Identification of Functional Single Nucleotide Polymorphisms in Porcine HSD17B14 Gene Associated with Estrus Behavior Difference between Large White and Mi Gilts
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Cell Culture, Cell Transfection, and Luciferase Assays
2.3. RNA Isolation and RT-qPCR
2.4. Promoter Prediction of the Porcine HSD17B14 Gene
2.5. Potential SNP Identification
2.6. Plasmid Construction
2.7. Western Blotting
2.8. E1 and E2 Detection
2.9. Cell Apoptosis Analysis
2.10. Statistical Analyses
3. Results
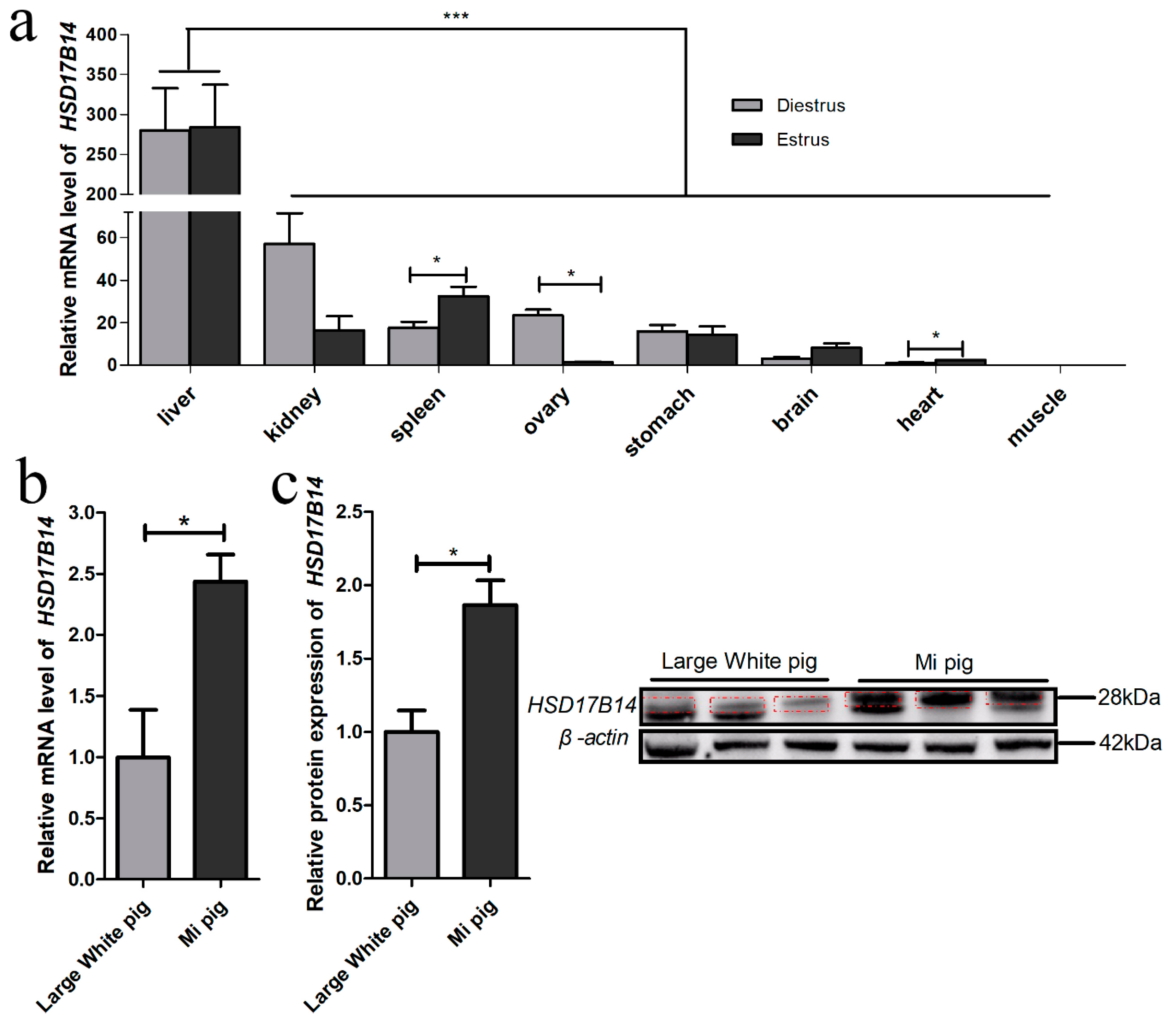
3.1. Tissue Expression Profile of HSD17B14 Gene in Gilts
3.2. Promoter Prediction of HSD17B14 Porcine Gene
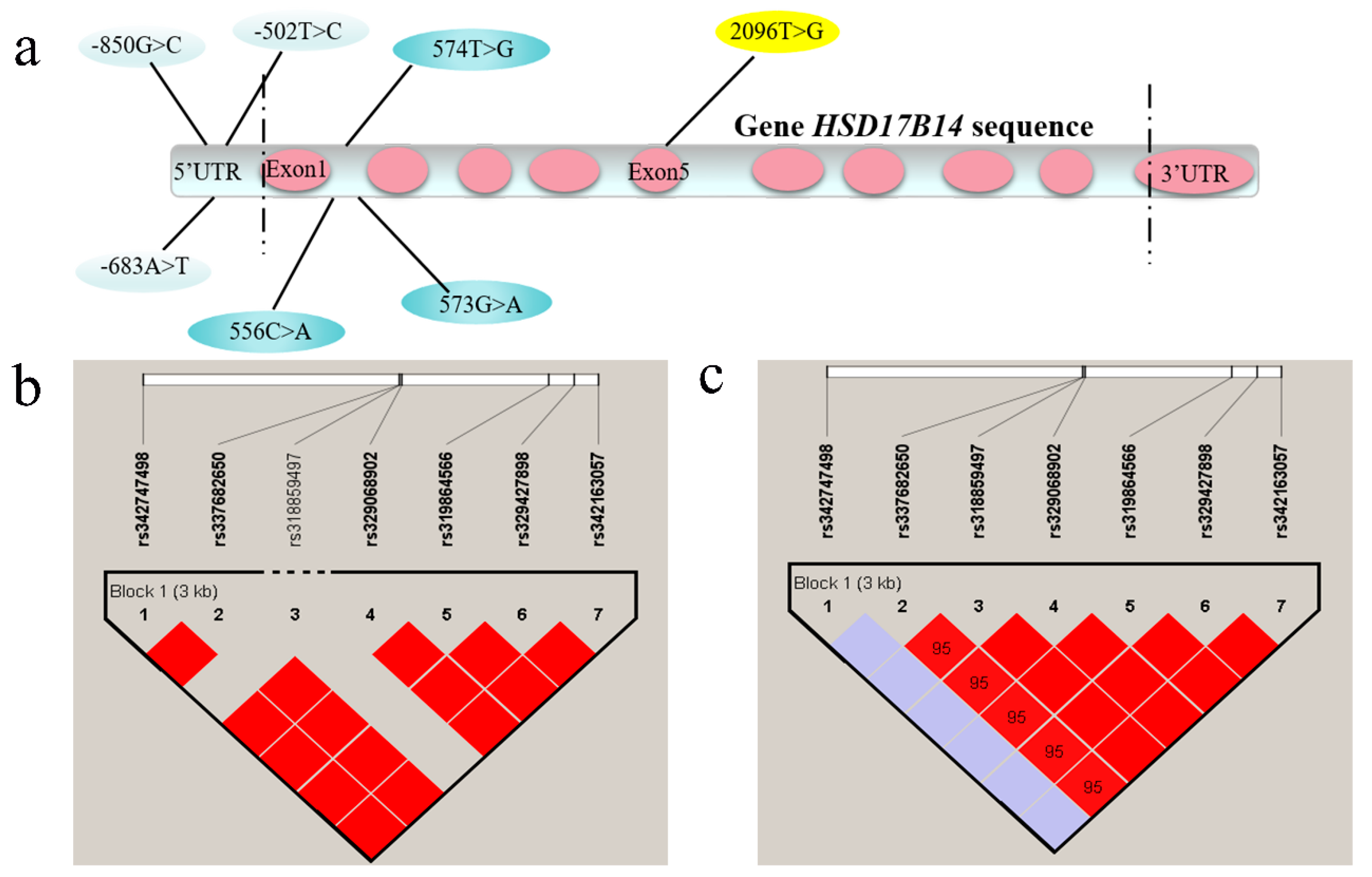
3.3. Genotypic Frequencies of HSD17B14 Gene in Large White and Mi Pigs and Linkage Disequilibrium (LD) Analyses
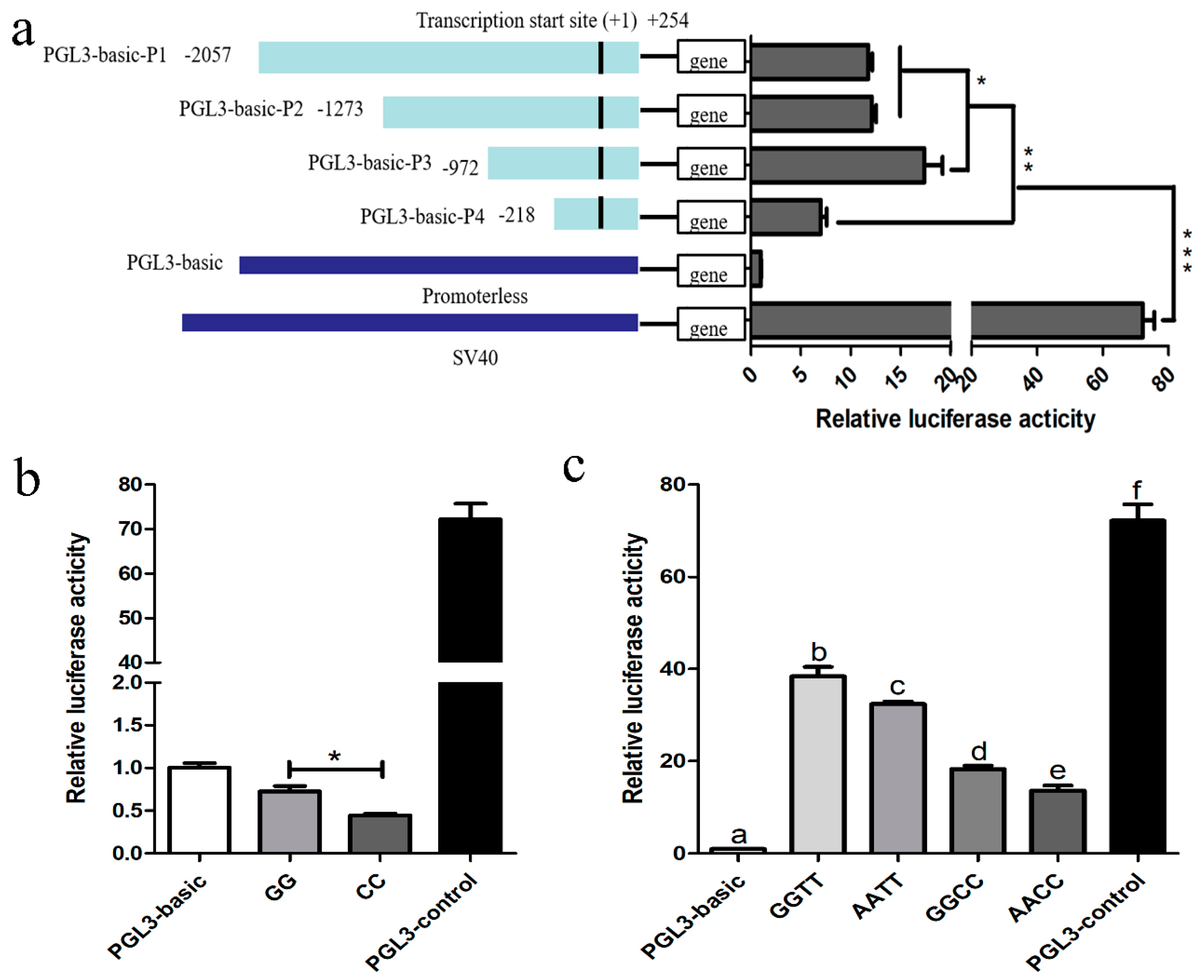
3.4. Promoter Identification of Porcine HSD17B14 Gene
3.5. Promoter Activity Analyses of Porcine HSD17B14 Gene
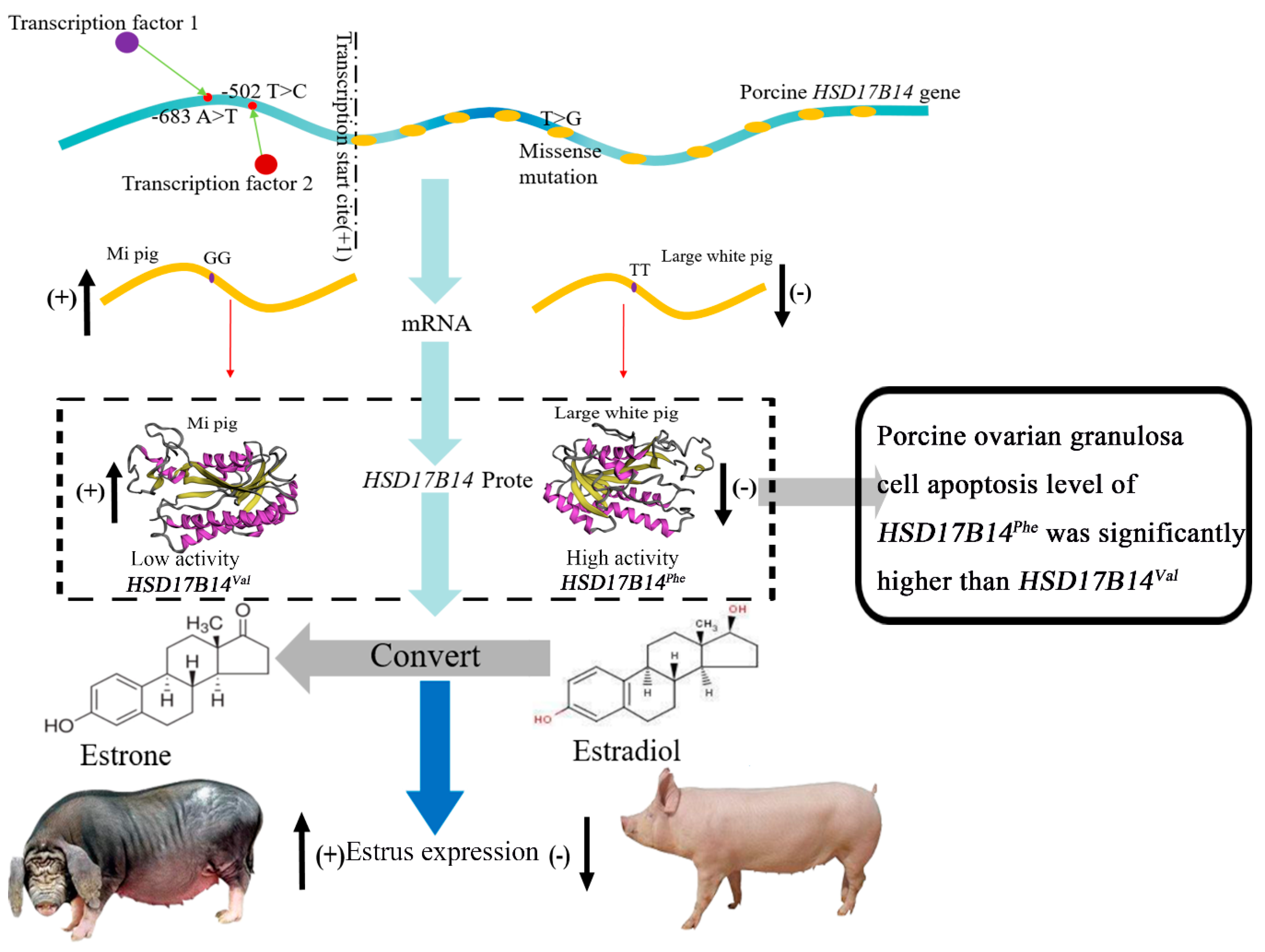
3.6. Differential Expression and Function of Phe73Val of HSD17B14 in Porcine Ovarian Granulosa Cells
3.7. Effect of Phe72Val of HSD17B14 Gene on Apoptosis of Porcine Ovarian Granulosa Cells
4. Discussion
4.1. Tissue Expression Profile of HSD17B14 Gene in Gilts
4.2. Prediction and Identification of the Core Promoter in the Porcine HSD17B14 Gene
4.3. Screening of SNPs in Porcine HSD17B14 Gene Associated with Estrus Behavior in Gilts
4.4. Promoter Activity Analyses of Porcine HSD17B14 Gene
4.5. Differential Expression and Function of Phe73Val of HSD17B14 in Porcine Ovarian Granulosa Cells
4.6. The Effect of HSD17B14 Phe73Val for Apoptosis of Ovarian Granulosa Cells by Degraded E2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nowak, R.A. Encyclopedia of Reproduction Estrous and Menstrual Cycles. In Encyclopedia of Reproduction (Second Edition); Elsevier: Urbana, IL, USA, 2018. [Google Scholar] [CrossRef]
