Mutant p53-Associated Molecular Mechanisms of ROS Regulation in Cancer Cells
Abstract
1. Introduction
2. Reactive Oxygen Species: Types and Formation
2.1. Main Antioxidant Enzymes Involved in Tumor Progression
2.2. Main Redox-Related Signaling Pathways and Transcription Factors Involved in Tumor Progression
2.3. Main Non-Enzymatic Antioxidant Systems Involved in Tumor Progression
3. Structure and Function of the Tumor Suppressor p53
P53 Mutations In Cancers: Gain-of-Function
4. Reactive Oxygen Species (ROS)-Related Mechanisms Regulated by Mutant p53
4.1. Regulation of Sestrins (SESN)/AMP-Activated Protein Kinase (AMPK) Axis by Mutant p53
4.2. Regulation of Akt/mTOR Signaling by Mutant p53 Proteins
4.3. Regulation of Glycolytic Metabolism by Mutant p53
4.4. Regulation of Antioxidant Systems or Enzymes by Mutant p53 Proteins
4.5. Regulation of ROS-Related Transcription Factors by Mutant p53 Proteins
4.6. Stimulation of Pro-Inflammatory Cytokines
5. Conclusions
Authors Contributions
Funding
Conflicts of Interest
References
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational Landscape and Significance across 12 Major Cancer Types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Badana, A.K.; Malla, R. Reactive Oxygen Species: A Key Constituent in Cancer Survival. Biomark. Insights 2018, 13, 117727191875539. [Google Scholar] [CrossRef] [PubMed]
- Malkin, D.; Li, F.; Strong, L.; Fraumeni, J.; Nelson, C.; Kim, D.; Kassel, J.; Gryka, M.; Bischoff, F.; Tainsky, M.; et al. Germ Line P53 Mutations in a Familial Syndrome of Breast Cancer, Sarcomas, and Other Neoplasms. Science. 1990, 250, 1233–1238. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of P53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef] [PubMed]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why Are There Hotspot Mutations in the TP53 Gene in Human Cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant P53 as a Guardian of the Cancer Cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef]
- He, C.; Li, L.; Guan, X.; Xiong, L.; Miao, X. Mutant P53 Gain of Function and Chemoresistance: The Role of Mutant P53 in Response to Clinical Chemotherapy. Chemotherapy 2017, 62, 43–53. [Google Scholar] [CrossRef]
- Cordani, M.; Butera, G.; Dando, I.; Torrens-Mas, M.; Butturini, E.; Pacchiana, R.; Oppici, E.; Cavallini, C.; Gasperini, S.; Tamassia, N.; et al. Mutant P53 Blocks SESN1/AMPK/PGC-1α/UCP2 Axis Increasing Mitochondrial O2−· Production in Cancer Cells. Br. J. Cancer 2018, 119, 994–1008. [Google Scholar] [CrossRef]
- Miller, D. Transition Metals as Catalysts of “Autoxidation” Reactions. Free Radic. Biol. Med. 1990, 8, 95–108. [Google Scholar] [CrossRef]
- Murphy, M.P. How Mitochondria Produce Reactive Oxygen Species. Biochem. J. 2009, 417, 1–13. [Google Scholar]
- Naidoo, K.; Birch-Machin, M. Oxidative Stress and Ageing: The Influence of Environmental Pollution, Sunlight and Diet on Skin. Cosmetics 2017, 4, 4. [Google Scholar] [CrossRef]
- Ludin, A.; Gur-Cohen, S.; Golan, K.; Kaufmann, K.B.; Itkin, T.; Medaglia, C.; Lu, X.-J.; Ledergor, G.; Kollet, O.; Lapidot, T. Reactive Oxygen Species Regulate Hematopoietic Stem Cell Self-Renewal, Migration and Development, As Well As Their Bone Marrow Microenvironment. Antioxid. Redox Signal. 2014, 21, 1605–1619. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Motohashi, H. Roles of Nrf2 in Cell Proliferation and Differentiation. Free Radic. Biol. Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Cruzat, V.F.; Keane, K.N.; Carlessi, R.; de Bittencourt, P.I.H. Molecular Mechanisms of ROS Production and Oxidative Stress in Diabetes. Biochem. J. 2016, 473, 4527–4550. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Kamendulis, L.M. The Role of Oxidative Stress in Carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 239–267. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Wang, Z. Oxidative Stress in Carcinogenesis. Curr. Opin. Toxicol. 2018, 7, 116–121. [Google Scholar] [CrossRef]
- Li, Y.R.; Trush, M. Defining ROS in Biology and Medicine. React. Oxyg. Species 2016, 1. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS Signalling in the Biology of Cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative Stress and Antioxidant Defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef]
- Mitra, S.; Nguyen, L.N.; Akter, M.; Park, G.; Choi, E.H.; Kaushik, N.K. Impact of ROS Generated by Chemical, Physical, and Plasma Techniques on Cancer Attenuation. Cancers (Basel). 2019, 11, 1030. [Google Scholar] [CrossRef] [PubMed]
- Donadelli, M.; Costanzo, C.; Beghelli, S.; Scupoli, M.T.; Dandrea, M.; Bonora, A.; Piacentini, P.; Budillon, A.; Caraglia, M.; Scarpa, A.; et al. Synergistic Inhibition of Pancreatic Adenocarcinoma Cell Growth by Trichostatin A and Gemcitabine. Biochim. Biophys. Acta - Mol. Cell Res. 2007, 1773, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide Dismutase Multigene Family: A Comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) Gene Structures, Evolution, and Expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide Dismutases: Dual Roles in Controlling ROS Damage and Regulating ROS Signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; He, T.; Domann, F.E. Decreased Expression of Manganese Superoxide Dismutase in Transformed Cells Is Associated with Increased Cytosine Methylation of the SOD2 Gene. DNA Cell Biol. 1999, 18, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Sarsour, E.H.; Kalen, A.L.; Goswami, P.C. Manganese Superoxide Dismutase Regulates a Redox Cycle Within the Cell Cycle. Antioxid. Redox Signal. 2014, 20, 1618–1627. [Google Scholar] [CrossRef]
