1. Introduction
Mass Spectrometry Imaging (MSI) is a label-free analytical technique that can locate chemical compounds (metabolites, peptides, lipids, or proteins) directly in a biological sample and give their concentration for every pixel. The most common analytical strategy is MALDI due to its soft ionization, fast analysis, high throughput, versatility, and selectivity [
1]. Other techniques, like desorption electrospray ionization (DESI), are becoming more popular because of the simplicity of their sample preparation [
2]. MSI is currently used in the fields of drug discovery and toxicology [
3,
4]. In most experiments, researchers use a targeted strategy, which consists of visualizing and (sometimes) quantifying the concentration of a particular compound, or a reduced set of compounds throughout the tissue. Many MSI software packages have been released [
5]. However, none of them provides an automated workflow for untargeted MSI applications since the end-user has to approach each MSI experiment data analysis in its unique manner.
Besides annotating and identifying the MS ions, one of the main challenges in untargeted MSI analysis is to determine the statistically differentiating ions in different regions of interest (ROIs) of the same tissue section or in different tissues of case versus control experiments. These key ions could be associated with biomarker candidates of disease or treatment efficacy. Previous studies have successfully used segmentation processes to find these key ions between clusters [
6,
7]. Most of these studies identify the key ions associated with a certain region by analysing the ions that most influence the segmenting process. In [
8], the authors applied a Non-negative Matrix Factorization multivariate analysis to select a reduced group of lipid MS signals associated with the metabolite profile of each component. The
t-test associated with segmentation with Spatial Shrunken Centroids can find the enriched and absent MS peaks for a particular region in a segmented image [
9,
10]. A technique based on deep unsupervised neural networks and parametric
t-SNE was used to detect metabolic hidden sub-regions [
11]. The same algorithm, linked to a significance analysis of microarrays (SAM), detected the protein subpopulations that can differentiate between
t-SNE segments in a dataset of breast cancer samples; interestingly, they used the selected ions for a kNN second segmentation step [
12]. Gorzolka et al. [
13] studied the space-time profiling of the barley germination process by carrying out an unsupervised joint segmentation of a high number of images and found the ion-associated profile for every segment. The Algorithm for MSI Analysis by Semi-supervised Segmentation (AMASS) was used to segment leech embryo samples [
14] and there is a complete analysis of the ions associated to every region according to its weighting factors. In all these references, no statistical significance test was conducted on the key ions found.
Another common strategy in MSI data analysis is to manually define the ROIs to be compared, guided by an annotated histology image [
15,
16,
17,
18]. In general, the ions are selected by means of statistical hypothesis testing and the fold change (FC) calculation of the ion concentrations between ROIs. These parameters are usually represented as volcano plots. By way of example, Hong et al. [
19] studied the global changes of phospholipids in brain samples from a mouse model of Alzheimer disease by performing ANOVA tests of ion concentrations in ROI. A common problem that MSI has in calculating statistical significance is that the
p-values are generally extremely low [
16]. This is because there are a large number of pixels within each ROI, which gives this parameter a low discrimination power.
Additionally, the statistical hypothesis testing (such as the
t-test) fails when is applied to compare the concentration of an ion between ROIs. The existence of morphological areas in the images is the responsible of a high pixel autocorrelation. This violates the assumption of observation independence necessary for statistical hypothesis testing. In order to find statistically significant ions between ROIs, Conditional Autoregressive (CAR) models, which take into account the auto-correlated nature of ion distribution concentration in MS image ROIs, are calculated to correct the
p-values [
20]. Nevertheless, the difficulty of calculating the autocorrelation models and the complexity of the computational approach hampers the inclusion of this strategy in a MSI workflow.
Another common situation in MS imaging is the elevated intensity differences of the ions’ concentration between pixels, due to the existence of several morphologic regions with different metabolic profiles [
21] and the ion shielding phenomena which takes place in MSI. It is also common to find a high proportion of pixels where a certain ion is not detected, for a given signal to noise ratio. This influences to a large extend the calculation of the
p-values and the FC.
