Deciphering the Role of Positions 145 and 165 in Fluorescence Lifetime Shortening in the EGFP Variants
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Site-Directed Mutagenesis
3.2. Spectroscopy and Fluorescence Brightness Evaluation
3.3. Fluorescence Lifetime Measurements
3.4. Photostability Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mishin, A.S.; Belousov, V.V.; Solntsev, K.M.; Lukyanov, K.A. Novel uses of fluorescent proteins. Curr. Opin. Chem. Biol. 2015, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Vu, C.Q.; Matsuda, T.; Nagai, T. Fluorescent Protein-Based Indicators for Functional Super-Resolution Imaging of Biomolecular Activities in Living Cells. Int. J. Mol. Sci. 2019, 20, 5784. [Google Scholar] [CrossRef] [PubMed]
- Specht, E.A.; Braselmann, E.; Palmer, A.E. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol. 2017, 79, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Gosnell, T.R. Fundamentals of Spectroscopy and Laser Physics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Sarder, P.; Maji, D.; Achilefu, S. Molecular Probes for Fluorescence Lifetime Imaging. Bioconjugate Chem. 2015, 26, 963–974. [Google Scholar] [CrossRef]
- Berezin, M.Y.; Achilefu, S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef]
- Merzlyak, E.M.; Goedhart, J.; Shcherbo, D.; Bulina, M.E.; Shcheglov, A.S.; Fradkov, A.F.; Gaintzeva, A.; Lukyanov, K.A.; Lukyanov, S.; Gadella, T.W.J.; et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 2007, 4, 555–557. [Google Scholar] [CrossRef]
- Goedhart, J.; van Weeren, L.; Hink, M.A.; Vischer, N.O.E.; Jalink, K.; Gadella, T.W.J. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods 2010, 7, 137–139. [Google Scholar] [CrossRef]
- Sarkisyan, K.S.; Goryashchenko, A.S.; Lidsky, P.V.; Gorbachev, D.A.; Bozhanova, N.G.; Gorokhovatsky, A.Y.; Pereverzeva, A.R.; Ryumina, A.P.; Zherdeva, V.V.; Savitsky, A.P.; et al. Green Fluorescent Protein with Anionic Tryptophan-Based Chromophore and Long Fluorescence Lifetime. Biophys. J. 2015, 109, 380–389. [Google Scholar] [CrossRef]