- Oberley, L.W.; Oberley, T.D.; Buettner, G.R. Cell Division in Normal and Transformed Cells: The Possible Role of Superoxide and Hydrogen Peroxide. Med. Hypotheses 1981, 7, 21–42. [Google Scholar] [CrossRef]
- Hempel, N.; M Carrico, P.; Melendez, J.A. Manganese Superoxide Dismutase (Sod2) and Redox-Control of Signaling Events That Drive Metastasis. Anticancer. Agents Med. Chem. 2011, 11, 191–201. [Google Scholar] [CrossRef]
- Ren, X.; Zou, L.; Zhang, X.; Branco, V.; Wang, J.; Carvalho, C.; Holmgren, A.; Lu, J. Redox Signaling Mediated by Thioredoxin and Glutathione Systems in the Central Nervous System. Antioxid. Redox Signal. 2017, 27, 989–1010. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of Its Protective Roles, Measurement, and Biosynthesis. Mol. Aspects Med. 2009, 30, 1–12. [Google Scholar] [CrossRef]
- Marí, M.; Morales, A.; Colell, A.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial Glutathione, a Key Survival Antioxidant. Antioxid. Redox Signal. 2009, 11, 2685–2700. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, R. GSH: A Double-Edged Sword. React. Oxyg. Species 2017, 3, 121–126. [Google Scholar] [CrossRef]
- Tew, K.D. Glutathione-Associated Enzymes in Anticancer Drug Resistance. Cancer Res. 1994, 54, 4313–4320. [Google Scholar] [CrossRef] [PubMed]
- Afanas’ev, I. Mechanisms of Superoxide Signaling in Epigenetic Processes: Relation to Aging and Cancer. Aging Dis. 2015, 6, 216. [Google Scholar] [CrossRef]
- Haddad, J.J. Antioxidant and Prooxidant Mechanisms in the Regulation of Redox(y)-Sensitive Transcription Factors. Cell. Signal. 2002, 14, 879–897. [Google Scholar] [CrossRef]
- England, K.; Cotter, T.G. Direct Oxidative Modifications of Signalling Proteins in Mammalian Cells and Their Effects on Apoptosis. Redox Rep. 2005, 10, 237–245. [Google Scholar] [CrossRef]
- Los, M.; Maddika, S.; Erb, B.; Schulze-Osthoff, K. Switching Akt: From Survival Signaling to Deadly Response. BioEssays 2009, 31, 492–495. [Google Scholar] [CrossRef]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS Signaling under Metabolic Stress: Cross-Talk between AMPK and AKT Pathway. Mol. Cancer 2017, 16, 79. [Google Scholar] [CrossRef]
- Emerling, B.M.; Weinberg, F.; Snyder, C.; Burgess, Z.; Mutlu, G.M.; Viollet, B.; Budinger, G.R.S.; Chandel, N.S. Hypoxic Activation of AMPK Is Dependent on Mitochondrial ROS but Independent of an Increase in AMP/ATP Ratio. Free Radic. Biol. Med. 2009, 46, 1386–1391. [Google Scholar] [CrossRef]
- Martínez-Sánchez, G.; Giuliani, A. Cellular Redox Status Regulates Hypoxia Inducible Factor-1 Activity. Role in Tumour Development. J. Exp. Clin. Cancer Res. 2007, 26, 39–50. [Google Scholar] [PubMed]
- Kissil, J.L.; Walmsley, M.J.; Hanlon, L.; Haigis, K.M.; Bender Kim, C.F.; Sweet-Cordero, A.; Eckman, M.S.; Tuveson, D.A.; Capobianco, A.J.; Tybulewicz, V.L.J.; et al. Requirement for Rac1 in a K-Ras–Induced Lung Cancer in the Mouse. Cancer Res. 2007, 67, 8089–8094. [Google Scholar] [CrossRef] [PubMed]
- Dando, I.; Fiorini, C.; Pozza, E.D.; Padroni, C.; Costanzo, C.; Palmieri, M.; Donadelli, M. UCP2 Inhibition Triggers ROS-Dependent Nuclear Translocation of GAPDH and Autophagic Cell Death in Pancreatic Adenocarcinoma Cells. Biochim. Biophys. Acta - Mol. Cell Res. 2013, 1833, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 Interface Provides Mechanistic Insight into Nrf2 Signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [PubMed]
- Cordani, M.; Sánchez-Álvarez, M.; Strippoli, R.; Bazhin, A.V.; Donadelli, M. Sestrins at the Interface of ROS Control and Autophagy Regulation in Health and Disease. Oxid. Med. Cell. Longev. 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Sánchez-Álvarez, M.; Strippoli, R.; Donadelli, M.; Bazhin, A.V.; Cordani, M. Sestrins as a Therapeutic Bridge between ROS and Autophagy in Cancer. Cancers (Basel). 2019, 11, 1415. [Google Scholar] [CrossRef]
- Bae, S.H.; Sung, S.H.; Oh, S.Y.; Lim, J.M.; Lee, S.K.; Park, Y.N.; Lee, H.E.; Kang, D.; Rhee, S.G. Sestrins Activate Nrf2 by Promoting P62-Dependent Autophagic Degradation of Keap1 and Prevent Oxidative Liver Damage. Cell Metab. 2013, 17, 73–84. [Google Scholar] [CrossRef]
- Eid, A.A.; Lee, D.-Y.; Roman, L.J.; Khazim, K.; Gorin, Y. Sestrin 2 and AMPK Connect Hyperglycemia to Nox4-Dependent Endothelial Nitric Oxide Synthase Uncoupling and Matrix Protein Expression. Mol. Cell. Biol. 2013, 33, 3439–3460. [Google Scholar] [CrossRef]
- Eid, A.A.; Ford, B.M.; Block, K.; Kasinath, B.S.; Gorin, Y.; Ghosh-Choudhury, G.; Barnes, J.L.; Abboud, H.E. AMP-Activated Protein Kinase (AMPK) Negatively Regulates Nox4-Dependent Activation of P53 and Epithelial Cell Apoptosis in Diabetes. J. Biol. Chem. 2010, 285, 37503–37512. [Google Scholar] [CrossRef]
- Lee, J.H.; Budanov, A.V.; Park, E.J.; Birse, R.; Kim, T.E.; Perkins, G.A.; Ocorr, K.; Ellisman, M.H.; Bodmer, R.; Bier, E.; et al. Sestrin as a Feedback Inhibitor of TOR That Prevents Age-Related Pathologies. Science 2010, 327, 1223–1228. [Google Scholar] [CrossRef]