- Zak, L.J.; Gaustad, A.H.; Bolarin, A.; Broekhuijse, M.; Walling, G.A.; Knol, E.F. Genetic control of complex traits, with a focus on reproduction in pigs. Mol. Reprod. Dev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Nonneman, D.J.; Schneider, J.F.; Lents, C.A.; Wiedmann, R.T.; Vallet, J.L.; Rohrer, G.A. Genome-wide association and identification of candidate genes for age at puberty in swine. BMC Genet. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Knauer, M.T.; Cassady, J.P.; Newcom, D.W.; See, M.T. Phenotypic and genetic correlations between gilt estrus, puberty, growth, composition, and structural conformation traits with first-litter reproductive measures. J. Anim. Sci. 2011, 89, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Koketsu, Y. Culling intervals and culling risks in four stages of the reproductive life of first service and reserviced female pigs in commercial herds. Theriogenology 2010, 73, 587–594. [Google Scholar] [CrossRef]
- Sharpe, R.M.; Franks, S. Environment, lifestyle and infertility-An inter-generational issue. Nat. Cell Biol. 2002, 4, 33–40. [Google Scholar] [CrossRef]
- Kuehn, L.A.; Nonneman, D.J.; Klindt, J.M.; Wise, T.H. Genetic relationships of body composition, serum leptin, and age at puberty in gilts. J. Anim. Sci. 2009, 87, 477–483. [Google Scholar] [CrossRef]
- Noguchi, M.; Ikedo, T.; Kawaguchi, H.; Tanimoto, A. Estrus synchronization in microminipig using estradiol dipropionate and prostaglandin F2α. J. Reprod. Dev. 2016. [Google Scholar] [CrossRef]
- Soede, N.M.; Langendijk, P.; Kemp, B. Reproductive cycles in pigs. Anim. Reprod. Sci. 2011. [Google Scholar] [CrossRef]
- Hossner, K. Hormonal Regulation of Farm Animal Growth; CABI Publishing: London, UK, 2005; p. 240. [Google Scholar] [CrossRef]
