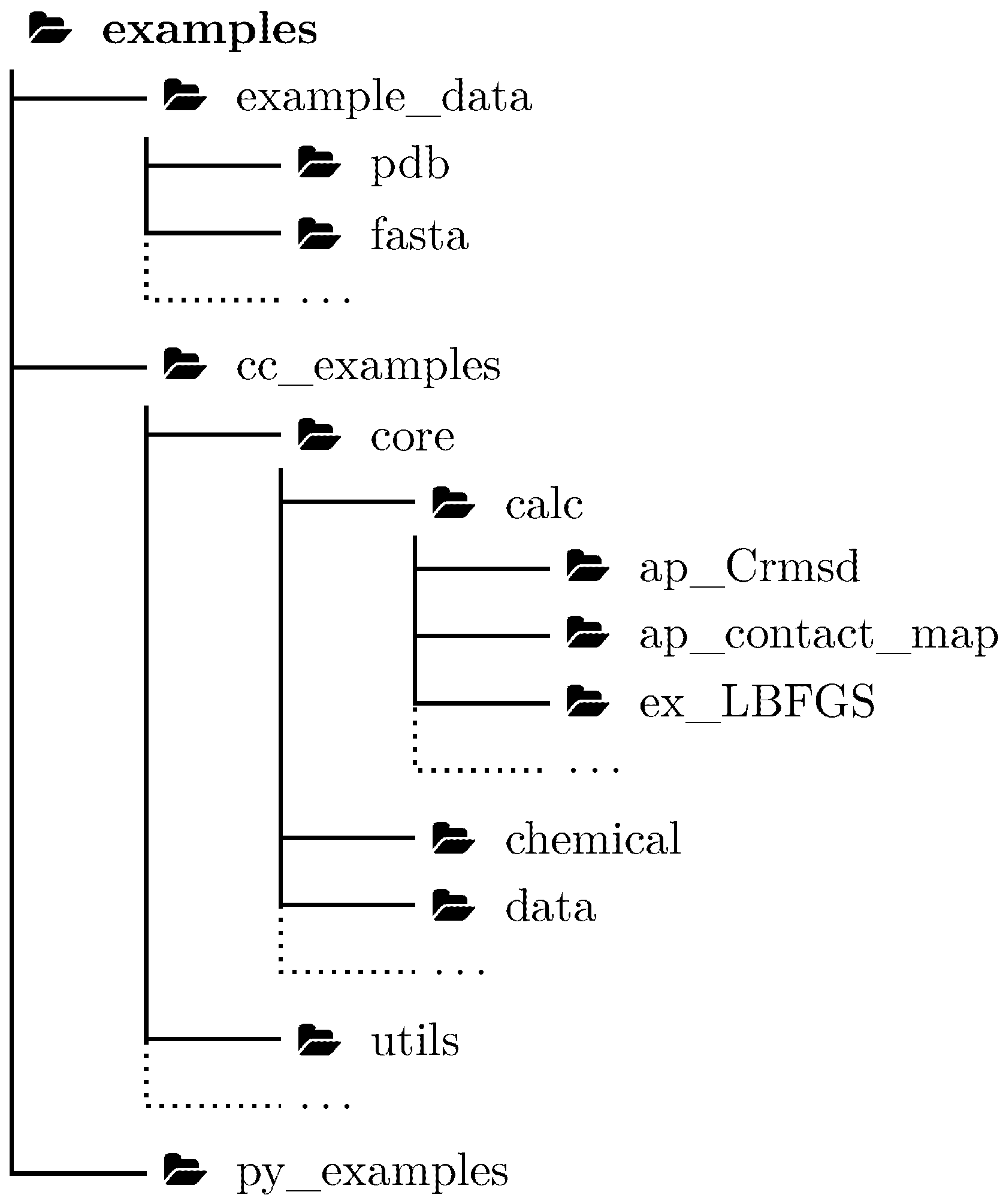
2.3. Python Library
3. Results
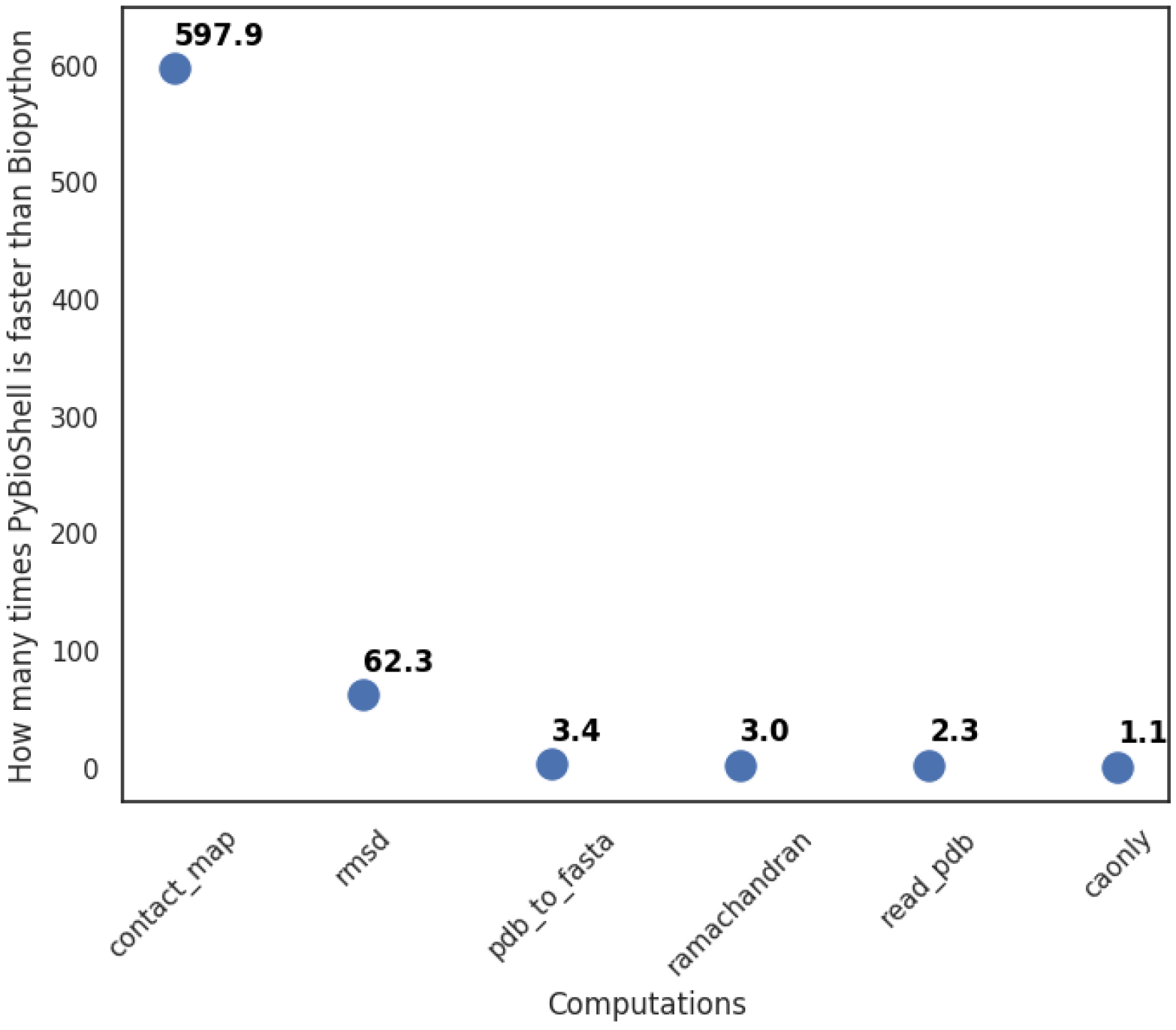
3.1. Improved Performance
3.2. Novel Testing Infrastructure
3.3. Test of Integration and Compatibility between Components
3.4. Unit Tests Serve as Examples for a C++ Library
3.5. Comparison with Biopython
- ca_only_multimodel—reads multiple PDB files with a single model and writes them into one file using only the C-alpha atoms’ coordinates;
- contact_map—checks which residues are within a given distance to each other and returns a list of neighbors with the number of contacts found in a multi-model PDB file;
- pdb_to_fasta—prepares biopolymer sequences in fasta format from a PDB file;
- ramachandran—returns information about phi and psi angles and amino acid type (Glycine, Pre-Proline, Proline or General);
- read_pdb—reads a pdb file;
- rmsd—calculates root mean square deviation between C-alpha atoms of two PDB files.