- Donadelli, M.; Dando, I.; Fiorini, C.; Palmieri, M. UCP2, a Mitochondrial Protein Regulated at Multiple Levels. Cell. Mol. Life Sci. 2014, 71, 1171–1190. [Google Scholar] [CrossRef]
- Donadelli, M.; Dando, I.; Pozza, E.D.; Palmieri, M. Mitochondrial Uncoupling Protein 2 and Pancreatic Cancer: A New Potential Target Therapy. World J. Gastroenterol. 2015, 21, 3232–3238. [Google Scholar] [CrossRef] [PubMed]
- Cordani, M.; Butera, G.; Pacchiana, R.; Donadelli, M. The Antioxidant Mitochondrial Protein UCP2 Promotes Cancer Development Connecting the Warburg Effect and Autophagy. Transl. Med. Reports 2017, 1, 6451. [Google Scholar] [CrossRef]
- Xu, Y.; Miriyala, S.; Fang, F.; Bakthavatchalu, V.; Noel, T.; Schell, D.M.; Wang, C.; St Clair, W.H.; St Clair, D.K. Manganese Superoxide Dismutase Deficiency Triggers Mitochondrial Uncoupling and the Warburg Effect. Oncogene 2015, 36, 4087. [Google Scholar] [CrossRef]
- Nowinski, S.M.; Solmonson, A.; Rundhaug, J.E.; Rho, O.; Cho, J.; Lago, C.U.; Riley, C.L.; Lee, S.; Kohno, S.; Dao, C.K.; et al. Mitochondrial Uncoupling Links Lipid Catabolism to Akt Inhibition and Resistance to Tumorigenesis. Nat. Commun. 2015, 6, 8137. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.; Bing, C.; McCulloch, P.; Williams, G. Muscle UCP-3 MRNA Levels Are Elevated in Weight Loss Associated with Gastrointestinal Adenocarcinoma in Humans. Br. J. Cancer 2002, 86, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.; Klumpp, D.; Hennenlotter, J.; Bedke, J.; Duranton, C.; Bleif, M.; Huber, S.M. UCP-3 Uncoupling Protein Confers Hypoxia Resistance to Renal Epithelial Cells and Is Upregulated in Renal Cell Carcinoma. Sci. Rep. 2015, 5, 13450. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Balaska, K.; Kalamida, D.; Kakouratos, C.; Sivridis, E.; Koukourakis, M.I. Thermogenic Protein UCP1 and UCP3 Expression in Nonsmall Cell Lung Cancer: Relation with Glycolysis and Anaerobic Metabolism. Cancer Biol. Med. 2017, 14, 396–404. [Google Scholar]
- Tanaka, T.; Yamashita, K.; Watanabe, M. Potential Therapeutic Targets of TP53 Gene in the Context of Its Classically Canonical Functions and Its Latest Non-Canonical Functions in Human Cancer. Oncotarget 2018, 9, 16234–16247. [Google Scholar] [CrossRef]
- Ma, B.; Pan, Y.; Zheng, J.; Levine, A.J.; Nussinov, R. Sequence Analysis of P53 Response-Elements Suggests Multiple Binding Modes of the P53 Tetramer to DNA Targets. Nucleic Acids Res. 2007, 35, 2986–3001. [Google Scholar] [CrossRef]
- Lane, D.P. P53, Guardian of the Genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Inga, A.; Storici, F.; Darden, T.A.; Resnick, M.A. Differential Transactivation by the P53 Transcription Factor Is Highly Dependent on P53 Level and Promoter Target Sequence. Mol. Cell. Biol. 2002, 22, 8612–8625. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.F.; Lal, A. MicroRNAs, Wild-Type and Mutant P53: More Questions than Answers. RNA Biol. 2012, 9, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Sablina, A.A.; Budanov, A.V.; Ilyinskaya, G.V.; Agapova, L.S.; Kravchenko, J.E.; Chumakov, P.M. The Antioxidant Function of the P53 Tumor Suppressor. Nat. Med. 2005, 11, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.M.; Bunz, F.; Yu, J.; Rago, C.; Chan, T.A.; Murphy, M.P.; Kelso, G.F.; Smith, R.A.J.; Kinzler, K.W.; Vogelstein, B. Ferredoxin Reductase Affects P53-Dependent, 5-Fluorouracil-Induced Apoptosis in Colorectal Cancer Cells. Nat. Med. 2001, 7, 1111–1117. [Google Scholar] [CrossRef] [PubMed]
- Ueno, M.; Masutani, H.; Arai, R.J.; Yamauchi, A.; Hirota, K.; Sakai, T.; Inamoto, T.; Yamaoka, Y.; Yodoi, J.; Nikaido, T. Thioredoxin-Dependent Redox Regulation of P53-Mediated P21 Activation. J. Biol. Chem. 1999, 274, 35809–35815. [Google Scholar] [CrossRef]
- Erster, S.; Mihara, M.; Kim, R.H.; Petrenko, O.; Moll, U.M. In Vivo Mitochondrial P53 Translocation Triggers a Rapid First Wave of Cell Death in Response to DNA Damage That Can Precede P53 Target Gene Activation. Mol. Cell. Biol. 2004, 24, 6728–6734. [Google Scholar] [CrossRef]
- Zhao, Y.; Chaiswing, L.; Velez, J.M.; Batinic-Haberle, I.; Colburn, N.H.; Oberley, T.D.; St., Clair, D.K. P53 Translocation to Mitochondria Precedes Its Nuclear Translocation and Targets Mitochondrial Oxidative Defense Protein-Manganese Superoxide Dismutase. Cancer Res. 2005, 65, 3745–3750. [Google Scholar] [CrossRef]
- Shi, D.; Gu, W. Dual Roles of MDM2 in the Regulation of P53: Ubiquitination Dependent and Ubiquitination Independent Mechanisms of MDM2 Repression of P53 Activity. Genes Cancer 2012, 3, 240–248. [Google Scholar] [CrossRef]
- Sparks, A.; Dayal, S.; Das, J.; Robertson, P.; Menendez, S.; Saville, M.K. The Degradation of P53 and Its Major E3 Ligase Mdm2 Is Differentially Dependent on the Proteasomal Ubiquitin Receptor S5a. Oncogene 2014, 33, 4685–4696. [Google Scholar] [CrossRef]
- Lavin, M.F.; Gueven, N. The Complexity of P53 Stabilization and Activation. Cell Death Differ. 2006, 13, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Bragado, P.; Armesilla, A.; Silva, A.; Porras, A. Apoptosis by Cisplatin Requires P53 Mediated P38α MAPK Activation through ROS Generation. Apoptosis 2007, 12, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Kurz, E.U.; Lees-Miller, S.P. DNA Damage-Induced Activation of ATM and ATM-Dependent Signaling Pathways. DNA Repair. 2004, 3, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Persons, D.L.; Yazlovitskaya, E.M.; Pelling, J.C. Effect of Extracellular Signal-Regulated Kinase on P53 Accumulation in Response to Cisplatin. J. Biol. Chem. 2000, 275, 35778–35785. [Google Scholar] [CrossRef] [PubMed]
