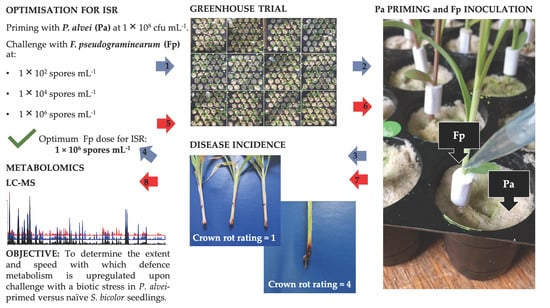
Differential Metabolic Reprogramming in Paenibacillus alvei-Primed Sorghum bicolor Seedlings in Response to Fusarium pseudograminearum Infection
Abstract
1. Introduction
2. Results
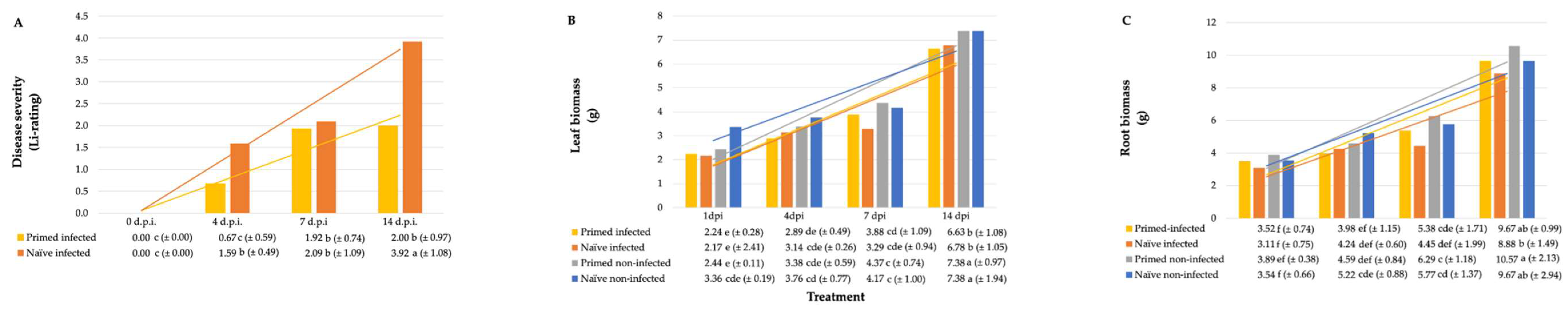
2.1. Plant Growth Parameters and Crown Rot Disease Severity
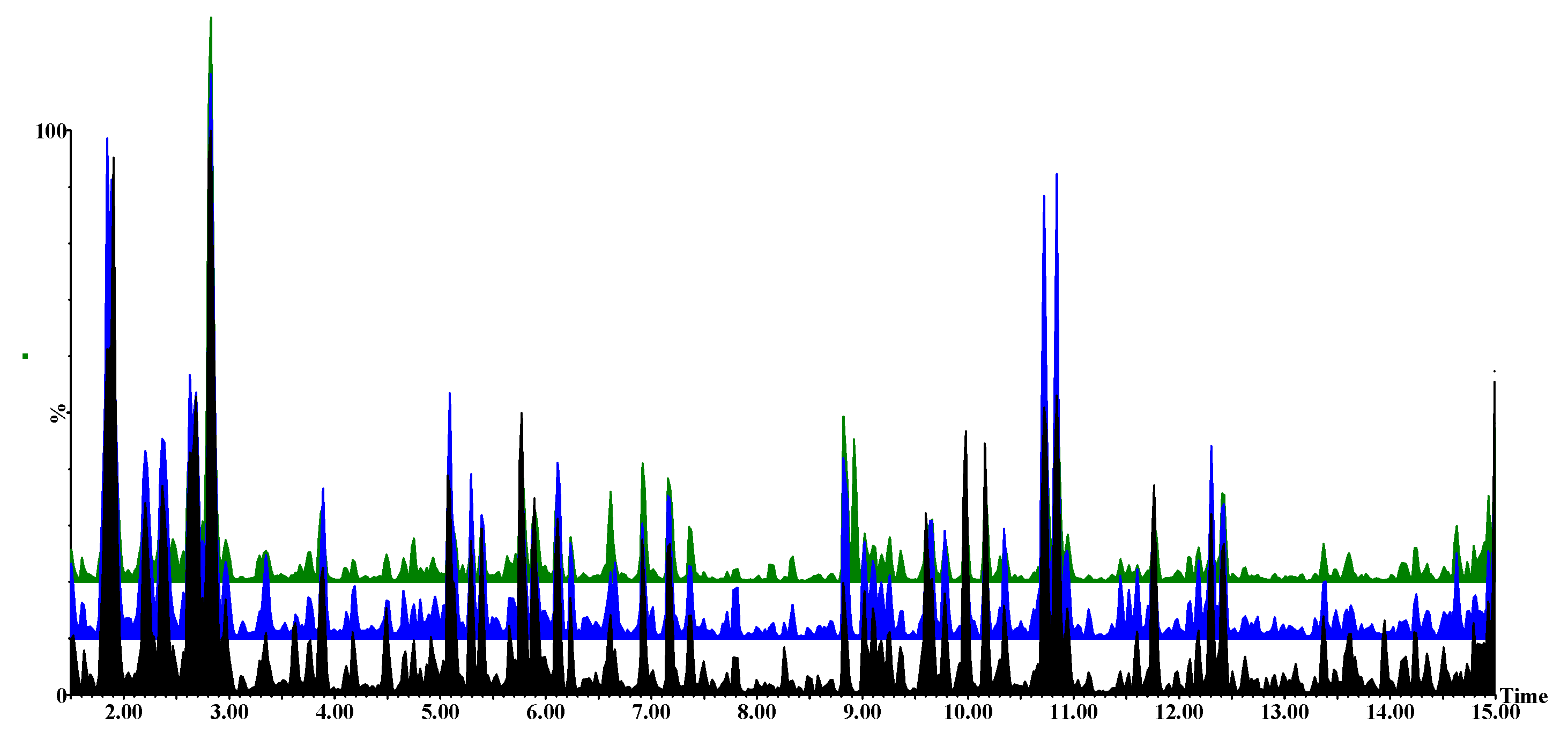
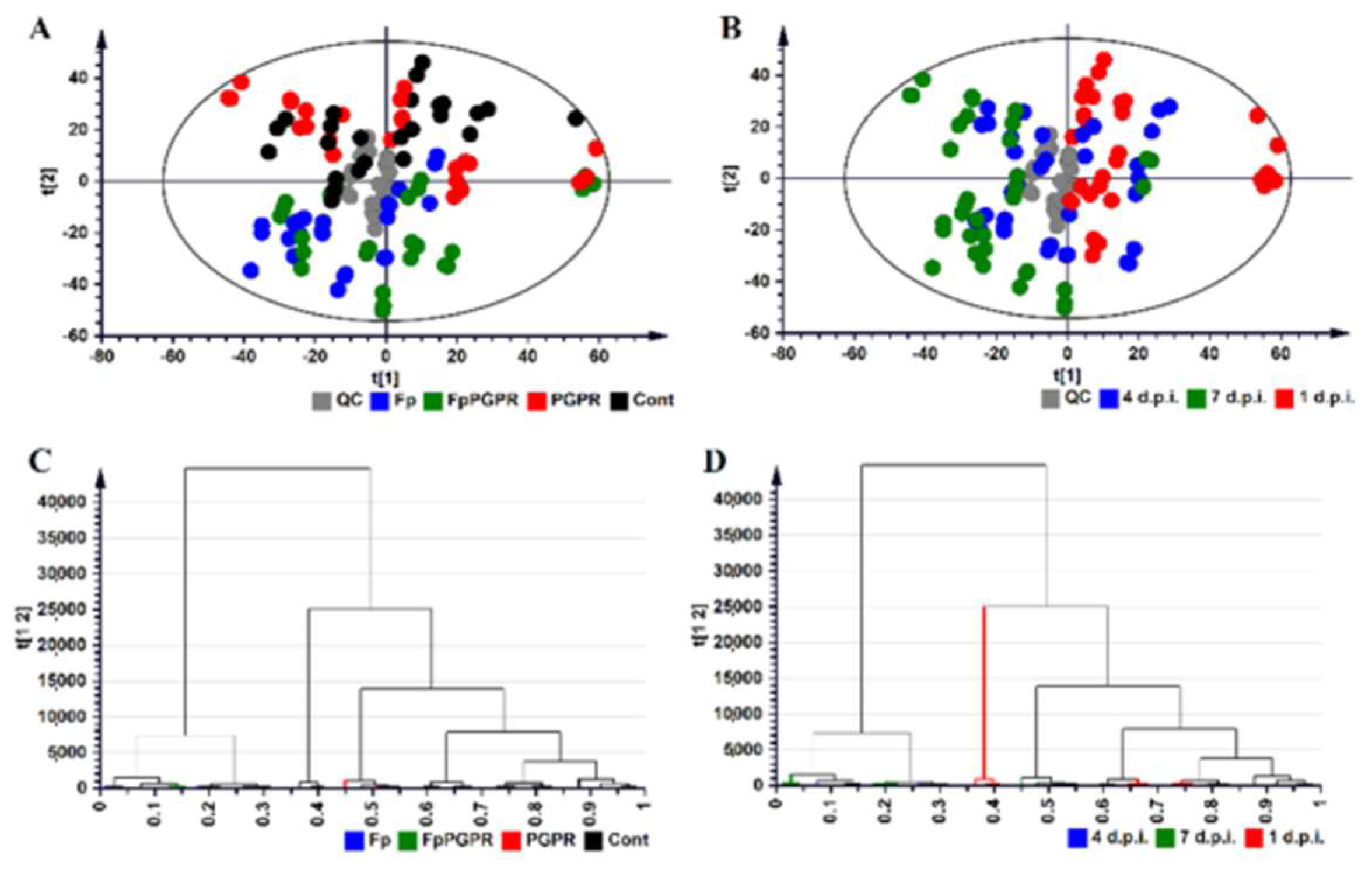
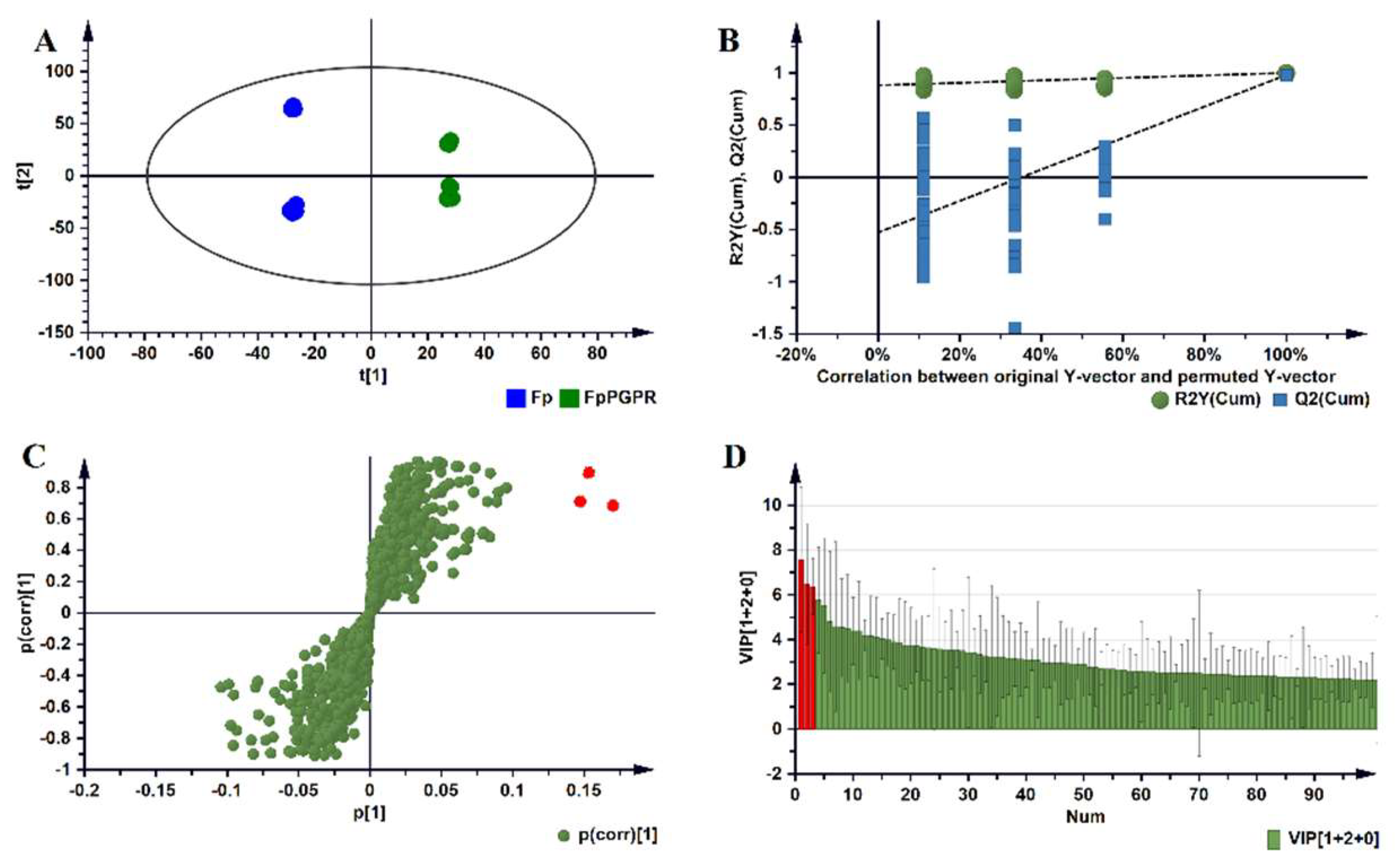
2.2. Metabolomic Profiles of Paenibacillus alvei-Primed and Naïve Fusarium pseudograminearum Infected Sorghum Plants
| Treatment | Crown Rot | Fresh Mass | Dry Mass | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. alvei | F. pseudograminearum | Li-Rating 3 | Isolations 4 | Leaves | Roots | Leaves | Roots | ||||||
| (cfu mL−1) 2 | (spores mL−1) | (%) | (g) | (g) | (g) | (g) | |||||||
| 0 | 0 | 0.00 (±0.00) | e | 0.00 (±0.00) | c | 0.68 (±0.16) | c | 0.73 (±0.30) | a | 0.14 (±0.07) | a | 0.04 (±0.01) | ab |
| 1 × 108 | 0 | 0.00 (±0.00) | e | 0.00 (±0.00) | c | 0.75 (±0.21) | ab | 0.80 (±0.35) | a | 0.14 (±0.06) | a | 0.05 (±0.02) | a |
| 0 | 1 × 102 | 1.14 (±1.03) | cd | 22.67 (±8.84) | ab | 0.65 (±0.21) | c | 0.74 (±0.36) | a | 0.11 (±0.07) | b | 0.05 (±0.02) | a |
| 0 | 1 × 104 | 1.90 (±0.96) | b | 24.00 (±9.66) | ab | 0.67 (±0.18) | c | 0.68 (±0.28) | a | 0.10 (±0.05) | b | 0.05 (±0.02) | a |
| 0 | 1 × 106 | 3.06 (±1.80) | a | 26.00 (±6.99) | a | 0.55 (±0.11) | d | 0.55 (±0.19) | a | 0.06 (±0.02) | c | 0.04 (±0.02) | ab |
| 1 × 108 | 1 × 102 | 0.57 (±0.86) | de | 21.33 (±10.60) | ab | 0.58 (±0.19) | d | 0.65 (±0.32) | a | 0.10 (±0.07) | b | 0.03 (±0.00) | b |
| 1 × 108 | 1 × 104 | 1.32 (±1.05) | bc | 21.00 (±11.01) | ab | 0.70 (±0.24) | bc | 0.78 (±0.32) | a | 0.11 (±0.06) | b | 0.05 (±0.03) | a |
| 1 × 108 | 1 × 106 | 1.67 (±1.35) | bc | 18.00 (±13.17) | b | 0.76 (±0.18) | a | 0.81 (±0.38) | a | 0.11 (±0.07) | b | 0.05 (±0.03) | a |