- Bindels, D.S.; Postma, M.; Haarbosch, L.; van Weeren, L.; Gadella, T.W.J. Multiparameter screening method for developing optimized red-fluorescent proteins. Nat. Protoc. 2020, 15, 450–478. [Google Scholar] [CrossRef]
- Interactive Chart of FP Properties. Available online: https://www.fpbase.org/chart/ (accessed on 20 June 2020).
- Mamontova, A.V.; Solovyev, I.D.; Savitsky, A.P.; Shakhov, A.M.; Lukyanov, K.A.; Bogdanov, A.M. Bright GFP with subnanosecond fluorescence lifetime. Sci. Rep. 2018, 8, 13224. [Google Scholar] [CrossRef]
- Piatkevich, K.D.; Malashkevich, V.N.; Morozova, K.S.; Nemkovich, N.A.; Almo, S.C.; Verkhusha, V.V. Extended Stokes Shift in Fluorescent Proteins: Chromophore–Protein Interactions in a Near-Infrared TagRFP675 Variant. Sci. Rep. 2013, 3, 1847. [Google Scholar] [CrossRef] [PubMed]
- Hense, A.; Prunsche, B.; Gao, P.; Ishitsuka, Y.; Nienhaus, K.; Nienhaus, G.U. Monomeric Garnet, a far-red fluorescent protein for live-cell STED imaging. Sci. Rep. 2015, 5, 18006. [Google Scholar] [CrossRef] [PubMed]
- Matela, G.; Gao, P.; Guigas, G.; Eckert, A.F.; Nienhaus, K.; Nienhaus, G.U. A far-red emitting fluorescent marker protein, mGarnet2, for microscopy and STED nanoscopy. Chem. Commun. 2017, 53, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar] [CrossRef]
- Kremers, G.-J.; Goedhart, J.; van den Heuvel, D.J.; Gerritsen, H.C.; Gadella, T.W.J. Improved Green and Blue Fluorescent Proteins for Expression in Bacteria and Mammalian Cells. Biochemistry 2007, 46, 3775–3783. [Google Scholar] [CrossRef]
- Scruggs, A.W.; Flores, C.L.; Wachter, R.; Woodbury, N.W. Development and Characterization of Green Fluorescent Protein Mutants with Altered Lifetimes. Biochemistry 2005, 44, 13377–13384. [Google Scholar] [CrossRef]