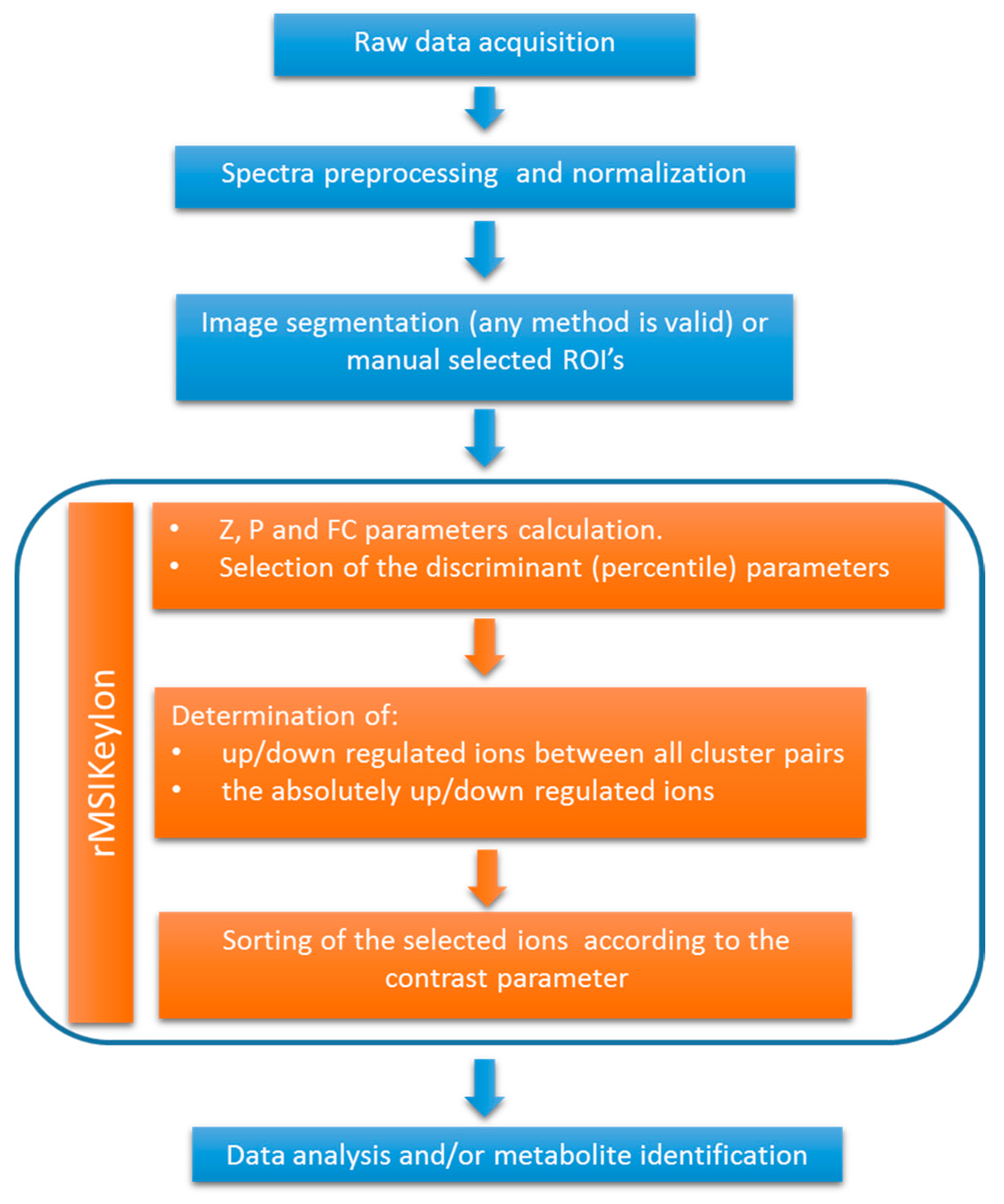
In this study, we describe the development of an ion filtering algorithm that is used in a workflow for the untargeted analysis of metabolomic MALDI-MS images. The workflow consists of a segmentation step, followed by the ion filtering procedure, independent of the segmentation process, that detects the up/down regulated ions between image segments. Our algorithm calculates and combines three parameters: (a) the Mann–Whitney U statistical test of the ion concentration between segments [
22]; (b) the FC in the ion concentration between segments; and (c) a new parameter that accounts for the proportion of pixels with undetected ions between segments. In addition, the data from which parameters (a) and (b) are derived is obtained by previously filtering out the undetected MS signals (null values). With this methodology, we can find the key ions between any segment pair in MSI datasets, from single or multiple tissue sections. We successfully applied this workflow to the analysis of mouse brain tissue sample and to study fatty liver disease in mice liver tissue samples.
3. Discussion
Here, we developed a new methodology for the untargeted analysis of MS images that can be used coupled with any segmentation process and an ion filtering algorithm based on the combination of three parameters: (a) The ratio of ions with a null concentration between the regions, (b) the U Mann–Whitney U Test, calculated by segregating the non-detected ions from the distribution, and (c) the FC between the medians of the distribution (the non-detected ions were also segregated from the distribution). This methodology has proved to be efficient at finding the up/down-expressed ions in an intra-image analysis or in the comparative analysis of groups of images. The presented workflow is different to previously released software tools due to two main reasons: (a) it is flexible and independent to the segmentation process, so the ion selection process can be applied to any clustering algorithm or manually drawn ROIs. (b) Our methodology provides a completely automated ion filtering approach enabling the fast detection of a morphological region characteristic ions.