3. Discussion
4. Materials and Methods
4.1. Greenhouse Assessment of Induced Systemic Resistance
4.1.1. Inoculum Preparation
4.1.2. Sorghum Cultivation
4.1.3. Treatment (Priming) with Paenibacillus alvei and Inoculation with Fusarium pseudograminearum
4.1.4. Crown Rot Disease Severity and Confirmation of Koch’s Postulates
4.1.5. Statistical Analyses of Growth Parameters and Disease Assessments
4.2. Metabolite Profiling
4.2.1. Sample Collection
4.2.2. Metabolite Extraction
4.2.3. Ultra-High Performance Liquid Chromatography-High Definition Mass Spectrometry Analysis
4.2.4. Data Analysis
4.2.5. Metabolite Annotation
4.2.6. Metabolic Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data availability
References
- Walters, D.; Walsh, D.; Newton, A.; Lyon, G. Induced resistance for plant disease control: Maximizing the efficacy of resistance elicitors. Phytopathology 2005, 95, 1368–1373. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Pieterse, C.M.J.; Mauch-Mani, B. Priming in plant—Pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; Métraux, J.P.; Van Loon, L.C. Inducing resistance: A summary of papers presented at the first international symposium on induced resistance to plant diseases, Corfu, May 2000. Eur. J. Plant Pathol. 2001, 107, 1–6. [Google Scholar] [CrossRef]
- Walters, D.; Heil, M. Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 2007, 71, 3–17. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Bakker, P.A.H.M.; Pieterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.; Van Wees, S.; Ton, J.; Pelt, J.A.; Van Loon, L.C. Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol. 2002, 4, 535–544. [Google Scholar] [CrossRef]
- Van Peer, R.; Niemann, G.J.; Schippers, B. Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Am. Phytopathol. Soc. 1991, 81, 728–734. [Google Scholar] [CrossRef]
