Virtual Screening for Reactive Natural Products and Their Probable Artifacts of Solvolysis and Oxidation
Abstract
1. Introduction
2. Materials and Methods
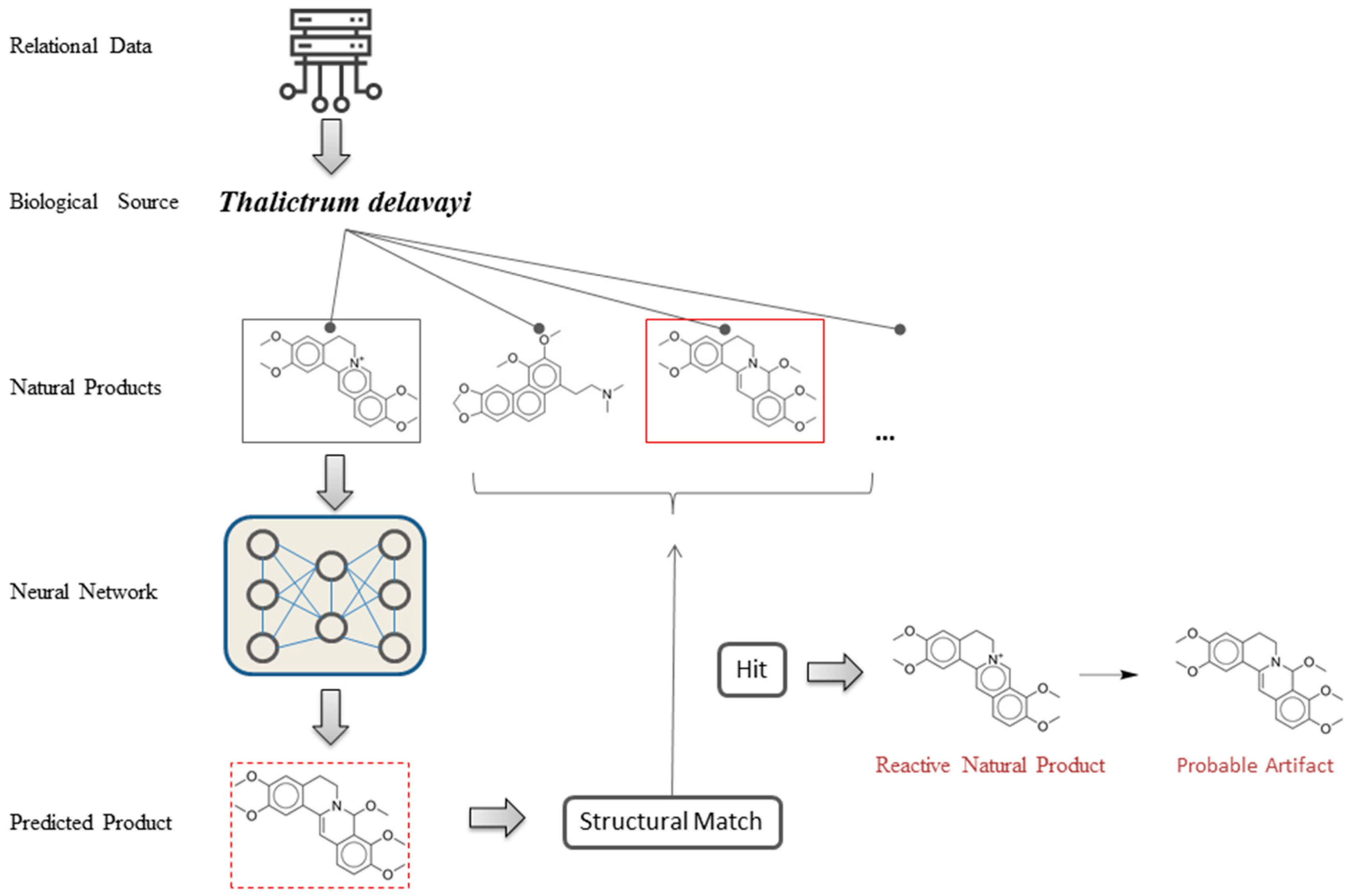
- Take a set of relational data (a specific biological source and all the natural products derived from it);
- Take one of the natural product molecules in this relational data set;
- Predict its solvolysis and oxidation products by neural network models;
- If predictions of the models are successful (or partially successful), match the predicted products with the other natural product molecules from the same biological source;
- If a predicted product matches one of the other natural product molecules, label the natural product and the predicted product as a potential case;
- Go through steps 2–5 with all the other molecules in the same relational data set;
- Go through steps 1–6 with all the other relational data sets and screen out all the potential cases in the data set.