- Geisert, R.D.; Sutvosky, P.; Lucy, M.C.; Bartol, F.F.; Meyer, A.E. Chapter 15-Reproductive physiology of swine. In Animal Agriculture; Bazer, F.W., Lamb, G.C., Wu, G., Eds.; Academic Press: Columbia, NY, USA, 2020. [Google Scholar] [CrossRef]
- Labrie, F.; Luuthe, V.; Lin, S.; Claude, L.; Simard, J.; Breton, R.; Belanger, A. The key role of 17β-hydroxysteroid dehydrogenases in sex steroid biology. Steroids 1997, 62, 148–158. [Google Scholar] [CrossRef]
- Mindnich, R.; Deluca, D.; Adamski, J. Identification and characterization of 17β-hydroxysteroid dehydrogenases in the zebrafish, Danio rerio. Mol. Cell. Endocrinol. 2004, 215, 19–30. [Google Scholar] [CrossRef]
- Lukacik, P.; Keller, B.; Bunkoczi, G.; Kavanagh, K.L.; Lee, W.H.; Adamski, J.; Oppermann, U. Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity. Biochem. J. 2007, 402, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Lukacik, P.; Kavanagh, K.L.; Oppermann, U. Structure and function of human 17β-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 2006. [Google Scholar] [CrossRef]
- Toran-Allerand, C.D. Estrogen and the brain: Beyond ER-α and ER-β. Exp. Gerontol. 2004, 39, 1579–1586. [Google Scholar] [CrossRef] [PubMed]
- Toran-Allerand, C.D.; Guan, X.; Maclusky, N.J.; Horvath, T.L.; Tinnikov, A.A. ER-X: A novel, plasma membrane-associated, putative estrogen receptor that is regulated during development and after ischemic brain injury. J. Neurosci. Off. J. Soc. Neurosci. 2002, 22, 8391–8401. [Google Scholar] [CrossRef]
- Bazer, F.W.; Thatcher, W.W.; Martinatbotte, F.; Terqui, M. Sexual maturation and morphological development of the reproductive tract in large white and prolific Chinese Meishan pigs. Reproduction 1988, 83, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Canario, L.; Billon, Y.; Caritez, J.C.; Bidanel, J.P.; Lalo, D. Comparison of sow farrowing characteristics between a Chinese breed and three French breeds. Livest. Ence 2009, 125, 132–140. [Google Scholar] [CrossRef]