- Heikal, A.A.; Hess, S.T.; Webb, W.W. Multiphoton molecular spectroscopy and excited-state dynamics of enhanced green fluorescent protein (EGFP): Acid-base specificity. Chem. Phys. 2001, 274, 37–55. [Google Scholar] [CrossRef]
- Sen, T.; Mamontova, A.; Titelmayer, A.; Shakhov, A.; Astafiev, A.; Acharya, A.; Lukyanov, K.; Krylov, A.; Bogdanov, A. Influence of the First Chromophore-Forming Residue on Photobleaching and Oxidative Photoconversion of EGFP and EYFP. IJMS 2019, 20, 5229. [Google Scholar] [CrossRef]
- Ganesan, S.; Ameer-beg, S.M.; Ng, T.T.C.; Vojnovic, B.; Wouters, F.S. A dark yellow fluorescent protein (YFP)-based Resonance Energy-Accepting Chromoprotein (REACh) for Forster resonance energy transfer with GFP. Proc. Natl. Acad. Sci. USA 2006, 103, 4089–4094. [Google Scholar] [CrossRef]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef]
- Strickler-Berg Equation Online-Calculator. Available online: https://huygens.science.uva.nl/Strickler_Berg/ (accessed on 3 August 2020).
- Arpino, J.A.J.; Rizkallah, P.J.; Jones, D.D. Crystal Structure of Enhanced Green Fluorescent Protein to 1.35 Å Resolution Reveals Alternative Conformations for Glu222. PLoS ONE 2012, 7, e47132. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Haggie, P.; Wachter, R.M.; Remington, S.J.; Verkman, A.S. Mechanism and Cellular Applications of a Green Fluorescent Protein-based Halide Sensor. J. Biol. Chem. 2000, 275, 6047–6050. [Google Scholar] [CrossRef] [PubMed]
- Strickler, S.J.; Berg, R.A. Relationship between Absorption Intensity and Fluorescence Lifetime of Molecules. J. Chem. Phys. 1962, 37, 814–822. [Google Scholar] [CrossRef]

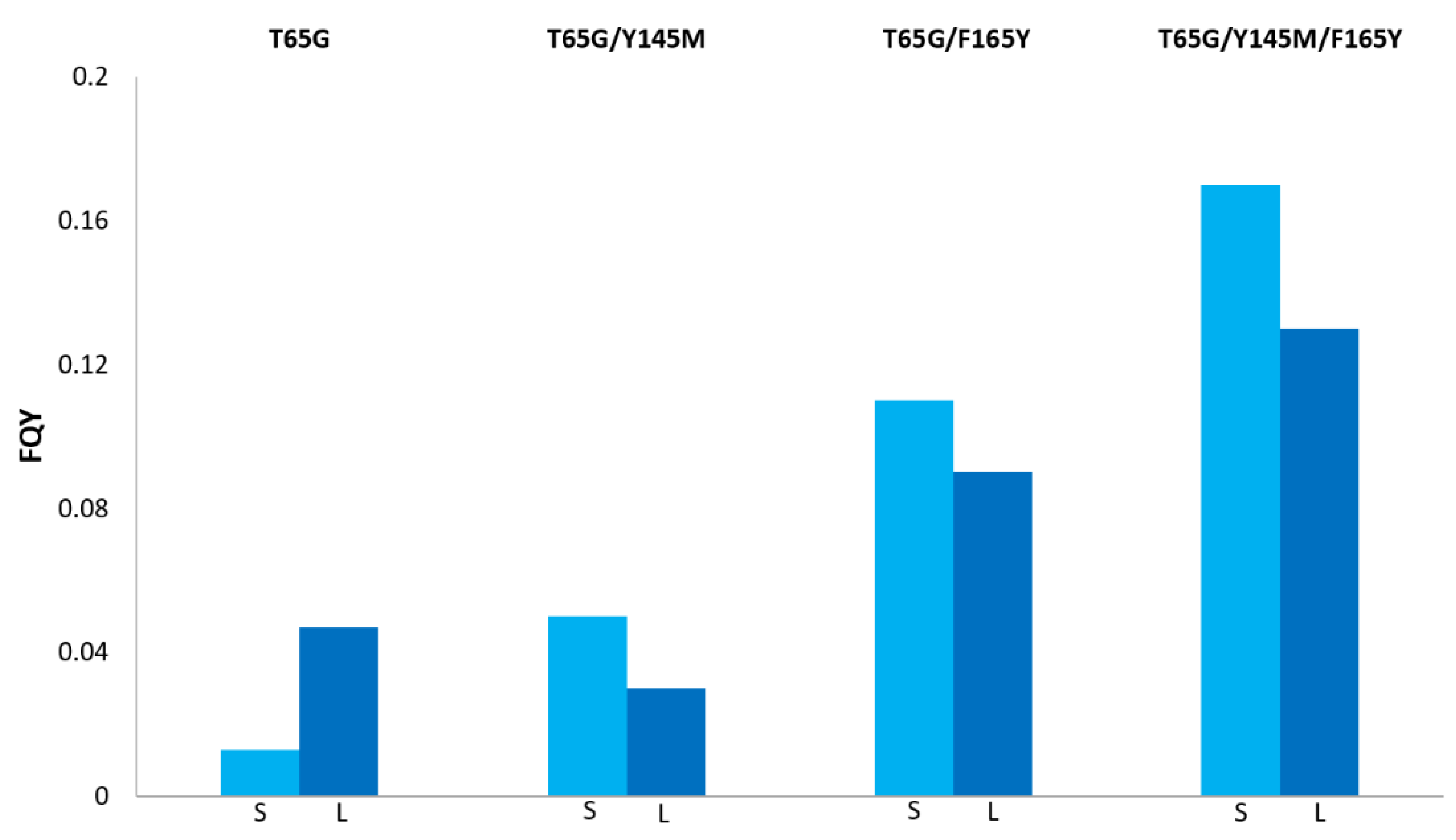
| Fluorescent Protein | λex/λem, nm | EC, M−1 cm−1 | FQY | Relative Brightness, % * | Data Source | |
|---|---|---|---|---|---|---|
| EGFP | 489/509 | 55,000 | 0.60 | 100 | [22] | |
| T65G | 488/508 | 70,000 | 0.06 | 13 | [12,20] | |