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maltese, F.; van der Kooy, F.; Verpoorte, R. Solvent derived artifacts in natural products chemistry. Nat. Prod. Commun. 2009, 4, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Marek, R.; Sečkářová, P.; Hulová, D.; Dostál, J.; Sklenář, V. Palmatine and berberine isolation artifacts. J. Nat. Prod. 2003, 66, 481–486. [Google Scholar] [CrossRef]
- Capon, R.J. Extracting value: Mechanistic insights into the formation of natural product artifacts–case studies in marine natural products. Nat. Prod. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Reimer, D.; Hughes, C.C. Thiol-based probe for electrophilic natural products reveals that most of the ammosamides are artifacts. J. Nat. Prod. 2017, 80, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; de Bruyn Kops, C.; Kirchmair, J. Data Resources for the Computer-Guided Discovery of Bioactive Natural Products. J. Chem. Inf. Model. 2017, 57, 2099–2111. [Google Scholar] [CrossRef]
- Medema, M.H.; Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015, 11, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.B.; Hodas, N.O.; Vishnu, A. Deep learning for computational chemistry. J. Comput. Chem. 2017, 38, 1291–1307. [Google Scholar] [CrossRef]
- Mater, A.C.; Coote, M.L. Deep learning in chemistry. J. Chem. Inf. Model. 2019, 59, 2545–2559. [Google Scholar] [CrossRef]
- Fooshee, D.; Mood, A.; Gutman, E.; Tavakoli, M.; Urban, G.; Liu, F.; Huynh, N.; Van Vranken, D.; Baldi, P. Deep learning for chemical reaction prediction. Mol. Syst. Des. Eng. 2018, 3, 442–452. [Google Scholar] [CrossRef]
- Zheng, S.; Rao, J.; Zhang, Z.; Xu, J.; Yang, Y. Predicting Retrosynthetic Reactions using Self-Corrected Transformer Neural Networks. J. Chem. Inf. Model. 2019, 60, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Ahneman, D.T.; Estrada, J.G.; Lin, S.; Dreher, S.D.; Doyle, A.G. Predicting reaction performance in C–N cross-coupling using machine learning. Science 2018, 360, 186–190. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J. Linking the neural machine translation and the prediction of organic chemistry reactions. arxiv 2016, arXiv:1612.09529. [Google Scholar]
- Kayala, M.A.; Baldi, P. ReactionPredictor: Prediction of complex chemical reactions at the mechanistic level using machine learning. J. Chem. Inf. Model. 2012, 52, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Schneider, N.; Lowe, D.M.; Sayle, R.A.; Landrum, G.A. Development of a novel fingerprint for chemical reactions and its application to large-scale reaction classification and similarity. J. Chem. Inf. Model. 2015, 55, 39–53. [Google Scholar] [CrossRef]
- NPBS Database. Available online: http://www.organchem.csdb.cn/scdb/NPBS (accessed on 30 July 2020).