- Tilton, J.E.; Biggs, C.; Hunter, M.G.; Foxcroft, G.R. Gonadotropin secretion in ovariectomized Chinese Meishan and hybrid large white gilts; responses to challenges with estradiol benzoate, gonadotropin-releasing hormone, or porcine follicular fluid. Alcohol 2018, 66, 963. [Google Scholar] [CrossRef][Green Version]
- Ding, W.; Wang, W.; Zhou, B.; Zhang, W.; Huang, P.; Shi, F.; Taya, K. Formation of primordial follicles and immunolocalization of PTEN, PKB and FOXO3A proteins in the ovaries of fetal and neonatal pigs. J. Reprod. Dev. 2010, 56, 162–168. [Google Scholar] [CrossRef]
- Hunter, M.G.; Biggs, C.; Foxcroft, G.R.; Mcneilly, A.S.; Tilton, J.E. Comparisons of endocrinology and behavioural events during the periovulatory period in Meishan and Large-White hybrid gilts. Reproduction 1993, 97, 475–480. [Google Scholar] [CrossRef]
- Chu, Q.; Liang, T.; Zhou, B. Genetic differences in oestrous signs and oestrogen metabolism-related genes between Chinese Mi and European Landrace-Large White pigs. Reprod. Domest. Anim. 2017. [Google Scholar] [CrossRef]
- Chu, Q.; Zhou, B.; Xu, F.; Chen, R.; Shen, C.; Liang, T.; Li, Y.; Schinckel, A.P. Genome-wide differential mRNA expression profiles in follicles of two breeds and at two stages of estrus cycle of gilts. Sci. Rep. 2017, 7, 5052. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Du, X.; Zhou, J.; Pan, Z.; Liu, H.; Li, Q. MicroRNA-26b Functions as a Proapoptotic Factor in Porcine Follicular Granulosa Cells by Targeting Sma-and Mad-Related Protein 41. Biol. Reprod. 2014, 91. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, S. Promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–361. [Google Scholar] [CrossRef]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Biol. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Nunez, O.; Martinez, J.; Alba, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Farre, D.; Roset, R.; Huerta, M.; Adsuara, J.; Rosello, L.; Alba, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef]