- Ongena, M.; Daayf, F.; Jacques, P.; Thonart, P.; Benhamou, N.; Paulitz, T.C.; Bélanger, R.R. Systemic induction of phytoalexins in cucumber in response to treatments with fluorescent pseudomonads. Plant Pathol. 2000, 49, 523–530. [Google Scholar] [CrossRef]
- Ramamoorthy, V.; Viswanathan, R.; Raguchander, T.; Prakasam, V.; Samiyappan, R. Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Prot. 2001, 20, 1–11. [Google Scholar] [CrossRef]
- Conrath, U. Priming of induced plant defense responses. Adv. Bot. Res. 2009, 51, 361–395. [Google Scholar]
- Garcion, C.; Lamotte, O.; Metraux, J.P. Mechanism of defence to pathogens: Biochemistry and physiology. In Induced Resistance for Plant Defence: A Sustainable Approach to Crop Protection; Walters, D., Newton, A., Lyon, G., Eds.; Blackwell Publishing: Oxford, UK, 2007; pp. 109–132. ISBN 9781405134477. [Google Scholar]
- Somers, E.; Vanderleyden, J.; Srinivasan, M. Rhizosphere bacterial signalling: A love parade beneath our feet. Crit. Rev. Microbiol. 2004, 30, 205–240. [Google Scholar] [CrossRef] [PubMed]
- DeBrito Alvarez, M.; Gagne, S.; Antoun, H. Effect of compost on rhizosphere microflora of the tomato and on the incidence of plant growth-promoting rhizobacteria. Appl. Environ. Microbiol. 1995, 61, 194–199. [Google Scholar]
- Bashan, Y.; De-Bashan, L.E. How the plant growth-promoting bacterium Azospirillum promotes plant growth—A critical assessment. In Advances in Agronomy; Sparks, D., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 108, pp. 77–136. ISBN 9780123810328. [Google Scholar]
- Luz, E.; Hernandez, J.; Bashan, Y.; Maier, R.M. Bacillus pumilus ES4: Candidate plant growth-promoting bacterium to enhance establishment of plants in mine tailings. Environ. Exp. Bot. 2010, 69, 343–352. [Google Scholar]