- Buzek, J. Redox State of Tumor Suppressor P53 Regulates Its Sequence-Specific DNA Binding in DNA-Damaged Cells by Cysteine 277. Nucleic Acids Res. 2002, 30, 2340–2348. [Google Scholar] [CrossRef] [PubMed]
- Pfister, N.T.; Prives, C. Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of P53. Cold Spring Harb. Perspect. Med. 2017, 7, a026054. [Google Scholar] [CrossRef] [PubMed]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the P53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef]
- Yue, X.; Zhao, Y.; Xu, Y.; Zheng, M.; Feng, Z.; Hu, W. Mutant P53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J. Mol. Biol. 2017, 429, 1595–1606. [Google Scholar] [CrossRef]
- Dittmer, D.; Pati, S.; Zambetti, G.; Chu, S.; Teresky, A.K.; Moore, M.; Finlay, C.; Levine, A.J. Gain of Function Mutations in P53. Nat. Genet. 1993, 4, 42–46. [Google Scholar] [CrossRef]
- Blandino, G.; Di Agostino, S. New Therapeutic Strategies to Treat Human Cancers Expressing Mutant P53 Proteins. J. Exp. Clin. Cancer Res. 2018, 37, 30. [Google Scholar] [CrossRef]
- Muller, P.A.J.; Vousden, K.H. Mutant P53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell. 2014, 25, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Budanov, A.V.; Karin, M. Sestrins Orchestrate Cellular Metabolism to Attenuate Aging. Cell Metabolism. 2013, 18, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, R.C.; Samborska, B.; Faubert, B.; Ma, E.H.; Gravel, S.P.; Andrzejewski, S.; Raissi, T.C.; Pause, A.; St., Pierre, J.; Jones, R.G. AMPK Maintains Cellular Metabolic Homeostasis through Regulation of Mitochondrial Reactive Oxygen Species. Cell Rep. 2017, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Budanov, A.V.; Karin, M. P53 Target Genes Sestrin1 and Sestrin2 Connect Genotoxic Stress and MTOR Signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Sanli, T.; Linher-Melville, K.; Tsakiridis, T.; Singh, G. Sestrin2 Modulates AMPK Subunit Expression and Its Response to Ionizing Radiation in Breast Cancer Cells. PLoS ONE 2012, 7, e32035. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, B.; Li, T.; Zhu, Y.; Luo, G.; Jiang, Y.; Tang, F.; Jian, Z.; Xiao, Y. AMPK Activation Serves a Critical Role in Mitochondria Quality Control via Modulating Mitophagy in the Heart under Chronic Hypoxia. Int. J. Mol. Med. 2018, 41, 69–76. [Google Scholar] [CrossRef]
- Tian, W.; Li, W.; Chen, Y.; Yan, Z.; Huang, X.; Zhuang, H.; Zhong, W.; Chen, Y.; Wu, W.; Lin, C.; et al. Phosphorylation of ULK1 by AMPK Regulates Translocation of ULK1 to Mitochondria and Mitophagy. FEBS Lett. 2015, 589, 1847–1854. [Google Scholar] [CrossRef]
- Joo, M.S.; Kim, W.D.; Lee, K.Y.; Kim, J.H.; Koo, J.H.; Kim, S.G. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol. Cell. Biol. 2016, 36, 1931–1942. [Google Scholar] [CrossRef]
- Cordani, M.; Oppici, E.; Dando, I.; Butturini, E.; Dalla Pozza, E.; Nadal-Serrano, M.; Oliver, J.; Roca, P.; Mariotto, S.; Cellini, B.; et al. Mutant P53 Proteins Counteract Autophagic Mechanism Sensitizing Cancer Cells to MTOR Inhibition. Mol. Oncol. 2016, 10, 1008–1029. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. MTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Yip, C.K.; Murata, K.; Walz, T.; Sabatini, D.M.; Kang, S.A. Structure of the Human MTOR Complex I and Its Implications for Rapamycin Inhibition. Mol. Cell 2010, 38, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Stuttfeld, E.; Aylett, C.H.S.; Imseng, S.; Boehringer, D.; Scaiola, A.; Sauer, E.; Hall, M.N.; Maier, T.; Ban, N. Architecture of the Human MTORC2 Core Complex. Elife 2018, 7, e33101. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sahra, I.; Hoxhaj, G.; Ricoult, S.J.H.; Asara, J.M.; Manning, B.D. MTORC1 Induces Purine Synthesis through Control of the Mitochondrial Tetrahydrofolate Cycle. Science 2016, 351, 728–733. [Google Scholar] [CrossRef]
- Selman, C.; Tullet, J.M.A.; Wieser, D.; Irvine, E.; Lingard, S.J.; Choudhury, A.I.; Claret, M.; Al-Qassab, H.; Carmignac, D.; Ramadani, F.; et al. Ribosomal Protein S6 Kinase 1 Signaling Regulates Mammalian Life Span. Science. 2009, 326, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Summer, R.; Shaghaghi, H.; Schriner, D.; Roque, W.; Sales, D.; Cuevas-Mora, K.; Desai, V.; Bhushan, A.; Ramirez, M.I.; Romero, F. Activation of the MTORC1/PGC-1 Axis Promotes Mitochondrial Biogenesis and Induces Cellular Senescence in the Lung Epithelium. Am. J. Physiol. Cell. Mol. Physiol. 2019, 316, L1049–L1060. [Google Scholar] [CrossRef]
- Morita, M.; Gravel, S.P.; Chénard, V.; Sikström, K.; Zheng, L.; Alain, T.; Gandin, V.; Avizonis, D.; Arguello, M.; Zakaria, C.; et al. MTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell Metab. 2013, 18, 698–711. [Google Scholar] [CrossRef]
- Morita, M.; Gravel, S.P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. MTOR Coordinates Protein Synthesis, Mitochondrial Activity. Cell Cycle. 2015, 14, 473–480. [Google Scholar] [CrossRef]
- Ozcan, U.; Ozcan, L.; Yilmaz, E.; Düvel, K.; Sahin, M.; Manning, B.D.; Hotamisligil, G.S. Loss of the Tuberous Sclerosis Complex Tumor Suppressors Triggers the Unfolded Protein Response to Regulate Insulin Signaling and Apoptosis. Mol. Cell 2008, 29, 541–551. [Google Scholar] [CrossRef]