- Kandathil, S.M.; Greener, J.G.; Jones, D.T. Prediction of interresidue contacts with DeepMetaPSICOV in CASP13. Proteins: Struct. Funct. Bioinform. 2019, 87, 1092–1099. [Google Scholar] [CrossRef]
- Li, Q.; Du, X.; Pan, Z.; Zhang, L.; Li, Q. The transcription factor SMAD4 and miR-10b contribute to E2 release and cell apoptosis in ovarian granulosa cells by targeting CYP19A1. Mol. Cell. Endocrinol. 2018, 476, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Garg, R.; Jain, M.K. A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol. J. 2013, 11, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Csukasi, F.; Zheng, Y.; Fei, Z.; Esther, V.D.K.; Catalá, C. Comprehensive Tissue-Specific Transcriptome Analysis Reveals Distinct Regulatory Programs during Early Tomato Fruit Development. Plant Physiol. 2015, 168, 1684–1701. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Vallender, E.J.; Miller, G.M. Functional characterization of the human TPH2 5′ regulatory region: Untranslated region and polymorphisms modulate gene expression in vitro. Hum. Genet. 2008, 122, 645–657. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Slobodin, B.; Agami, R. Transcription Initiation Determines Its End. Mol. Cell 2015. [Google Scholar] [CrossRef]
- Thomas, M.C.; Chiang, C.-M. The General Transcription Machinery and General Cofactors. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 105–178. [Google Scholar] [CrossRef]
- Ardlie, K.G.; Kruglyak, L.; Seielstad, M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002, 3, 299–309. [Google Scholar] [CrossRef]
- Ponzoni, L.; Bahar, I. Structural dynamics is a determinant of the functional significance of missense variants. Proc. Natl. Acad. Sci. USA 2018, 115, 4164–4169. [Google Scholar] [CrossRef]