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stajich, J.E.; Lapp, H. Open source tools and toolkits for bioinformatics: Significance, and where are we? Briefings Bioinform. 2006, 7, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Pocock, M.; Down, T.; Hubbard, T. BioJava: Open source components for bioinformatics. ACM SIGBIO Newsl. 2000, 20, 10–12. [Google Scholar] [CrossRef]
- Holland, R.C.G.; Down, T.A.; Pocock, M.; Prlić, A.; Huen, D.; James, K.; Foisy, S.; Dräger, A.; Yates, A.; Heuer, M.; et al. BioJava: An open-source framework for bioinformatics. Bioinformatics 2008, 24, 2096–2097. [Google Scholar] [CrossRef] [PubMed]
- Chapman, B.; Chang, J. Biopython: Python tools for computational biology. SIGBIO Newsl. 2000, 20, 15–19. [Google Scholar] [CrossRef]
- Hamelryck, T.; Manderick, B. PDB file parser and structure class implemented in Python. Bioinformatics 2003, 19, 2308–2310. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Block, D.; Boulez, K.; Brenner, S.E.; Chervitz, S.A.; Dagdigian, C.; Fuellen, G.; Gilbert, J.G.; Korf, I.; Lapp, H.; et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002, 12, 1611–1618. [Google Scholar] [CrossRef]
- Gront, D.; Kolinski, A. BioShell—A package of tools for structural biology computations. Bioinformatics 2006, 22, 621–622. [Google Scholar] [CrossRef]
- Gront, D.; Kolinski, A. Utility library for structural bioinformatics. Bioinformatics 2008, 24, 584–585. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.; Prins, P.; Nakao, M.; Bonnal, R.; Aerts, J.; Katayama, T. BioRuby: Bioinformatics software for the Ruby programming language. Bioinformatics 2010, 26, 2617–2619. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.D.; Sarkar, A.K.; Lahiri, A. Effect of Inactivating Mutations on Peptide Conformational Ensembles: The Plant Polypeptide Hormone Systemin. J. Chem. Inf. Model. 2016, 56, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, Ó.; Fernández-Martínez, J.L.; Fernández-Brillet, C.; Cernea, A.; Fernández-Muñiz, Z.; Kloczkowski, A. Principal component analysis in protein tertiary structure prediction. J. Bioinf. Comput. Biol. 2018, 16, 1850005. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, Ó.; Fernández-Martínez, J.L.; Corbeanu, A.C.; Fernández-Muñiz, Z.; Kloczkowski, A. Predicting protein tertiary structure and its uncertainty analysis via particle swarm sampling. J. Mol. Model. 2019, 25, 79. [Google Scholar] [CrossRef] [PubMed]
- Geidl, S.; SvobodováVařeková, R.; Bendová, V.; Petrusek, L.; Ionescu, C.M.; Jurka, Z.; Abagyan, R.; Koča, J. How does the methodology of 3D structure preparation influence the quality of pKa prediction? J. Chem. Inf. Model. 2015, 55, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Gront, D.; Hansmann, U.H.E.H.; Kolinski, A. Exploring protein energy landscapes with hierarchical clustering. Int. J. Quantum Chem. 2005, 105, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Gront, D.; Kolinski, A. HCPM—Program for hierarchical clustering of protein models. Bioinformatics 2005, 21, 3179–3180. [Google Scholar] [CrossRef] [PubMed]
- Torres, E. Inadequate Software Testing Can Be Disastrous [Essay]. IEEE Potentials 2018, 37, 9–47. [Google Scholar] [CrossRef]
Sample Availability: The source code is available at: https://bitbucket.org/dgront/bioshell. The website https://bioshell.readthedocs.io contains full reference documentation. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macnar, J.M.; Szulc, N.A.; Kryś, J.D.; Badaczewska-Dawid, A.E.; Gront, D. BioShell 3.0: Library for Processing Structural Biology Data. Biomolecules 2020, 10, 461. https://doi.org/10.3390/biom10030461
Macnar JM, Szulc NA, Kryś JD, Badaczewska-Dawid AE, Gront D. BioShell 3.0: Library for Processing Structural Biology Data. Biomolecules. 2020; 10(3):461. https://doi.org/10.3390/biom10030461
Chicago/Turabian StyleMacnar, Joanna M., Natalia A. Szulc, Justyna D. Kryś, Aleksandra E. Badaczewska-Dawid, and Dominik Gront. 2020. "BioShell 3.0: Library for Processing Structural Biology Data" Biomolecules 10, no. 3: 461. https://doi.org/10.3390/biom10030461
APA StyleMacnar, J. M., Szulc, N. A., Kryś, J. D., Badaczewska-Dawid, A. E., & Gront, D. (2020). BioShell 3.0: Library for Processing Structural Biology Data. Biomolecules, 10(3), 461. https://doi.org/10.3390/biom10030461