- Ghyselinck, J.; Velivelli, S.L.S.; Heylen, K.; O’Herlihy, E.; Franco, J.; Rojas, M.; De Vos, P.; Prestwich, B.D. Bioprospecting in potato fields in the central Andean highlands: Screening of rhizobacteria for plant growth-promoting properties. Syst. Appl. Microbiol. 2013, 36, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Harish, S.; Kavino, M.; Kumar, N.; Saravanakumar, D.; Soorianathasundaram, K.; Samiyappan, R. Biohardening with plant growth promoting rhizosphere and endophytic bacteria induces systemic resistance against banana bunchy top virus. Appl. Soil Ecol. 2008, 39, 187–200. [Google Scholar] [CrossRef]
- Walters, D.R. Are plants in the field already induced? Implications for practical disease control. Crop Prot. 2009, 28, 459–465. [Google Scholar] [CrossRef]
- Mareya, C.; Tugizimana, F.; Piater, L.; Madala, N.; Steenkamp, P.; Dubery, I. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 2019, 9, 8. [Google Scholar] [CrossRef]
- Van Wyk, P.S.; Scholtz, D.S. A selective medium for the isolation of Fusarium spp. from soil debris. Phytophylactica 1995, 18, 67–69. [Google Scholar]
- Li, X.; Liu, C.; Chakraborty, S.; Manners, J.M.; Kazan, K. A simple method for the assessment of crown rot disease severity in wheat seedlings inoculated with Fusarium pseudograminearum. J. Phytopathol. 2008, 156, 751–754. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Vos, C.M.F.; De Cremer, K.; Cammue, B.P.A.; De Coninck, B. The toolbox of Trichoderma spp. in the biocontrol of Botrytis cinerea disease. Mol. Plant Pathol. 2015, 16, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Buensanteai, N.; Yuen, G.Y.; Prathuangwong, S. Priming, signaling, and protein production associated with induced resistance by Bacillus amyloliquefaciens KPS46. World J. Microbiol. Biotechnol. 2009, 25, 1275–1286. [Google Scholar] [CrossRef]