- Olsen, R.K.J.; Broner, S.; Sabaratnam, R.; Doktor, T.K.; Andersen, H.S.; Bruun, G.H.; Gahrn, B.; Stenbroen, V.; Olpin, S.E.; Dobbie, A. The ETFDH c.158A>G Variation Disrupts the Balanced Interplay of ESE- and ESS-Binding Proteins thereby Causing Missplicing and Multiple Acyl-CoA Dehydrogenation Deficiency. Hum. Mutat. 2014, 35, 86–95. [Google Scholar] [CrossRef]
- Bongiorni, S.; Tilesi, F.; Bicorgna, S.; Iacoponi, F.; Willems, D.; Gargani, M.; Dandrea, M.; Pilla, F.; Valentini, A. Promoter polymorphisms in genes involved in porcine myogenesis influence their transcriptional activity. Bmc Genet. 2014, 15, 119. [Google Scholar] [CrossRef] [PubMed]
- Juneja, M.; Ilm, K.; Schlag, P.M.; Stein, U. Promoter identification and transcriptional regulation of the metastasis gene MACC1 in colorectal cancer. Mol. Oncol. 2013, 7, 929–943. [Google Scholar] [CrossRef]
- Parker, J. Encyclopedia of Genetics || Editing and Proofreading in Translation. Brenners Encycl. Genet. 2001, 601–602. [Google Scholar] [CrossRef]
- Badran, M.J.; Bertoletti, N.; Keils, A.; Heine, A.; Klebe, G.; Marchais-Oberwinkler, S. Mutational and structural studies uncover crucial amino acids determining activity and stability of 17β-HSD14. J. Steroid Biochem. Mol. Biol. 2019, 189, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Ciesiółka, S.; Budna, J.; Karol, J.; Jopek, K.; Bryja, A.; Kranc, W.; Borys, S.; Jeseta, M.; Chachuła, A.; Ziółkowska, A.; et al. Time- and Dose-Dependent Effects of 17 Beta-Estradiol on Short-Term, Real-Time Proliferation and Gene Expression in Porcine Granulosa Cells. Biomed Res. Int. 2017, 2017, 9738640. [Google Scholar] [CrossRef]
- Tasaki, H.; Iwata, H.; Sato, D.; Monji, Y.; Kuwayama, T. Estradiol has a major role in antrum formation of porcine preantral follicles cultured in vitro. Theriogenology 2013, 79, 809–814. [Google Scholar] [CrossRef]