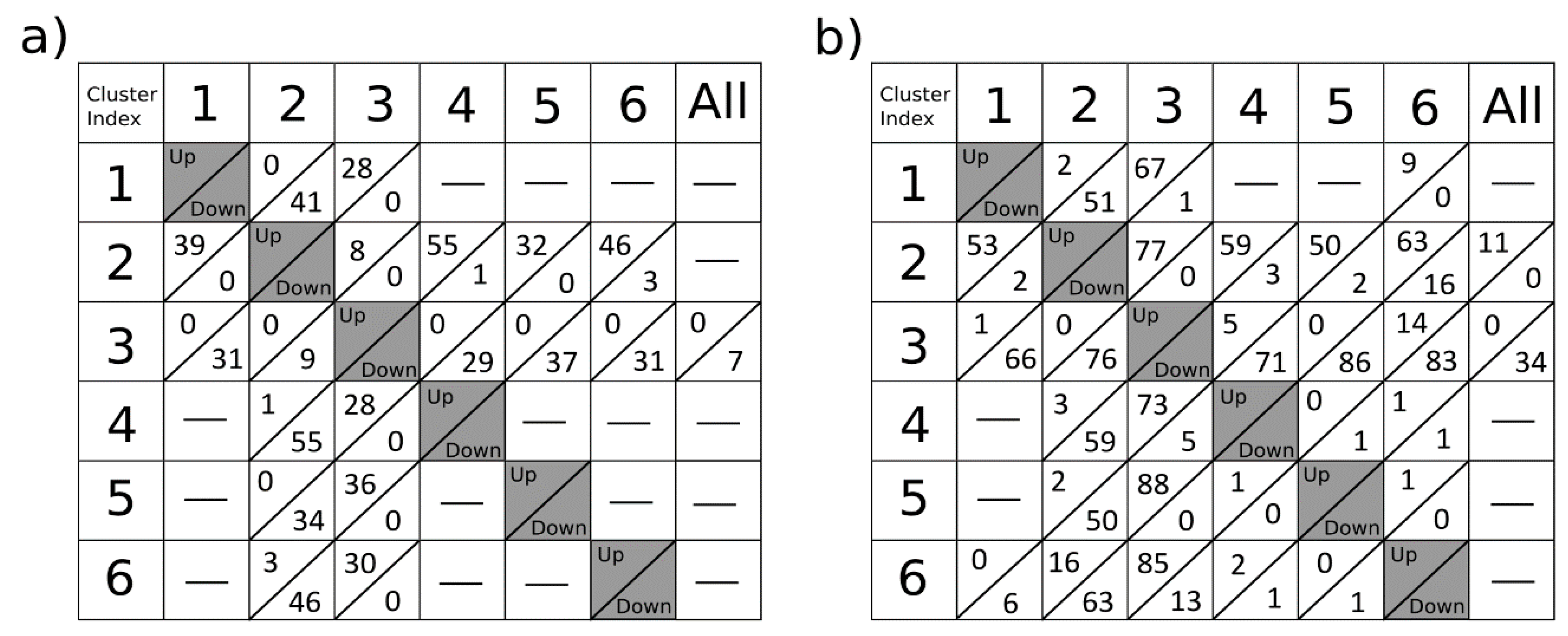
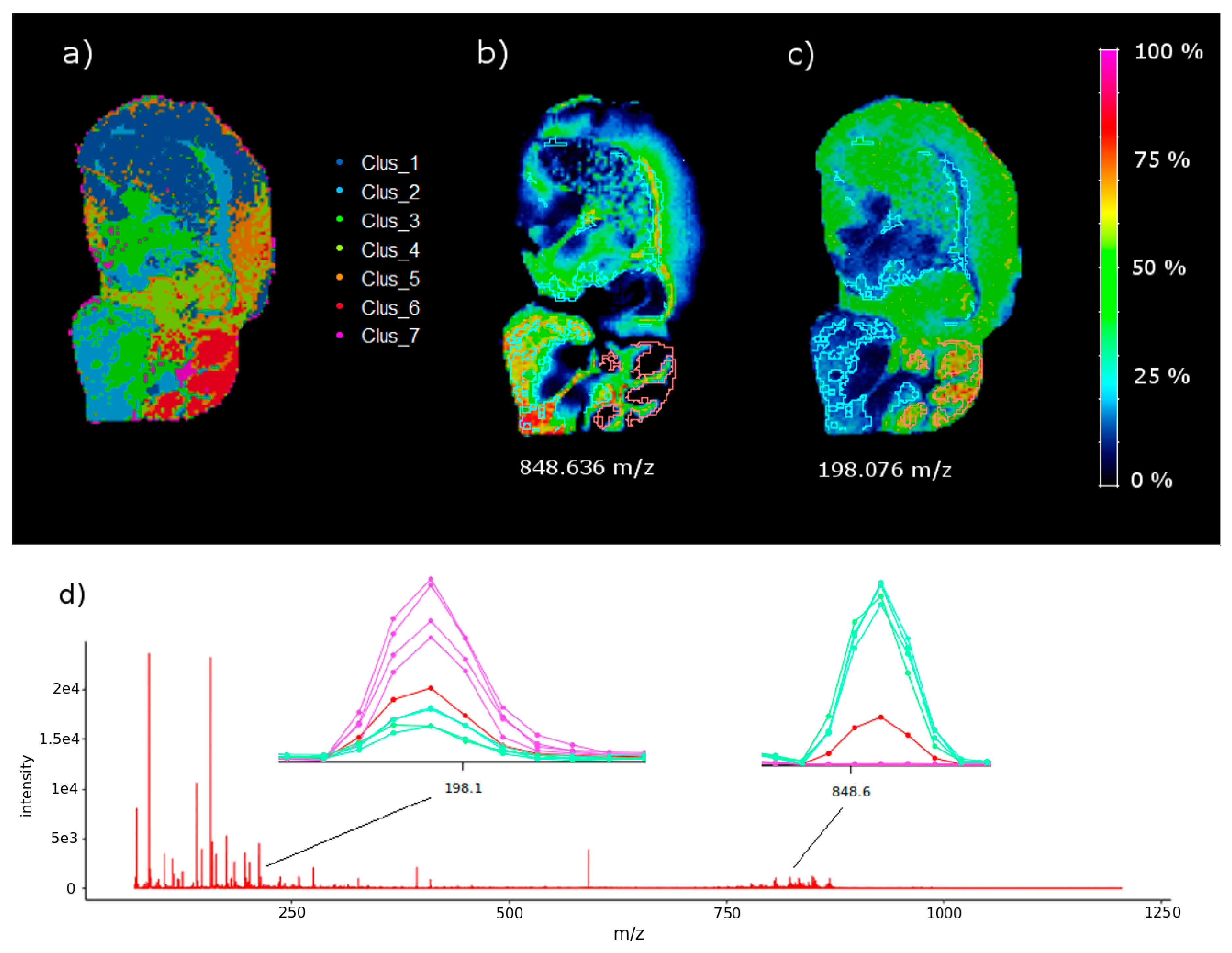
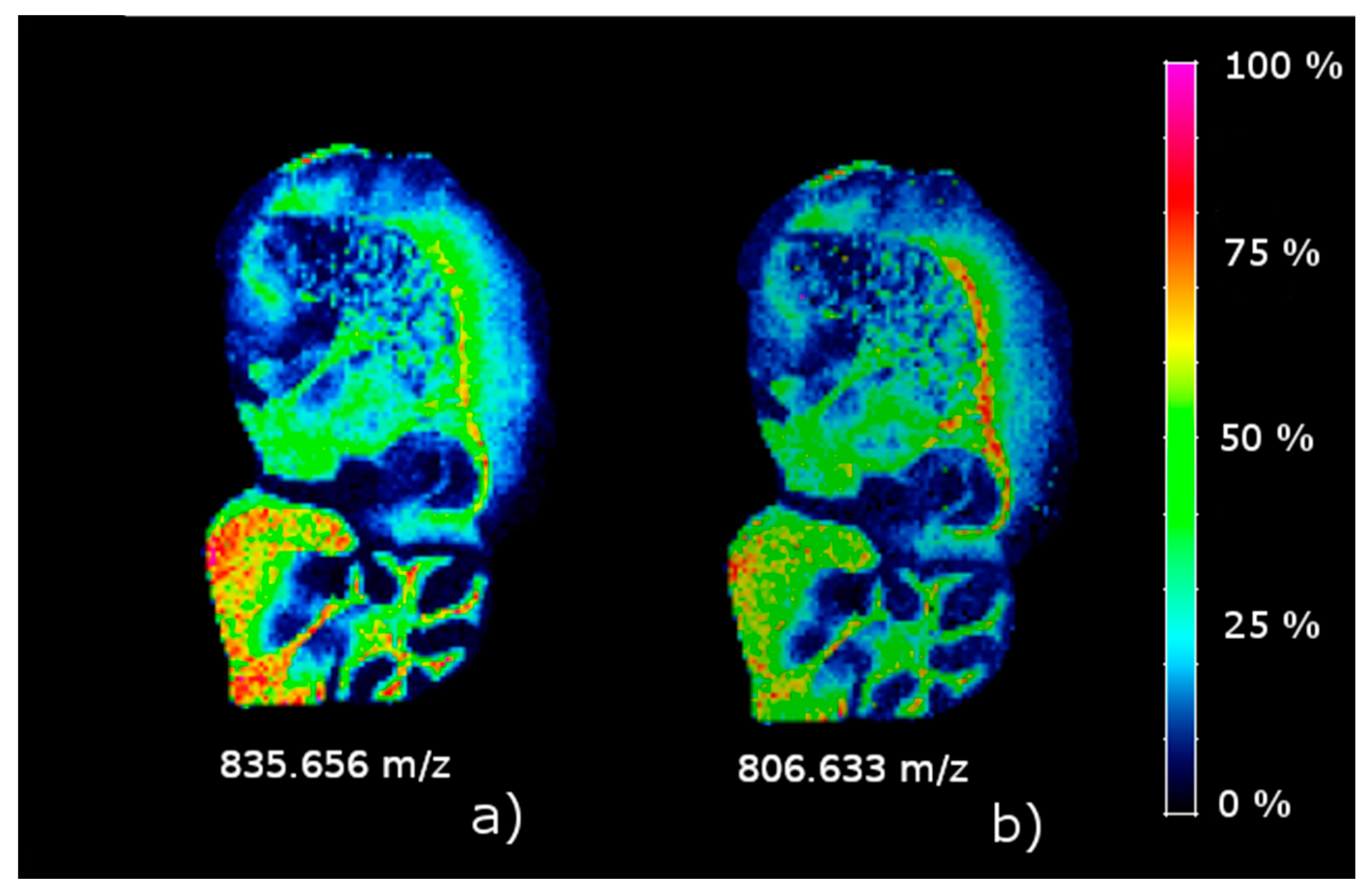
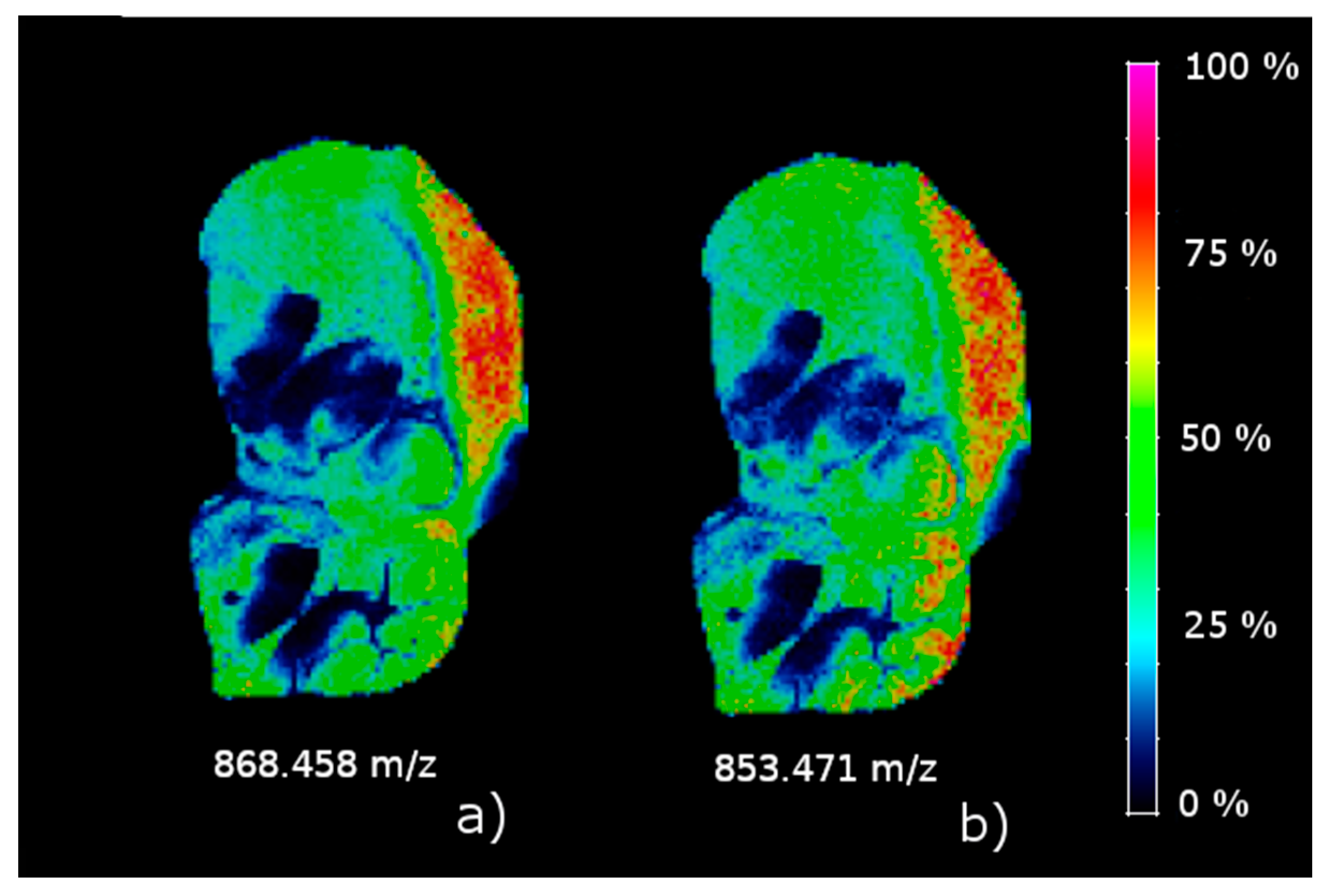
The results on the sagittal mouse brain sample show that an unsupervised clustering process followed by the rMSIKeyIon algorithm is able to select the (possible) up/down-regulated ions between any pair of clusters, in a holistic approach, and between one cluster and the rest. The concentration maps of the selected ions, ordered by the contrast parameter, depicts faithfully the morphology of the brain. These ions are probably biologically relevant and could be interesting to identify.
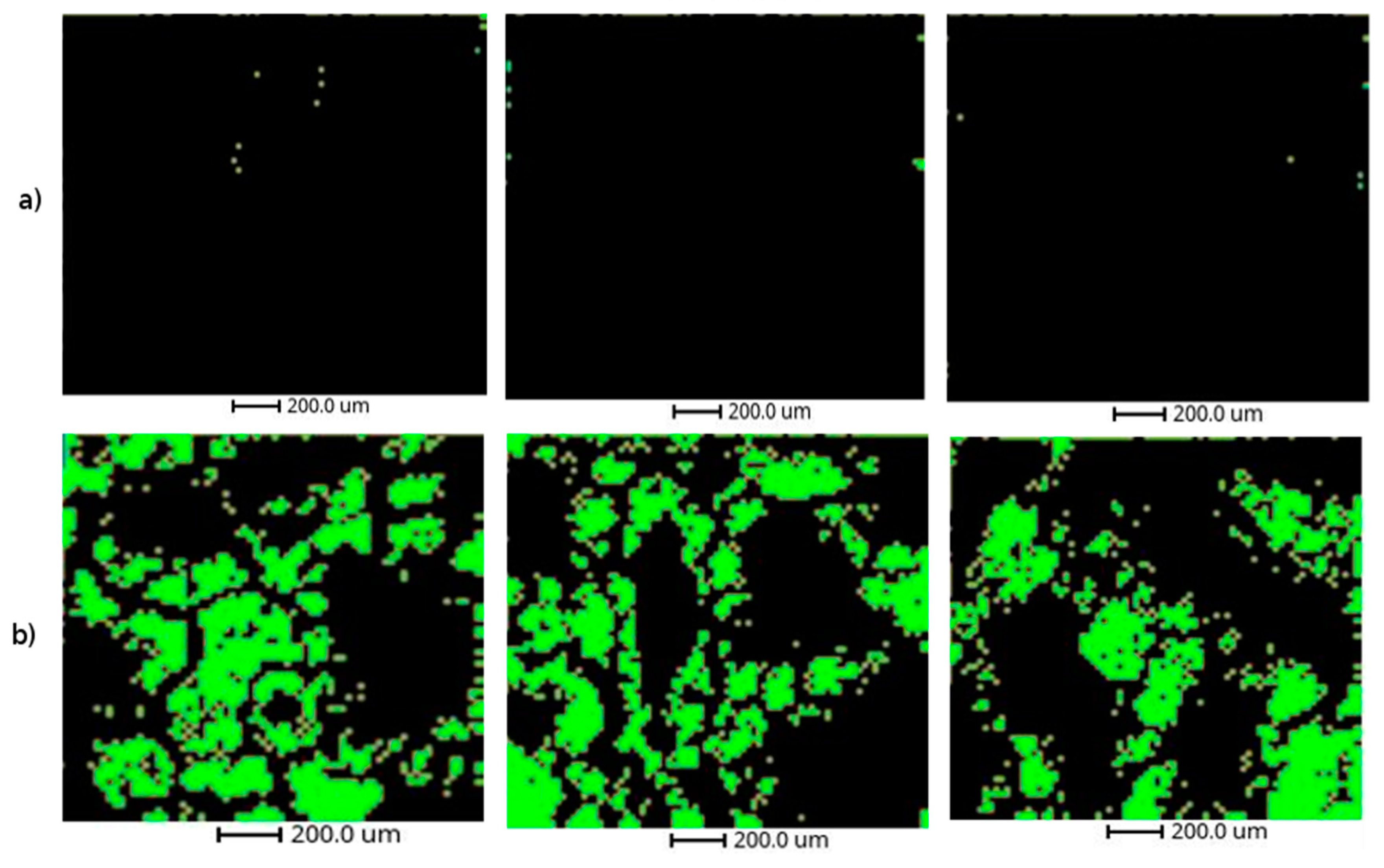
Using the described methodology, we have been able to detect the regions containing the lipid droplets in the liver samples from mouse exposed to THS. The putative identification of the key up-regulated ions in the cluster 2, mainly triglycerides and phosphatidylcholines, confirm that THS exposure conducts to the apparition of fatty liver disease in mice [
23].
Untargeted metabolomics data analysis workflows are associated to standard analytical platforms (LC-MS, GC-MS, and NMR) [
24]. These analyses compare the concentrations of chemical compounds in a CASE and a CONTROL group in order to discover features that they express differently and which could be used as biomarkers or in biological pathway analysis. In general, the number of samples (n) of each