- Gutiérrez-Mañero, F.J.; Ramos-Solano, B.; Probanza, A.; Mehouachi, J.; Tadeo, F.R.; Talon, M. The plant-growth-promoting rhizobacteria Bacillus pumilus and Bacillus licheniformis produce high amounts of physiologically active gibberellins. Physiol. Plant. 2001, 111, 206–211. [Google Scholar] [CrossRef]
- Bottini, R.; Cassán, F.; Piccoli, P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 2004, 65, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Brotman, Y.; Lisec, J.; Meret, M.; Chet, I.; Willmitzer, L.; Viterbo, A. Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology 2012, 158, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.N.; Wallsgrove, R.M. Secondary metabolites in plant defence mechanisms. New Phytol. 1994, 127, 617–633. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef]
- Carrasco, A.; Boudet, A.M.; Marigo, G. Enhanced resistance of tomato plants to Fusarium by controlled stimulation of their natural phenolic production. Physiol. Plant Pathol. 1978, 12, 225–232. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 2014, 83, 40–50. [Google Scholar] [CrossRef]
- Skadhauge, B.; Thomsen, K.K.; Wettstein, D. The role of the barley testa layer and its flavonoid content in resistance to Fusarium infections. Hereditas 2004, 126, 147–160. [Google Scholar] [CrossRef]
- Bollina, V.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol. Biol. 2011, 77, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Iranshahi, M.; Rezaee, R.; Parhiz, H.; Roohbakhsh, A.; Soltani, F. Protective effects of flavonoids against microbes and toxins: The cases of hesperidin and hesperetin. Life Sci. 2015, 137, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Kostyn, K.; Czemplik, M.; Kulma, A.; Bortniczuk, M.; Skała, J.; Szopa, J. Genes of phenylpropanoid pathway are activated in early response to Fusarium attack in flax plants. Plant Sci. 2012, 190, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Buśko, M.; Góral, T.; Ostrowska, A.; Matysiak, A.; Walentyn-Góral, D.; Perkowski, J. The effect of Fusarium inoculation and fungicide application on concentrations of flavonoids (apigenin, kaempferol, luteolin, naringenin, quercetin, rutin, vitexin) in winter wheat cultivars. Am. J. Plant Sci. 2014, 5, 3727–3736. [Google Scholar] [CrossRef]
- Steinkellner, S.; Mammerler, R. Effect of flavonoids on the development of Fusarium oxysporum f. sp. lycopersici. J. Plant Interact. 2007, 2, 17–23. [Google Scholar] [CrossRef]
- Merkl, R.; Hrádková, I.; Filip, V.; Šmidrkal, J. Antimicrobial and antioxidant properties of phenolic acids alkyl esters. Czech J. Food Sci. 2010, 28, 275–279. [Google Scholar] [CrossRef]
- Beimen, A.; Witte, L.; Barz, W. Growth characteristics and elicitor-induced reactions of photosynthetically active and heterotrophic cell suspension cultures of Lycopersicon peruvianum (Mill.). Bot. Acta 1992, 105, 152–160. [Google Scholar] [CrossRef]
- Kumaraswamy, K.G.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against Fusarium head blight (Fusarium graminearum). J. Chem. Ecol. 2011, 37, 846–856. [Google Scholar] [CrossRef]
- Berkey, R.; Bendigeri, D.; Xiao, S. Sphingolipids and plant defense/disease: The “death” connection and beyond. Front. Plant Sci. 2012, 3, 1–22. [Google Scholar] [CrossRef]
- Brodersen, P.; Petersen, M.; Pike, H.M.; Olszak, B.; Skov, S.; Ødum, N.; Jørgensen, L.B.; Brown, R.E.; Mundy, J. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002, 16, 490–502. [Google Scholar] [CrossRef]
- Boubakri, H.; Gargouri, M.; Mliki, A.; Brini, F.; Chong, J.; Jbara, M. Vitamins for enhancing plant resistance. Planta 2016, 244, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Asensi-Fabado, M.A.; Munné-Bosch, S. Vitamins in plants: Occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 2010, 15, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Nahrstedt, A. Cyanogenic compounds as protecting agents for organisms. Plant Syst. Evol. 1985, 150, 35–47. [Google Scholar] [CrossRef]