- Casarini, L.; Riccetti, L.; De Pascali, F.; Gilioli, L.; Marino, M.; Vecchi, E.; Morini, D.; Nicoli, A.; La Sala, G.B.; Simoni, M. Estrogen Modulates Specific Life and Death Signals Induced by LH and hCG in Human Primary Granulosa Cells In Vitro. Int. J. Mol. Sci. 2017, 18, 926. [Google Scholar] [CrossRef]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular Growth and Atresia in Mammalian Ovar 6ies: Regulation by Survival and Death of Granulosa Cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef]
- Li, J.; Gao, H.; Tian, Z.; Wu, Y.; Zeng, S. Effects of chronic heat stress on granulosa cell apoptosis and follicular atresia in mouse ovary. J. Anim. Ence Biotechnol. 2016, 7, 57–66. [Google Scholar] [CrossRef]


| SNPs | Region | SNP Site | Allele | Large White Pigs/Mi Pigs | ꭓ2 | p-Value |
|---|---|---|---|---|---|---|
| rs342163057 | 5’UTR | G > C | G | 0.477/0.720 | 5.369 | 0.020 * |
| C | 0.523/0.280 | |||||
| rs329427898 | 5’UTR | A > G | A | 0.363/0.767 | 14.415 | 0.000 *** |
| G | 0.637/0.233 | |||||
| rs319864566 | 5’UTR | T > C | T | 0.400/0.090 | 12.141 | 0.000 *** |
| C | 0.600/0.910 | |||||
| rs329068902 | Intron 1 | C > A | C | 0.700/0.750 | 0.198 | 0.656 |
| A | 0.300/0.250 | |||||
| rs318859497 | Intron 1 | C > T | C | 1.000/0.750 | 14.326 | 0.000 *** |
| T | 0.000/0.250 | |||||
| rs337682650 | Intron 1 | G > A | G | 0.820/0.170 | 42.688 | 0.000 *** |
| A | 0.180/0.830 | |||||
| rs342747498 | Exon 5 | T > G | T | 0.400/0.078 | 14.411 | 0.000 *** |
| G | 0.600/0.922 |
| SNPs | Mutation Position | ObsHET | PredHET | HW P | %Geno | MAF |
|---|---|---|---|---|---|---|
| rs342163057 | 6:54145324 | 0.54 | 0.484 | 0.6479 | 100 | 0.41 |
| rs329427898 | 6:54145156 | 0.54 | 0.484 | 0.6479 | 100 | 0.41 |
| rs319864566 | 6:54144975 | 0.54 | 0.000 | 1.0000 | 100 | 0.41 |
| rs329068902 | 6:54143919 | 0.54 | 0.476 | 0.5589 | 100 | 0.39 |
| rs318859497 | 6:54143903 | 0.00 | 0.000 | 1.0000 | 100 | 0.00 |
| rs337682650 | 6:54143902 | 0.54 | 0.476 | 0.5589 | 100 | 0.39 |
| rs342747498 | 6:54142047 | 0.54 | 0.476 | 0.5589 | 100 | 0.39 |