- Young, R.M.; Ackerman, D.; Quinn, Z.L.; Mancuso, A.; Gruber, M.; Liu, L.; Giannoukos, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Keith, B.; et al. Dysregulated MTORC1 Renders Cells Critically Dependent on Desaturated Lipids for Survival under Tumor-like Stress. Genes Dev. 2013, 27, 1115–1131. [Google Scholar] [CrossRef]
- Santos, C.X.C.; Tanaka, L.Y.; Wosniak, J.; Laurindo, F.R.M. Mechanisms and Implications of Reactive Oxygen Species Generation during the Unfolded Protein Response: Roles of Endoplasmic Reticulum Oxidoreductases, Mitochondrial Electron Transport, and NADPH Oxidase. Antioxidants & redox signaling. 2009, 11, 2409–2427. [Google Scholar]
- Kritsiligkou, P.; Rand, J.D.; Weids, A.J.; Wang, X.; Kershaw, C.J.; Grant, C.M. Endoplasmic Reticulum (ER) Stress–Induced Reactive Oxygen Species (ROS) Are Detrimental for the Fitness of a Thioredoxin Reductase Mutant. J. Biol. Chem. 2018, 293, 11984–11995. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, H.M.A.; Lee, G.H.; Kim, H.R.; Chae, H.J. Endoplasmic Reticulum Stress and Associated ROS. International Journal of Molecular Sciences. 2016, 17, 327. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.A.; Alvarez, M.S.; Sailem, H.; Bousgouni, V.; Sero, J.; Bakal, C. Differential RNAi Screening Provides Insights into the Rewiring of Signalling Networks during Oxidative Stress. Mol. Biosyst. 2012, 8, 2605–2613. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Bell, C.M.; Taylor, S.M.; Moran, R.G. P53 Deletion or Hotspot Mutations Enhance MTORC1 Activity by Altering Lysosomal Dynamics of TSC2 and Rheb. Mol. Cancer Res. 2016, 14, 66–77. [Google Scholar] [CrossRef]
- Chatterjee, S.; Browning, E.A.; Hong, N.; DeBolt, K.; Sorokina, E.M.; Liu, W.; Birnbaum, M.J.; Fisher, A.B. Membrane Depolarization Is the Trigger for PI3K/Akt Activation and Leads to the Generation of ROS. Am. J. Physiol. Circ. Physiol. 2012, 302, H105–H114. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Phosphoinositide 3-Kinase/Akt Signaling and Redox Metabolism in Cancer. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef]
- Li, C.; Li, Y.; He, L.; Agarwal, A.R.; Zeng, N.; Cadenas, E.; Stiles, B.L. PI3K/AKT Signaling Regulates Bioenergetics in Immortalized Hepatocytes. Free Radic. Biol. Med. 2013, 60, 29–40. [Google Scholar] [CrossRef]
- Tan, B.S.; Tiong, K.H.; Choo, H.L.; Fei-Lei Chung, F.; Hii, L.W.; Tan, S.H.; Yap, I.K.S.; Pani, S.; Khor, N.T.W.; Wong, S.F.; et al. Mutant P53-R273H Mediates Cancer Cell Survival and Anoikis Resistance through AKT-Dependent Suppression of BCL2-Modifying Factor (BMF). Cell Death Dis. 2015, 6, e1826. [Google Scholar] [CrossRef]
- Guo, F.; Chen, H.; Chang, J.; Zhang, L. Mutation R273H Confers P53 a Stimulating Effect on the IGF-1R-AKT Pathway via MiR-30a Suppression in Breast Cancer. Biomed. Pharmacother. 2016, 78, 335–341. [Google Scholar] [CrossRef]
- Butera, G.; Pacchiana, R.; Mullappilly, N.; Margiotta, M.; Bruno, S.; Conti, P.; Riganti, C.; Donadelli, M. Mutant P53 Prevents GAPDH Nuclear Translocation in Pancreatic Cancer Cells Favoring Glycolysis and 2-Deoxyglucose Sensitivity. Biochim. Biophys. Acta - Mol. Cell Res. 2018, 1865, 1914–1923. [Google Scholar] [CrossRef]
- Adorno, M.; Cordenonsi, M.; Montagner, M.; Dupont, S.; Wong, C.; Hann, B.; Solari, A.; Bobisse, S.; Rondina, M.B.; Guzzardo, V.; et al. A Mutant-P53/Smad Complex Opposes P63 to Empower TGFβ-Induced Metastasis. Cell 2009, 137, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Sauer, L.; Gitenay, D.; Vo, C.; Baron, V.T. Mutant P53 Initiates a Feedback Loop That Involves Egr-1/EGF Receptor/ERK in Prostate Cancer Cells. Oncogene 2010, 29, 2628–2637. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, B.; Miao, L.; Me, Y.; Wu, M. Mutant P53-R273H Gains New Function in Sustained Activation of EGFR Signaling via Suppressing MiR-27a Expression. Cell Death Dis. 2013, 4, e574. [Google Scholar] [CrossRef] [PubMed]
- Grugan, K.D.; Vega, M.E.; Wong, G.S.; Diehl, J.A.; Bass, A.J.; Wong, K.K.; Nakagawa, H.; Rustgi, A.K. A Common P53 Mutation (R175H) Activates c-Met Receptor Tyrosine Kinase to Enhance Tumor Cell Invasion. Cancer Biol. Ther. 2013, 14, 853–859. [Google Scholar] [CrossRef]
- Muller, P.A.J.; Trinidad, A.G.; Timpson, P.; Morton, J.P.; Zanivan, S.; Van Den Berghe, P.V.E.; Nixon, C.; Karim, S.A.; Caswell, P.T.; Noll, J.E.; et al. Mutant P53 Enhances MET Trafficking and Signalling to Drive Cell Scattering and Invasion. Oncogene 2013, 32, 1252–1265. [Google Scholar] [CrossRef]
- Fadaka, A.; Ajiboye, B.; Ojo, O.; Adewale, O.; Olayide, I.; Emuowhochere, R. Biology of Glucose Metabolization in Cancer Cells. J. Oncol. Sci. 2017, 3, 45–51. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Liang, Y.; Wu, R.; Zhao, Y.; Hong, X.; Lin, M.; Yu, H.; Liu, L.; Levine, A.J.; et al. Tumour-Associated Mutant P53 Drives the Warburg Effect. Nat. Commun. 2013, 4, 2935. [Google Scholar] [CrossRef]
- Hernández-Reséndiz, I.; Gallardo-Pérez, J.C.; López-Macay, A.; Robledo-Cadena, D.X.; García-Villa, E.; Gariglio, P.; Saavedra, E.; Moreno-Sánchez, R.; Rodríguez-Enríquez, S. Mutant P53 R248Q Downregulates Oxidative Phosphorylation and Upregulates Glycolysis under Normoxia and Hypoxia in Human Cervix Cancer Cells. J. Cell. Physiol. 2019, 234, 5524–5536. [Google Scholar] [CrossRef]