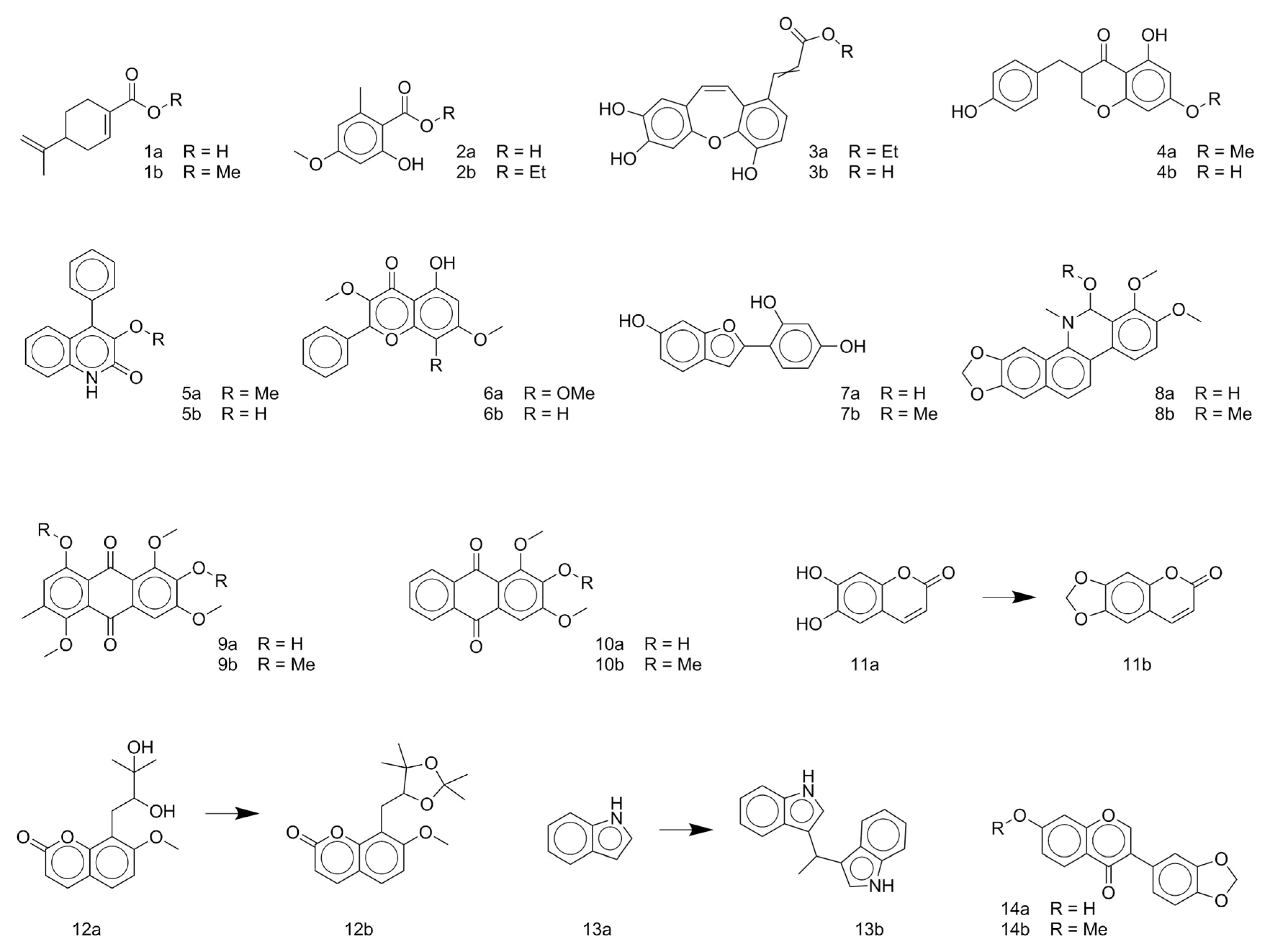
- Li, M.; Chen, X.; Tang, Q.M.; Zhang, J. Isoquinoline alkaloids from Thalictrum delavayi. Planta. Med. 2001, 67, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ali, Y.E.; Sharaf, M.; Wong, L.K.; Fu, E.W.; Lin, F.; Duah, F.K.; Schiff, P.L. Alkaloids of Thalictrum delavayi. Phytochemistry 1990, 29, 1895–1897. [Google Scholar] [CrossRef]
- Chemistry Database. Available online: http://www.organchem.csdb.cn (accessed on 30 July 2020).