- Adeyanju, A.; Yu, J.; Little, C.; Rooney, W.; Klein, P.; Burke, J.; Tesso, T. Sorghum RILs segregating for stay-green QTL and leaf dhurrin content show differential reaction to stalk rot diseases. Crop Sci. 2016, 56, 1–9. [Google Scholar] [CrossRef]
- Tugizimana, F.; Djami-Tchatchou, A.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2019, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Johnson, S.K.; Bornman, J.F.; Bennett, S.J.; Fang, Z. Changes in whole grain polyphenols and antioxidant activity of six sorghum genotypes under different irrigation treatments. Food Chem. 2017, 214, 199–207. [Google Scholar] [CrossRef]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1846. [Google Scholar] [CrossRef]
- Fong, Y.K.; Anuar, S.; Lim, H.P.; Tham, F.Y.; Sanderson, F.R. A modified filter paper technique for long-term preservation of some fungal cultures. Mycologist 2000, 14, 127–130. [Google Scholar] [CrossRef]
- Bai, G.-H.; Shaner, G. Variation in Fusarium graminearum and cultivar resistance to Wheat Scab. Plant Dis. 1996, 80, 975–979. [Google Scholar] [CrossRef]
- Tugizimana, F. Metabolomic Studies of Induced Defense-Related Changes in Sorghum Bicolor in Response to the Pathogen Colletotrichum Sublineolum. Ph.D. Thesis, University of Johannesburg, Johannesburg, South Africa, 2017. [Google Scholar]
- Mitter, V.; Zhang, M.C.; Liu, C.J.; Ghosh, R.; Ghosh, M.; Chakraborty, S. A high-throughput glasshouse bioassay to detect crown rot resistance in wheat germplasm. Plant Pathol. 2006, 55, 433–441. [Google Scholar] [CrossRef]
- Xi, K.; Turkington, T.K.; Chen, M.H. Systemic stem infection by Fusarium species in barley and wheat. Can. J. Plant Pathol. 2008, 30, 588–594. [Google Scholar] [CrossRef]