- Faubert, B.; Boily, G.; Izreig, S.; Griss, T.; Samborska, B.; Dong, Z.; Dupuy, F.; Chambers, C.; Fuerth, B.J.; Viollet, B.; et al. AMPK Is a Negative Regulator of the Warburg Effect and Suppresses Tumor Growth in Vivo. Cell Metab. 2013, 17, 113–124. [Google Scholar] [CrossRef]
- Kamat, C.D.; Green, D.E.; Warnke, L.; Thorpe, J.E.; Ceriello, A.; Ihnat, M.A. Mutant P53 Facilitates Pro-Angiogenic, Hyperproliferative Phenotype in Response to Chronic Relative Hypoxia. Cancer Lett. 2007, 249, 209–219. [Google Scholar] [CrossRef]
- Dando, I.; Cordani, M.; Donadelli, M. Mutant P53 and MTOR/PKM2 Regulation in Cancer Cells. IUBMB Life 2016, 68, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Chen, X.; Ma, J.; Peng, H.; Wang, F.; Zha, X.; Wang, Y.; Jing, Y.; Yang, H.; Chen, R.; et al. Mammalian Target of Rapamycin Up-Regulation of Pyruvate Kinase Isoenzyme Type M2 Is Critical for Aerobic Glycolysis and Tumor Growth. Proc. Natl. Acad. Sci. USA 2011, 108, 4129–4134. [Google Scholar] [CrossRef] [PubMed]
- Desagher, S.; Glowinski, J.; Prémont, J. Pyruvate Protects Neurons against Hydrogen Peroxide-Induced Toxicity. J. Neurosci. 1997, 17, 9060–9067. [Google Scholar] [CrossRef] [PubMed]
- Puntel, R.L.; Roos, D.H.; Grotto, D.; Garcia, S.C.; Nogueira, C.W.; Batista Teixeira Rocha, J. Antioxidant Properties of Krebs Cycle Intermediates against Malonate Pro-Oxidant Activity in Vitro: A Comparative Study Using the Colorimetric Method and HPLC Analysis to Determine Malondialdehyde in Rat Brain Homogenates. Life Sci. 2007, 81, 51–62. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, S.M.; Mahmoud, A.A.; El Sawy, S.A.; Abdelaal, E.A.; Fouad, A.M.; Yousif, R.S.; Hashim, M.S.; Hemdan, S.B.; Kadry, Z.M.; Abdelmoaty, M.A.; et al. Warburg Effect Increases Steady-State ROS Condition in Cancer Cells through Decreasing Their Antioxidant Capacities (Anticancer Effects of 3-Bromopyruvate through Antagonizing Warburg Effect). Med. Hypotheses 2013, 81, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Liemburg-Apers, D.C.; Willems, P.H.; Koopman, W.J.H.; Grefte, S. Interactions between Mitochondrial Reactive Oxygen Species and Cellular Glucose Metabolism. Arch. Toxicol. 2015, 89, 1209–1226. [Google Scholar] [CrossRef] [PubMed]
- Kozlovsky, N.; Rudich, A.; Potashnik, R.; Ebina, Y.; Murakami, T.; Bashan, N. Transcriptional Activation of the Glut1 Gene in Response to Oxidative Stress in L6 Myotubes. J. Biol. Chem. 1997, 272, 33367–33372. [Google Scholar] [CrossRef]
- Milićević, Z.; Kasapović, J.; Gavrilović, L.; Milovanović, Z.; Bajić, V.; Spremo-Potparević, B. Mutant P53 Protein Expression and Antioxidant Status Deficiency in Breast Cancer. EXCLI J. 2014, 13, 691–708. [Google Scholar]
- Nisimoto, Y.; Diebold, B.A.; Constentino-Gomes, D.; Lambeth, J.D. Nox4: A Hydrogen Peroxide-Generating Oxygen Sensor. Biochemistry 2014, 53, 5111–5120. [Google Scholar] [CrossRef]
- Boudreau, H.E.; Casterline, B.W.; Burke, D.J.; Leto, T.L. Wild-Type and Mutant P53 Differentially Regulate NADPH Oxidase 4 in TGF-β-Mediated Migration of Human Lung and Breast Epithelial Cells. Br. J. Cancer 2014, 110, 2569–2582. [Google Scholar] [CrossRef]
- Torrens-Mas, M.; Cordani, M.; Mullappilly, N.; Pacchiana, R.; Riganti, C.; Palmieri, M.; Pons, D.G.; Roca, P.; Oliver, J.; Donadelli, M. Mutant P53 Induces SIRT3/MnSOD Axis to Moderate ROS Production in Melanoma Cells. Arch. Biochem. Biophys. 2020, 679, 108219. [Google Scholar] [CrossRef] [PubMed]
- Weisz, L.; Oren, M.; Rotter, V. Transcription Regulation by Mutant P53. Oncogene 2007, 26, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Kalo, E.; Kogan-Sakin, I.; Solomon, H.; Bar-Nathan, E.; Shay, M.; Shetzer, Y.; Dekel, E.; Goldfinger, N.; Buganim, Y.; Stambolsky, P.; et al. Mutant P53R273H Attenuates the Expression of Phase 2 Detoxifying Enzymes and Promotes the Survival of Cells with High Levels of Reactive Oxygen Species. J. Cell Sci. 2012, 125, 5578–5586. [Google Scholar] [CrossRef] [PubMed]
- Lisek, K.; Campaner, E.; Ciani, Y.; Walerych, D.; Del, G. Mutant P53 Tunes the NRF2-Dependent Antioxidant Response to Support Survival of Cancer Cells. Oncotarget 2018, 9, 20508–20523. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.S.; Duong, C.P.; Haupt, S.; Montgomery, K.G.; House, C.M.; Azar, W.J.; Pearson, H.B.; Fisher, O.M.; Read, M.; Guerra, G.R.; et al. Inhibiting the System XC−/Glutathione Axis Selectively Targets Cancers with Mutant-P53 Accumulation. Nat. Commun. 2017, 8, 14844. [Google Scholar] [CrossRef]
- Handschin, C.; Spiegelman, B.M. Peroxisome Proliferator-Activated Receptor γ Coactivator 1 Coactivators, Energy Homeostasis, and Metabolism. Endocr. Rev. 2006, 27, 728–735. [Google Scholar] [CrossRef]
- Basu, S.; Gnanapradeepan, K.; Barnoud, T.; Kung, C.P.; Tavecchio, M.; Scott, J.; Watters, A.; Chen, Q.; Kossenkov, A.V.; Murphy, M.E. Mutant P53 Controls Tumor Metabolism and Metastasis by Regulating PGC-1α. Genes Dev. 2018, 32, 230–243. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Crusz, S.M.; Balkwill, F.R. Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 2015, 12, 584–596. [Google Scholar] [CrossRef]
- Vandercappellen, J.; Van Damme, J.; Struyf, S. The Role of CXC Chemokines and Their Receptors in Cancer. Cancer Letters 2008, 267, 226–244. [Google Scholar] [CrossRef]