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Attention-Based Sequence-to-Sequence in Keras. Available online: https://wanasit.github.io/attention-based-sequence-to-sequence-in-keras.html (accessed on 30 July 2020).
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. Adv. NIPS 2014, 3104–3112. [Google Scholar]
- Keras. Sequence-To-Sequence Example in KERAS (Character-Level). Available online: https://github.com/keras-team/keras/blob/master/examples/cnn_seq2seq.py (accessed on 30 July 2020).
- Python. Available online: https://www.python.org (accessed on 30 July 2020).
- Keras. Available online: https://keras.io (accessed on 30 July 2020).
- Tensorflow. Available online: https://github.com/tensorftow/tensorftow (accessed on 30 July 2020).
- Landrum, G. RDKit: Open-Source Cheminformatics Software. Available online: http://www.rdkit.org. (accessed on 30 July 2020).
- da Silva, M.H.; Andrade, E.H.; Maia, J.G. The essential oil of Pectis elongate Kunth occurring in North Brazil. Flavour Fragr. J. 2005, 20, 462–464. [Google Scholar] [CrossRef]
- Nishitoba, Y.; Nishimura, H.; Nishiyama, T.; Mizutani, J. Lichen acids, plant growth inhibitors from Usnea longissima. Phytochemistry 1987, 26, 3181–3185. [Google Scholar] [CrossRef]
- Sun, H.; Lin, Z.; Shen, X.; Niu, F.; Zhou, C. Chemical constituents of seven species of lichen plants in Yunnan. Yunnan Zhiwu Yanjiu 1986, 8, 483–488. [Google Scholar]
- Lin, Y.; Chang, Y.; Kuo, Y.; Shiao, M. Anti-Lipid-Peroxidative Principles from Tournefortia sarmentosa. J. Nat. Prod. 2002, 65, 745–747. [Google Scholar] [CrossRef]
- Mutanyatta, J.; Matapa, B.G.; Shushu, D.D.; Abegaz, B.M. Homoisoflavonoids and xanthones from the tubers of wild and in vitro regenerated Ledebouria graminifolia and cytotoxic activities of some of the homoisoflavonoids. Phytochemistry 2003, 62, 797–804. [Google Scholar] [CrossRef]
- Hodge, R.P.; Harris, C.M.; Harris, T.M. Verrucofortine, a major metabolite of Penicillium verrucosum var. cyclopium, the fungus that produces the mycotoxin verrucosidin. J. Nat. Prod. 1988, 51, 66–73. [Google Scholar] [CrossRef]
- Chou, T.; Chen, J.; Lee, S.; Chiang, M.Y.; Yang, C.; Chen, I. Cytotoxic Flavonoids from the Leaves of Cryptocarya chinensis. J. Nat. Prod. 2010, 73, 1470–1475. [Google Scholar] [CrossRef]
- Tanaka, H.; Hattori, H.; Sato, M.; Yamaguchi, R.; Fukai, T.; Tanaka, T.; Sakai, E. New constituents from the roots of Erythrina x bidwillii. Heterocycles 2007, 71, 1779–1785. [Google Scholar] [CrossRef]
- Xu, L.; Niu, S.; Wu, Z.; Liu, X.; Shi, F. Benzophenanthridine alkaloids from Zanthoxylum nitidum (Roxb.) DC. Zhongcaoyao 2009, 40, 538–540. [Google Scholar]
- Tang, Y.; Hu, J.; Wang, J.; Lou, F. A new coumaronochromone from Sophora japonica. J. Asian Nat. Prod. Res. 2002, 4, 1–5. [Google Scholar] [CrossRef]