| SNPs | Mutation Position | ObsHET | PredHET | HW P | %Geno | MAF |
|---|---|---|---|---|---|---|
| rs342163057 | 6:54145324 | 0.714 | 0.493 | 0.0043 | 100 | 0.439 |
| rs329427898 | 6:54145156 | 0.714 | 0.493 | 0.0043 | 100 | 0.439 |
| rs319864566 | 6:54144975 | 0.714 | 0.493 | 0.0043 | 100 | 0.439 |
| rs329068902 | 6:54143919 | 0.714 | 0.493 | 0.0043 | 100 | 0.439 |
| rs318859497 | 6:54143903 | 0.755 | 0.499 | 9 × 10−4 | 100 | 0.480 |
| rs337682650 | 6:54143902 | 0.714 | 0.493 | 0.0043 | 100 | 0.439 |
| rs342747498 | 6:54142047 | 0.020 | 0.020 | 1.0000 | 100 | 0.010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, S.; Tao, R.; Tong, X.; Xu, Q.; Zhao, J.; Guo, Y.; Schinckel, A.P.; Zhou, B. Identification of Functional Single Nucleotide Polymorphisms in Porcine HSD17B14 Gene Associated with Estrus Behavior Difference between Large White and Mi Gilts. Biomolecules 2020, 10, 1545. https://doi.org/10.3390/biom10111545
Gao S, Tao R, Tong X, Xu Q, Zhao J, Guo Y, Schinckel AP, Zhou B. Identification of Functional Single Nucleotide Polymorphisms in Porcine HSD17B14 Gene Associated with Estrus Behavior Difference between Large White and Mi Gilts. Biomolecules. 2020; 10(11):1545. https://doi.org/10.3390/biom10111545
Chicago/Turabian StyleGao, Siyuan, Ruixin Tao, Xian Tong, Qinglei Xu, Jing Zhao, Yanli Guo, Allan P. Schinckel, and Bo Zhou. 2020. "Identification of Functional Single Nucleotide Polymorphisms in Porcine HSD17B14 Gene Associated with Estrus Behavior Difference between Large White and Mi Gilts" Biomolecules 10, no. 11: 1545. https://doi.org/10.3390/biom10111545
APA StyleGao, S., Tao, R., Tong, X., Xu, Q., Zhao, J., Guo, Y., Schinckel, A. P., & Zhou, B. (2020). Identification of Functional Single Nucleotide Polymorphisms in Porcine HSD17B14 Gene Associated with Estrus Behavior Difference between Large White and Mi Gilts. Biomolecules, 10(11), 1545. https://doi.org/10.3390/biom10111545