experimental group are similar, the distribution is normal (for large n values), and the principle of independent measures is assumed. However, in spatial metabolomics, the number of samples in every group (i.e., the number of pixels in an ROI) is not determined a priori, as in metabolomics studies.
Untargeted image analysis has two main applications:
(a) The comparison of two regions inside the same tissue section (intra-image analysis) to find the relevant ions. This could be used to discover cancer biomarkers by comparing the ion profile of the tumorous area with a non-tumorous area from the same sample. In general, the areas to be compared are determined by a histopathologist annotating a consecutive tissue section. The size of the ROIs in which we will compare the ions is determined manually.
(b) For several reasons, the analysis of morphologically equivalent regions in different tissues in a case-control experiment is much more complicated. First of all, the tissue samples to be compared between groups are equivalent but not similar because of the biological differences between the animals and the intrinsic difficulty of achieving identical tissue sections. Consequently, it is not straightforward to delimit the areas to be compared. The ROIs to be compared can be determined by histological annotation (supervised process), or automatically by means of a segmentation process (unsupervised process). In both cases, there are not established rules, and the following steps in the statistical analysis of the ions between ROIs can be highly affected by this fact.
In both cases, it is very common to find skewed ion distributions and a high percentage of null values, a high degree of autocorrelation between pixels, and a very high number of observations (pixels). This leads to extremely low
p-values when classical parametric or non-parametric statistical tests are used [
25], so these tests are not appropriate for this kind of analysis. For all the above reasons, the untargeted analysis of images remains a challenge. However, the results shown by rMSIKeyIon R package have been revealed to be very useful to find the most differential ions between ROIs. The biological relevance of these ions has been validated in a fatty liver study with animal models.
Author Contributions
X.C. and E.C. designed and conducted the research. M.M.-G. designed the animal model experiments, and generated and collected the mice samples, and M.S. processed the liver and brain samples. P.R. acquired the images and processed the data, N.R. supervised the biological interpretation and S.T. worked on the putative identification of the metabolites in the liver samples. E.d.C. and L.S. programmed the ion filtering routine software. E.d.C., X.C. and N.R. wrote the article and L.S. was in charge of the illustrations. All the authors revised the manuscript for important intellectual content and read and approved the final manuscript.
Funding
This study has been supported by the Spanish Ministry of Economy and Competitiveness through projects TEC2015-69076-P and RTI2018-096061-B-I00, PR’s predoctoral grant No. BES-2013-065572 and the General Directorate of Research of the Government of Catalonia through project 2017 SGR 1119. Animal model development was funded by the Tobacco Research Disease Related Program (TRDRP) of the University of California under projects 22RT-0121 and 23DT-0103.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Calculation of the Similarity Parameters between ROIs
In order to determine the ions that are expressed differently in two given ROIs, we calculate three parameters:
(a) The null concentration parameter (Z parameter)
The
parameter is calculated according to Equation (A1):
where
is the parameter that accounts for the null values (i.e., the non-detected values) of the
i ion when comparing the
j and
k ROIs;
and
are the number of pixels with null values of the
i ion in
j and
k ROIs, respectively;
and
are the total number of ROI pixels in
j and
k, respectively;
I is the set of ions and
Sp is the set of ROIs.
The equation calculates the ratio between the null values of a particular ion in the two ROIs. A value of ( being a positive value greater than 1) means that the i ion is more expressed in k ROI than in j ROI, while ( being a positive value much lower than 1) means that the i ion is less expressed in k ROI than in j ROI.