- Schulze-Osthoff, K.; Beyaert, R.; Vandevoorde, V.; Haegeman, G.; Fiers, W. Depletion of the Mitochondrial Electron Transport Abrogates the Cytotoxic and Gene-Inductive Effects of TNF. EMBO J. 1993, 12, 3095–3104. [Google Scholar] [CrossRef] [PubMed]
- Corda, S.; Laplace, C.; Vicaut, E.; Duranteau, J. Rapid Reactive Oxygen Species Production by Mitochondria in Endothelial Cells Exposed to Tumor Necrosis Factor-α Is Mediated by Ceramide. Am. J. Respir. Cell Mol. Biol. 2001, 24, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.F.; Caramona, M.M.; Carvalho, A.P.; Lopes, M.C. Hydrogen Peroxide Mediates Interleukin-1β-Induced AP-1 Activation in Articular Chondrocytes: Implications for the Regulation of INOS Expression. Cell Biol. Toxicol. 2003, 19, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.S.; Jeong, M.; Park, J.S.; Kim, M.H.; Lee, D.B.; Shin, B.A.; Mukaida, N.; Ellis, L.M.; Kim, H.R.; Ahn, B.W.; et al. Interleukin-1β Stimulates IL-8 Expression through MAP Kinase and ROS Signaling in Human Gastric Carcinoma Cells. Oncogene 2004, 23, 6603–6611. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Dhaunsi, G.S.; Turner, R.B. Interleukin-1 and Nitric Oxide Increase NADPH Oxidase Activity in Human Coronary Artery Smooth Muscle Cells. Med. Princ. Pract. 2004, 13, 26–29. [Google Scholar] [CrossRef]
- Watanabe, Y.; Suzuki, O.; Haruyama, T.; Akaike, T. Interferon-γ Induces Reactive Oxygen Species and Endoplasmic Reticulum Stress at the Hepatic Apoptosis. J. Cell. Biochem. 2003, 89, 244–253. [Google Scholar] [CrossRef]
- Yang, D.; Elner, S.G.; Bian, Z.M.; Till, G.O.; Petty, H.R.; Elner, V.M. Pro-Inflammatory Cytokines Increase Reactive Oxygen Species through Mitochondria and NADPH Oxidase in Cultured RPE Cells. Exp. Eye Res. 2007, 85, 462–472. [Google Scholar] [CrossRef]
- Lugrin, J.; Rosenblatt-Velin, N.; Parapanov, R.; Liaudet, L. The Role of Oxidative Stress during Inflammatory Processes. Biol. Chem. 2014, 395, 203–230. [Google Scholar] [CrossRef]
- Shao, J.; Fujiwara, T.; Kadowaki, Y.; Fukazawa, T.; Waku, T.; Itoshima, T.; Yamatsuji, T.; Nishizaki, M.; Roth, J.A.; Tanaka, N. Overexpression of the Wild-Type P53 Gene Inhibits NF-ΚB Activity and Synergizes with Aspirin to Induce Apoptosis in Human Colon Cancer Cells. Oncogene 2000, 19, 726–736. [Google Scholar] [CrossRef]
- Murphy, S.H.; Suzuki, K.; Downes, M.; Welch, G.L.; De Jesus, P.; Miraglia, L.J.; Orth, A.P.; Chanda, S.K.; Evans, R.M.; Verma, I.M. Tumor Suppressor Protein (p)53, Is a Regulator of NF-κ;B Repression by the Glucocorticoid Receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 17117–17122. [Google Scholar] [CrossRef]
- Dijsselbloem, N.; Goriely, S.; Albarani, V.; Gerlo, S.; Francoz, S.; Marine, J.-C.; Goldman, M.; Haegeman, G.; Berghe, W.V. A Critical Role for P53 in the Control of NF-ΚB-Dependent Gene Expression in TLR4-Stimulated Dendritic Cells Exposed to Genistein. J. Immunol. 2007, 178, 5048–5057. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Karin, M. NF-B, Inflammation, Immunity and Cancer: Coming of Age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.A.; Christopherson, K.W.; Bhat-Nakshatri, P.; Goulet, R.J.; Broxmeyer, H.E.; Kopelovich, L.; Nakshatri, H. Negative Regulation of Chemokine Receptor CXCR4 by Tumor Suppressor P53 in Breast Cancer Cells: Implications of P53 Mutation or Isoform Expression on Breast Cancer Cell Invasion. Oncogene 2007, 26, 3329–3337. [Google Scholar] [CrossRef] [PubMed]
- Moskovits, N.; Kalinkovich, A.; Bar, J.; Lapidot, T.; Oren, M. P53 Attenuates Cancer Cell Migration and Invasion through Repression of SDF-1/CXCL12 Expression in Stromal Fibroblasts. Cancer Res. 2006, 66, 10671–10676. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.; Shi, J.; Li, S.; Li, M.; Zhang, L.; Zheng, L.; Zheng, D.; Tang, F.; Zhang, X.; et al. P53 Predominantly Regulates IL-6 Production and Suppresses Synovial Inflammation in Fibroblast-like Synoviocytes and Adjuvant-Induced Arthritis. Arthritis Res. Ther. 2016, 18, 271. [Google Scholar] [CrossRef]
- Cordani, M.; Pacchiana, R.; Butera, G.; D’Orazi, G.; Scarpa, A.; Donadelli, M. Mutant P53 Proteins Alter Cancer Cell Secretome and Tumour Microenvironment: Involvement in Cancer Invasion and Metastasis. Cancer Letters. 2016, 376, 303–309. [Google Scholar] [CrossRef]
- Yeudall, W.A.; Vaughan, C.A.; Miyazaki, H.; Ramamoorthy, M.; Choi, M.Y.; Chapman, C.G.; Wang, H.; Black, E.; Bulysheva, A.A.; Deb, S.P.; et al. Gain-of-Function Mutant P53 Upregulates CXC Chemokines and Enhances Cell Migration. Carcinogenesis 2012, 33, 442–451. [Google Scholar] [CrossRef]
- Weisz, L.; Damalas, A.; Liontos, M.; Karakaidos, P.; Fontemaggi, G.; Maor-Aloni, R.; Kalis, M.; Levrero, M.; Strano, S.; Gorgoulis, V.G.; et al. Mutant P53 Enhances Nuclear Factor ΚB Activation by Tumor Necrosis Factor α in Cancer Cells. Cancer Res. 2007, 67, 2396–2401. [Google Scholar] [CrossRef]
- Rahnamoun, H.; Lu, H.; Duttke, S.H.; Benner, C.; Glass, C.K.; Lauberth, S.M. Mutant P53 Shapes the Enhancer Landscape of Cancer Cells in Response to Chronic Immune Signaling. Nat. Commun. 2017, 8, 754. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Tarcic, O.; Solomon, H.; Schetter, A.J.; Wilder, S.; Lozano, G.; Pikarsky, E.; Forshew, T.; Rozenfeld, N.; et al. Mutant P53 Prolongs NF-ΚB Activation and Promotes Chronic Inflammation and Inflammation-Associated Colorectal Cancer. Cancer Cell 2013, 23, 634–646. [Google Scholar] [CrossRef]