- Tang, Y.; Lou, F.; Wang, J. Isoflavonoids from the peel of Sophora japonica. Zhongcaoyao 2002, 33, 20–21. [Google Scholar]
- Barba, B.; Diaz, J.G.; Herz, W. Cassanes and anthraquinones from Chamaecrista greggii. Phytochemistry 1994, 37, 837–845. [Google Scholar] [CrossRef]
- Halim, A.F.; Abd El-Fattah, H.; El-Gamal, A.A.; Thomson, R.H. Anthraquinones from Galium sinaicum. Phytochemistry 1992, 31, 355–356. [Google Scholar] [CrossRef]
- Adfa, M.; Yoshimura, T.; Komura, K.; Koketsu, M. Antitermite Activities of Coumarin Derivatives and Scopoletin from Protium javanicum Burm. f. J. Chem. Ecol. 2010, 36, 720–726. [Google Scholar] [CrossRef]
- Gu, S.; Song, X.; Su, W. Coumarins from Citrus grandis. Zhongcaoyao 2005, 36, 341–343. [Google Scholar]
- Bell, R.; Carmeli, S.; Sar, N. Vibrindole A, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostracion cubicus. J. Nat. Prod. 1994, 57, 1587–1590. [Google Scholar] [CrossRef]
- Barros-Filho, B.A.; Nunes, F.M.; de Oliveira, M.C.F.; Mafezoli, J.; Andrade-Neto, M.; Silveira, R.; Pirani, J.R. Volatile constituents from Esenbeckia almawillia (Rutaceae). Biochem. Syst. Ecol. 2004, 32, 817–821. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Urano, A.; Sudo, H.; Kitajima, M.; Takayama, H.; Yamazaki, M.; Aimi, N.; Saito, K. Metabolite profiling of alkaloids and strictosidine synthase activity in camptothecin producing plants. Phytochemistry 2003, 62, 461–470. [Google Scholar] [CrossRef]
- Zhang, M.; Deng, Y.; Zhang, H.; Su, X.; Chen, H.; Yu, T.; Guo, P. Two new coumarins from Herpetospermum caudigerum. Chem. Pharm. Bull. 2008, 56, 192–193. [Google Scholar] [CrossRef]
- Takemura, Y.; Kawaguchi, H.; Maki, S.; Ju-ichi, M.; Omura, M.; Ito, C.; Furukawa, H. Studies on the constituents of Yalaha. Structures of a new acridone alkaloid and two new coumarins. Chem. Pharm. Bull. 1996, 44, 804–809. [Google Scholar] [CrossRef]
- Hussein, A.A.; Rodriguez, B. Isopimarane Diterpenoids from Lycopus europaeus. J. Nat. Prod. 2000, 63, 419–421. [Google Scholar] [CrossRef]
- Lin, J. Cinnamamide derivatives from Clausena lansium. Phytochemistry 1989, 28, 621–622. [Google Scholar] [CrossRef]
- Oketch-Rabah, H.A.; Mwangi, J.W.; Lisgarten, J.; Mberu, E.K. A new antiplasmodial coumarin from Toddalia asiatica roots. Fitoterapia 2000, 71, 636–640. [Google Scholar] [CrossRef]
- Diaz, F.; Medina, J.D. Furanonaphthoquinones from Tabebuia ochracea ssp. neochrysanta. J. Nat. Prod. 1996, 59, 423–424. [Google Scholar] [CrossRef]
- Xu, T.; Chen, W.; Zhou, J.; Dai, J.; Li, Y.; Zhao, Y. Neural machine translation of chemical nomenclature between English and Chinese. J. Cheminform. 2020, 12, 50. [Google Scholar] [CrossRef]