The importance of this parameter is assessed in
Figure S7. For clusters 1 to 7, we plotted, the percentage of pixels that have null concentration for every ion.
The and values are calculated by following these steps:
- (1)
The Z values of all ions, for all cluster-pairs, are calculated according to Equation (A1).
- (2)
An ordered rank list of all the Z values is created.
- (3)
is determined considering that this value is a certain percentile PZ of the rank list of Z values.
- (4)
is determined considering that this value is a certain percentile 100 − PZ of the rank list of Z values.
(b) Non-null concentration parameters (V parameters)
Provided that the distribution of the ions concentration is non-normal, we considered the U Mann–Whitney U test and the concentration FC between two ROIs, as a non-null concentration parameters.
Generally speaking, if
Nj and
Nk are high, the random variable
U can be regarded as normally distributed [
22]. The
parameter is then normalized following Equation (A2):
where
and
are the average and standard deviation of zero
and
is a random variable with a normalized Gaussian distribution. If
V has values close to 1 the similarity between the distributions is high, while values close to zero indicate disparate distributions. The value obtained for
V indicates the similarity between the distributions of two ROIs for an ion.
Another parameter often used to compare sets of magnitudes is the FC, defined as the ion median concentration quotient between two ROIs Equation (A3):
where
is the distribution median of the
i ion in
j ROI and
is the same for
k ROI. For every
i ion, the
parameter is calculated between the
j and
k ROIs. For a pair of ROIs, a Volcano plot [
31] can be drawn from the
V and
FC parameters.
In this representation, the position occupied by the ions is important: the ions located in the top corners generate very different distributions in the two ROIs. The ions at the top left are under-expressed () and the ions at the top right are over-expressed ().
The values , and are calculated following the same steps as for and , but with a difference in the percentile value. The ions located in the areas of interest must satisfy the probability of being within a range associated with two random variables; that is to say:
for under-expressed ions and for over-expressed ions. Assuming that these are independent random variables, we obtain . That is, the percentile that has to be used to determine the cutoff values in the volcano plot should be
References
- Karas, M.; Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988, 60, 2299–2301. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, J.M.; Ifa, D.R.; Song, Q.; Cooks, R.G. Tissue imaging at atmospheric pressure using Desorption Electrospray Ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. 2006, 45, 7188–7192. [Google Scholar] [CrossRef] [PubMed]
- Morosi, L.; Zucchetti, M.; D’Incalci, M.; Davoli, E. Imaging mass spectrometry: Challenges in visualization of drug distribution in solid tumors. Curr. Opin. Pharmacol. 2013, 13, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Greer, T.; Sturm, R.; Li, L. Mass spectrometry imaging for drugs and metabolites. J. Proteom. 2011, 74, 2617–2631. [Google Scholar] [CrossRef] [PubMed]
- Ràfols, P.; Vilalta, D.; Brezmes, J.; Cañellas, N.; del Castillo, E.; Yanes, O.; Ramírez, N.; Correig, X. Signal preprocessing, multivariate analysis and software tools for MA(LDI)-TOF mass spectrometry imaging for biological applications. Mass Spectrom. Rev. 2018, 37, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, T. MALDI imaging mass spectrometry: Statistical data analysis and current computational challenges. BMC Bioinform. 2012, 13, S11. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.A.; Deininger, S.O.; Hogendoorn, P.C.; Deelder, A.M.; McDonnell, L.A. Imaging mass spectrometry statistical analysis. J. Proteom. 2012, 75, 4962–4989. [Google Scholar] [CrossRef]
- Lee, D.Y.; Platt, V.; Bowen, B.; Louie, K.; Canaria, C.A.; McMurray, C.T.; Northen, T. Resolving brain regions using nanostructure initiator mass spectrometry imaging of phospholipids. Integr. Biol. 2012, 4, 693–699. [Google Scholar] [CrossRef][Green Version]
- Bemis, K.D.; Harry, A.; Eberlin, L.S.; Ferreira, C.R.; van de Ven, S.M.; Mallick, P.; Stolowitz, M.; Vitek, O. Probabilistic Segmentation of Mass Spectrometry (MS) Images Helps Select Important Ions and Characterize Confidence in the Resulting Segments. Mol. Cell. Proteom. 2016, 15, 1761–1772. [Google Scholar] [CrossRef]
- Bemis, K.D.; Harry, A.; Eberlin, L.S.; Ferreira, C.; van de Ven, S.M.; Mallick, P.; Stolowitz, M.; Vitek, O. Cardinal: An R package for statistical analysis of mass spectrometry-based imaging experiments. Bioinformatics 2015, 31, 2418–2420. [Google Scholar] [CrossRef]