- Ham, S.W.; Jeon, H.Y.; Jin, X.; Kim, E.J.; Kim, J.K.; Shin, Y.J.; Lee, Y.; Kim, S.H.; Lee, S.Y.; Seo, S.; et al. TP53 Gain-of-Function Mutation Promotes Inflammation in Glioblastoma. Cell Death Differ. 2019, 26, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Di Minin, G.; Bellazzo, A.; DalFerro, M.; Chiaruttini, G.; Nuzzo, S.; Bicciato, S.; Piazza, S.; Rami, D.; Bulla, R.; Sommaggio, R.; et al. Mutant P53 Reprograms TNF Signaling in Cancer Cells through Interaction with the Tumor Suppressor DAB2IP. Mol. Cell 2014, 56, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Valentino, E.; Bellazzo, A.; Di Minin, G.; Sicari, D.; Apollonio, M.; Scognamiglio, G.; Di Bonito, M.; Botti, G.; Del Sal, G.; Collavin, L. Mutant P53 Potentiates the Oncogenic Effects of Insulin by Inhibiting the Tumor Suppressor DAB2IP. Proc. Natl. Acad. Sci. USA 2017, 114, 7623–7628. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Chen, X. Identification of GRO1 as a Critical Determinant for Mutant P53 Gain of Function. J. Biol. Chem. 2009, 284, 12178–12187. [Google Scholar] [CrossRef] [PubMed]
- Ubertini, V.; Norelli, G.; D’Arcangelo, D.; Gurtner, A.; Cesareo, E.; Baldari, S.; Gentileschi, M.P.; Piaggio, G.; Nisticò, P.; Soddu, S.; et al. Mutant P53 Gains New Function in Promoting Inflammatory Signals by Repression of the Secreted Interleukin-1 Receptor Antagonist. Oncogene 2015, 34, 2493–2504. [Google Scholar] [CrossRef]
- Hussain, S.P.; Amstad, P.; He, P.; Robles, A.; Lupold, S.; Kaneko, I.; Ichimiya, M.; Sengupta, S.; Mechanic, L.; Okamura, S.; et al. P53-Induced Up-Regulation of MnSOD and GPx but Not Catalase Increases Oxidative Stress and Apoptosis. Cancer Res. 2004, 64, 2350–2356. [Google Scholar] [CrossRef]
- Budanov, A.V.; Sablina, A.A.; Feinstein, E.; Koonin, E.V.; Chumakov, P.M. Regeneration of Peroxiredoxins by P53-Regulated Sestrins, Homologs of Bacterial AhpD. Science. 2004, 304, 596–600. [Google Scholar] [CrossRef]
- O’Connor, J.C.; Wallace, D.M.; O’Brien, C.J.; Cotter, T.G. A Novel Antioxidant Function for the Tumor-Suppressor Gene P53 in the Retinal Ganglion Cell. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4237–4244. [Google Scholar] [CrossRef]
- Kang, M.Y.; Kim, H.B.; Piao, C.; Lee, K.H.; Hyun, J.W.; Chang, I.Y.; You, H.J. The Critical Role of Catalase in Prooxidant and Antioxidant Function of P53. Cell Death Differ. 2013, 20, 117–129. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Hu, W.; Feng, Z. Tumor Suppressor P53 and Its Mutants in Cancer Metabolism. Cancer Letters 2015, 356, 197–203. [Google Scholar] [CrossRef]
- Madan, E.; Gogna, R.; Bhatt, M.; Pati, U.; Kuppusamy, P.; Mahdi, A.A. Regulation of Glucose Metabolism by P53: Emerging New Roles for the Tumor Suppressor. Oncotarget. 2011, 2, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.A.; Post, S.M.; Elizondo-Fraire, A.C.; Maccio, D.R.; Jackson, J.G.; El-Naggar, A.K.; Van Pelt, C.; Terzian, T.; Lozano, G. Multiple Stress Signals Activate Mutant P53 in Vivo. Cancer Res. 2011, 71, 7168–7175. [Google Scholar] [CrossRef] [PubMed]
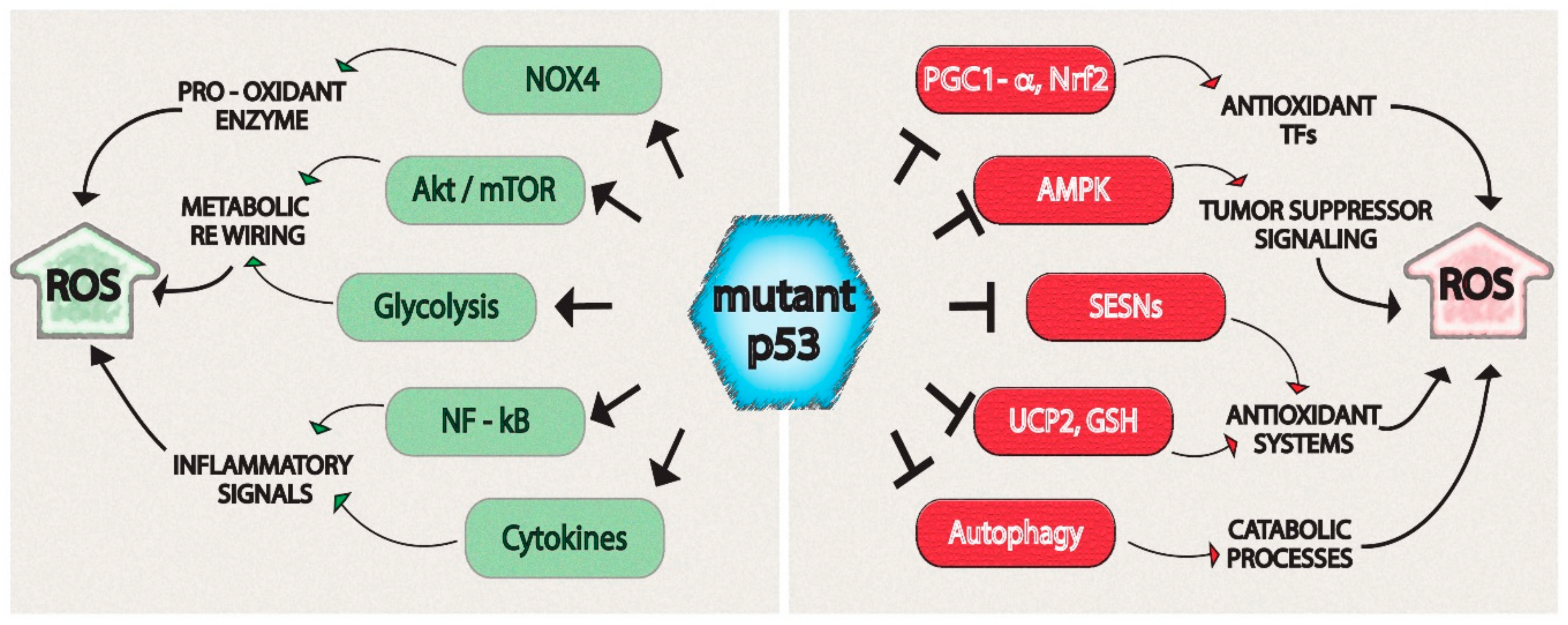
| p53 Mutant Isoforms | Target/Biological Process | Tumor Type |
|---|---|---|
| R175H, R280K | NOX4 | Lung and breast cancer [130] |
| R273H, R280K, R280T | Akt | Nasopharyngeal carcinoma, breast and pancreatic cancer [108,109,110] |
| R175H, R248W, R273H | mTOR | Colon carcinoma, lung carcinoma, pancreatic and breast cancer [89,104,121] |
| R175H, R248Q, R273H | Glycolysis | Lung carcinoma, breast and pancreatic cancer [110,117,118,120,121] |
| R175H, R248W, R273H | NF-kB | Lung, pancreatic, breast and colon cancer [158,159,160,161,162] |
| R175H, R281G, R273H | Cytokines | Lung, breast, pancreatic and colon cancer [157,164,165] |
| R175H, R273H | PGC1-α | Lung, colon and pancreatic cancer [8,137] |
| R175H, R280K, R273H | NRF2 | Colon carcinoma, oesophageal adenocarcinoma, lung and breast cancer [133,134,135] |
| R175H, R273H | AMPK | Pancreatic and breast cancer [8,89,110] |
| R175H, R248H, R273H | SESNs | Breast and pancreatic cancer [8,89] |
| R175H, R273H | UCP2 | Lung, pancreatic and breast cancer [8] |
| R175H, R273H | GSH | Oesophageal adenocarcinoma, pancreatic and breast cancer [128,135] |
| R175H, R273H | Autophagy | Lung carcinoma, pancreatic and breast cancer [89] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordani, M.; Butera, G.; Pacchiana, R.; Masetto, F.; Mullappilly, N.; Riganti, C.; Donadelli, M. Mutant p53-Associated Molecular Mechanisms of ROS Regulation in Cancer Cells. Biomolecules 2020, 10, 361. https://doi.org/10.3390/biom10030361
Cordani M, Butera G, Pacchiana R, Masetto F, Mullappilly N, Riganti C, Donadelli M. Mutant p53-Associated Molecular Mechanisms of ROS Regulation in Cancer Cells. Biomolecules. 2020; 10(3):361. https://doi.org/10.3390/biom10030361
Chicago/Turabian StyleCordani, Marco, Giovanna Butera, Raffaella Pacchiana, Francesca Masetto, Nidula Mullappilly, Chiara Riganti, and Massimo Donadelli. 2020. "Mutant p53-Associated Molecular Mechanisms of ROS Regulation in Cancer Cells" Biomolecules 10, no. 3: 361. https://doi.org/10.3390/biom10030361
APA StyleCordani, M., Butera, G., Pacchiana, R., Masetto, F., Mullappilly, N., Riganti, C., & Donadelli, M. (2020). Mutant p53-Associated Molecular Mechanisms of ROS Regulation in Cancer Cells. Biomolecules, 10(3), 361. https://doi.org/10.3390/biom10030361