| No. | Biological Source | Natural Product |
|---|---|---|
| 1 | Thalictrum delavayi | 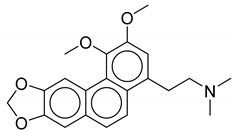 |
| 2 | Thalictrum delavayi | 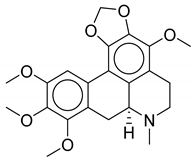 |
| 3 | Thalictrum delavayi | 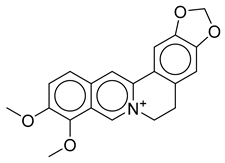 |
| 4 | Thalictrum delavayi | 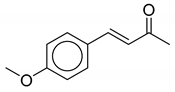 |
| 5 | Thalictrum delavayi | 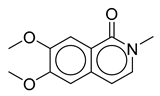 |
| 6 | Thalictrum delavayi | 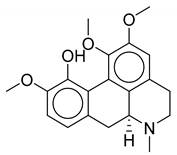 |
| 7 | Thalictrum delavayi | 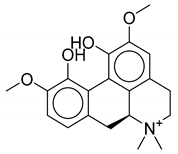 |
| 8 | Thalictrum delavayi |  |
| 9 | Thalictrum delavayi | 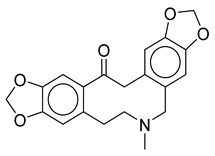 |
| 10 | Thalictrum delavayi | 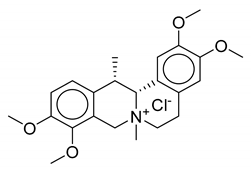 |
| Class of CNN Model | Batch Size | Epoch | Latent Dimensionality of Encoding Space | Latent Dimensionality of Decoding Space | Optimizer |
|---|---|---|---|---|---|
| Solvolysis of methanol | 64 | 100 | 256 | 64 | Adam |
| Solvolysis of ethanol | 64 | 500 | 256 | 64 | Adam |
| Solvolysis of acetone | 64 | 100 | 256 | 64 | Adam |
| Solvolysis of dichloromethane | 64 | 500 | 256 | 64 | Adam |
| Solvolysis of chloroform | 64 | 1000 | 256 | 64 | Adam |
| Solvolysis of water | 64 | 500 | 256 | 64 | Adam |
| Oxidation | 64 | 500 | 256 | 64 | Adam |
| Class of CNN Model | Success | Concordance | Accuracy |
|---|---|---|---|
| Solvolysis of methanol | 88.21% | 0.93 | 75.72% |
| Solvolysis of ethanol | 86.80% | 0.87 | 78.27% |
| Solvolysis of acetone | 98.18% | 0.97 | 87.91% |
| Solvolysis of dichloromethane | 95.00% | 0.97 | 89.64% |
| Solvolysis of chloroform | 88.64% | 0.96 | 85.23% |
| Solvolysis of water | 82.33% | 0.86 | 70.40% |
| Oxidation | 86.86% | 0.85 | 71.07% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.; Chen, W.; Zhou, J.; Dai, J.; Li, Y.; Zhao, Y. Virtual Screening for Reactive Natural Products and Their Probable Artifacts of Solvolysis and Oxidation. Biomolecules 2020, 10, 1486. https://doi.org/10.3390/biom10111486
Xu T, Chen W, Zhou J, Dai J, Li Y, Zhao Y. Virtual Screening for Reactive Natural Products and Their Probable Artifacts of Solvolysis and Oxidation. Biomolecules. 2020; 10(11):1486. https://doi.org/10.3390/biom10111486
Chicago/Turabian StyleXu, Tingjun, Weiming Chen, Junhong Zhou, Jingfang Dai, Yingyong Li, and Yingli Zhao. 2020. "Virtual Screening for Reactive Natural Products and Their Probable Artifacts of Solvolysis and Oxidation" Biomolecules 10, no. 11: 1486. https://doi.org/10.3390/biom10111486
APA StyleXu, T., Chen, W., Zhou, J., Dai, J., Li, Y., & Zhao, Y. (2020). Virtual Screening for Reactive Natural Products and Their Probable Artifacts of Solvolysis and Oxidation. Biomolecules, 10(11), 1486. https://doi.org/10.3390/biom10111486