- Inglese, P.; McKenzie, J.S.; Mroz, A.; Kinross, J.; Veselkov, K.; Holmes, E.; Takats, Z.; Nicholson, J.K.; Glen, R. Deep learning and 3D-DESI imaging reveal the hidden metabolic heterogeneity of cancer. Chem. Sci. 2017, 8, 3500–3511. [Google Scholar] [CrossRef] [PubMed]
- Abdelmoula, W.M.; Balluff, B.; Englert, S.; Dijkstra, J.; Reinders, M.J.; Walch, A.; McDonnell, L.A.; Lelieveldt, B.P. Data-Driven Identification of Prognostic Tumor Subpopulations Using Spatially Mapped t-SNE of Mass Spectrometry Imaging Data. Proc. Natl. Acad. Sci. USA 2016, 113, 12244–12249. [Google Scholar] [CrossRef] [PubMed]
- Gorzolka, K.; Kölling, J.; Nattkemper, T.W.; Niehaus, K. Spatio-Temporal metabolite profiling of the barley germination process by MALDI MS imaging. PLoS ONE 2016, 11, e0150208. [Google Scholar] [CrossRef] [PubMed]
- Bruand, J.; Alexandrov, T.; Sistla, S.; Wisztorski, M.; Meriaux, C.; Becker, M.; Salzet, M.; Fournier, I.; Macagno, E.; Bafna, V. AMASS: Algorithm for MSI analysis by semi-supervised segmentation. J. Proteome Res. 2011, 10, 4734–4743. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Gordaliza, E.; Esteban-Fernández, D.; Lázaro, A.; Aboulmagd, S.; Humanes, B.; Tejedor, A.; Linscheid, M.W.; Gómez-Gómez, M.M. Lipid imaging for visualizing cilastatin amelioration of cisplatin-induced nephrotoxicity. J. Lipid Res. 2018, 59, 1561–1574. [Google Scholar] [CrossRef] [PubMed]
- Yajima, Y.; Hiratsuka, T.; Kakimoto, Y.; Ogawa, S.; Shima, K.; Yamazaki, Y.; Yoshikawa, K.; Tamaki, K.; Tsuruyama, T. Region of Interest analysis using mass spectrometry imaging of mitochondrial and sarcomeric proteins in acute cardiac infarction tissue. Sci. Rep. 2018, 8, 7493. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Han, J.; Hardie, D.B.; Yang, J.; Pan, J.; Borchers, C.H. Metabolomic profiling of prostate cancer by matrix assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry imaging using Matrix Coating Assisted by an Electric Field (MCAEF). Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 755–767. [Google Scholar] [CrossRef]
- Otsuka, Y.; Satoh, S.; Naito, J.; Kyogaku, M.; Hashimoto, H. Visualization of cancer-related chemical components in mouse pancreas tissue by tapping-mode scanning probe electrospray ionization mass spectrometry. J. Mass Spectrom. 2015, 50, 1157–1162. [Google Scholar] [CrossRef]
- Hong, J.H.; Kang, J.W.; Kim, D.K.; Baik, S.H.; Kim, K.H.; Shanta, S.R.; Jung, J.H.; Mook-Jung, I.; Kim, K.P. Global changes of phospholipids identified by MALDI imaging mass spectrometry in a mouse model of Alzheimer’s disease. J. Lipid Res. 2016, 57, 36–45. [Google Scholar] [CrossRef]
- Cassese, A.; Ellis, S.R.; Ogrinc Potočnik, N.; Burgermeister, E.; Ebert, M.; Walch, A.; Van Den Maagdenberg, A.M.; McDonnell, L.A.; Heeren, R.M.; Balluff, B. Spatial Autocorrelation in Mass Spectrometry Imaging. Anal. Chem. 2016, 88, 5871–5878. [Google Scholar] [CrossRef]
- Chernyavsky, I.; Nikolenko, S.; von Eggeling, F.; Alexandrov, T.; Becker, M. Analysis and Interpretation of Imaging Mass Spectrometry Data by Clustering Mass-to-Charge Images According to Their Spatial Similarity. Anal. Chem. 2013, 85, 11189–11195. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Martins-Green, M.; Adhami, N.; Frankos, M.; Valdez, M.; Goodwin, B.; Lyubovitsky, J.; Dhall, S.; Garcia, M.; Egiebor, I.; Martinez, B.; et al. Cigarette smoke toxins deposited on surfaces: Implications for human health. PLoS ONE 2014, 9, e86391. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Fagerland, M.W. t-tests, non-parametric tests, and large studies—A paradox of statistical practice? BMC Med. Res. Methodol. 2012, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Adhami, N.; Starck, S.R.; Flores, C.; Green, M.M. A health threat to bystanders living in the homes of smokers: How smoke toxins deposited on surfaces can cause insulin resistance. PLoS ONE 2016, 11, e0149510. [Google Scholar] [CrossRef]
- Ràfols, P.; Vilalta, D.; Torres, S.; Calavia, R.; Heijs, B.; McDonnell, L.A.; Brezmes, J.; del Castillo, E.; Yanes, O.; Ramírez, N.; et al. Assessing the potential of sputtered gold nanolayers in mass spectrometry imaging for metabolomics applications. PLoS ONE 2018, 13, e0208908. [Google Scholar] [CrossRef] [PubMed]
- Ràfols, P.; Torres, S.; Ramírez, N.; Del Castillo, E.; Yanes, O.; Brezmes, J.; Correig, X. rMSI: An R package for MS imaging data handling and visualization. Bioinformatics 2017, 33, 2427–2428. [Google Scholar] [CrossRef] [PubMed]
- Ràfols, P.; del Castillo, E.; Yanes, O.; Brezmes, J.; Correig, X. Novel automated workflow for spectral alignment and mass calibration in MS imaging using a sputtered Ag nanolayer. Anal. Chim. Acta 2018, 1022, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Mak, T.D.; Laiakis, E.C.; Goudarzi, M.; Fornace, A.J. MetaboLyzer: A Novel Statistical Workflow for Analyzing Postprocessed LC–MS Metabolomics Data. Anal. Chem. 2014, 86, 506–513. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).