| Y145M | 486/508 | 46,100 | 0.65 | 91 | this paper | |
| F165Y | 489/509 | 55,600 | 0.57 | 96 | this paper | |
| T65G/Y145M | 488/508 | 84,500 | 0.08 | 20 | [12] | |
| T65G/F165Y | 488/509 | 84,000 | 0.20 | 51 | this paper | |
| Y145M/F165Y | 488/509 | 56,300 | 0.72 | 123 | this paper | |
| T65G/Y145M/F165Y (BrUSLEE) | 487/509 | 86,000 | 0.30 | 78 | [12] |
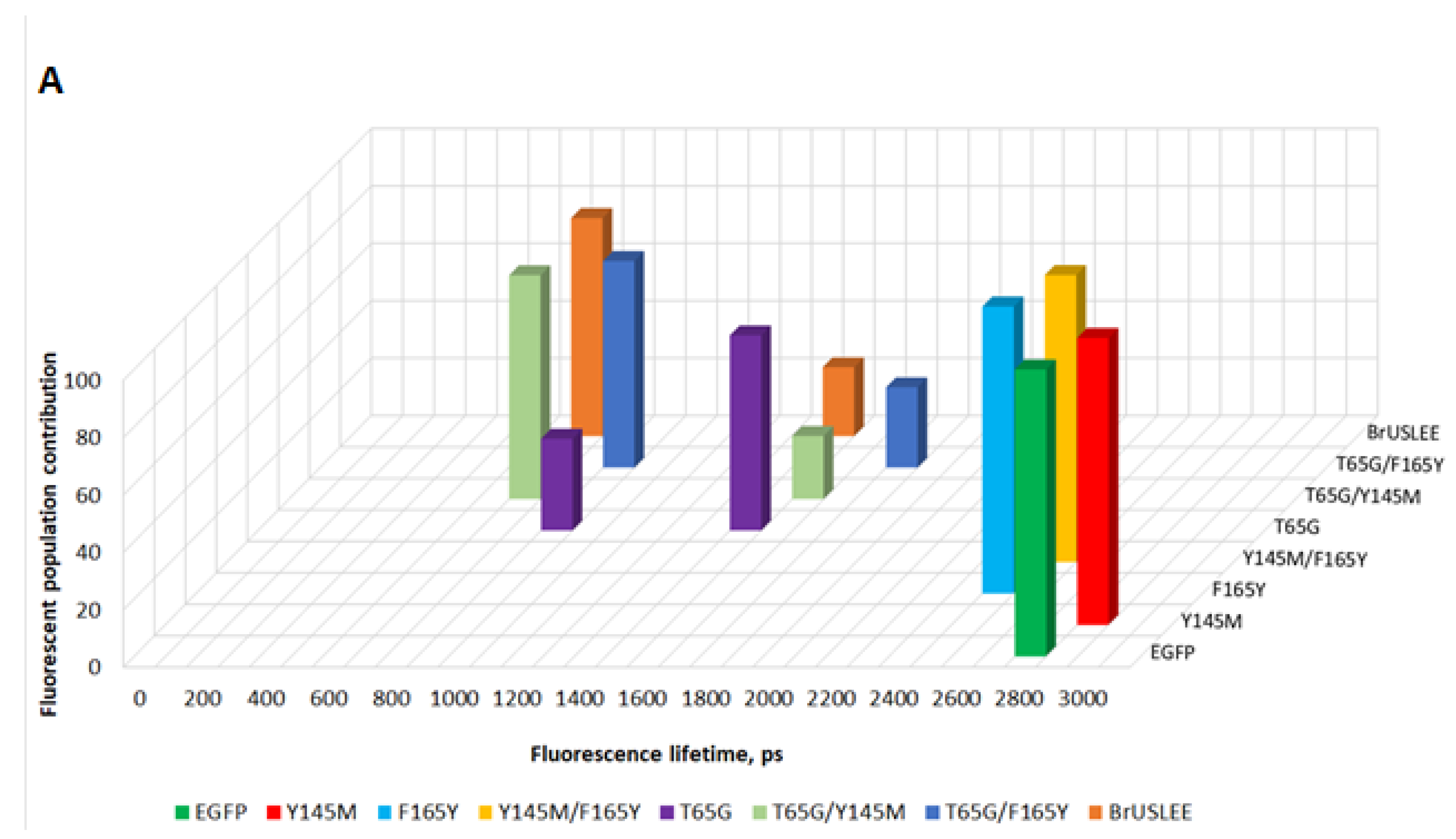
| Protein | |||||
|---|---|---|---|---|---|
| EGFP | 2830 ± 20 | 100 | - | - | 2.08 |
| T65G | 1530 ± 120 | 68 | 890 ± 210 | 32 | 0.18 |
| Y145M | 2935 ± 24 | 100 | - | - | 2.57 |
| F165Y | 2490 ± 15 | 100 | - | - | 1.72 |
| T65G/Y145M | 690 ± 30 | 78 | 1575 ± 95 | 22 | 0.18 |
| T65G/F165Y | 905 ± 35 | 72 | 1795 ± 95 | 28 | 0.51 |
| Y145M/F165Y | 2595 ± 12 | 100 | - | - | 2.16 |
| BrUSLEE * | 660 ± 35 | 76 | 1500 ± 80 | 24 | 0.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamontova, A.V.; Shakhov, A.M.; Lukyanov, K.A.; Bogdanov, A.M. Deciphering the Role of Positions 145 and 165 in Fluorescence Lifetime Shortening in the EGFP Variants. Biomolecules 2020, 10, 1547. https://doi.org/10.3390/biom10111547
Mamontova AV, Shakhov AM, Lukyanov KA, Bogdanov AM. Deciphering the Role of Positions 145 and 165 in Fluorescence Lifetime Shortening in the EGFP Variants. Biomolecules. 2020; 10(11):1547. https://doi.org/10.3390/biom10111547
Chicago/Turabian StyleMamontova, Anastasia V., Aleksander M. Shakhov, Konstantin A. Lukyanov, and Alexey M. Bogdanov. 2020. "Deciphering the Role of Positions 145 and 165 in Fluorescence Lifetime Shortening in the EGFP Variants" Biomolecules 10, no. 11: 1547. https://doi.org/10.3390/biom10111547
APA StyleMamontova, A. V., Shakhov, A. M., Lukyanov, K. A., & Bogdanov, A. M. (2020). Deciphering the Role of Positions 145 and 165 in Fluorescence Lifetime Shortening in the EGFP Variants. Biomolecules, 10(11), 1547. https://doi.org/10.3390/biom10111547