- Akinsanmi, O.A.; Mitter, V.; Simpfendorfer, S.; Backhouse, D.; Chakraborty, S. Identity and pathogenicity of Fusarium spp. isolated from wheat fields in Queensland and northern New South Wales. Aust. J. Agric. Res. 2004, 55, 97–107. [Google Scholar] [CrossRef]
- Brown, M.; Dunn, W.B.; Dobson, P.; Patel, Y.; Winder, C.L.; Francis-Mcintyre, S.; Begley, P.; Carroll, K.; Broadhurst, D.; Tseng, A.; et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst 2009, 134, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Wedge, D.C.; Goodacre, R.; Kell, D.B.; Baker, P.N.; Kenny, L.C.; Mamas, M.A.; Neyses, L.; Dunn, W.B. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics 2011, 27, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Chemspider. Available online: http://www.chemspider.com (accessed on 10 June 2018).
- SorghumbicolorCyc. Available online: http://www.plantcyc.org (accessed on 30 September 2018).
- Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.kegg.jp (accessed on 5 June 2018).
- Dykes, L.; Rooney, L.W. Sorghum and millet phenols and antioxidants. J. Cereal Sci. 2006, 44, 236–251. [Google Scholar] [CrossRef]
- Kang, J.; Price, W.E.; Ashton, J.; Tapsell, L.C.; Johnson, S. Identification and characterization of phenolic compounds in hydromethanolic extracts of sorghum wholegrains by LC-ESI-MSn. Food Chem. 2016, 211, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Samuel, T.; Noble, R.; Gmbh, S.D.; Barrett, D.; Beale, M.H.; Hardy, N. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- MetaboAnalyst. Available online: http://www.metaboanalyst.ca (accessed on 15 October 2018).
- Aoki, T.; O’Donnell, K. Morphological characterization of Gibberella coronicola sp. nov., obtained through mating experiments of Fusarium pseudograminearum. Mycoscience 1999, 40, 443–453. [Google Scholar] [CrossRef]
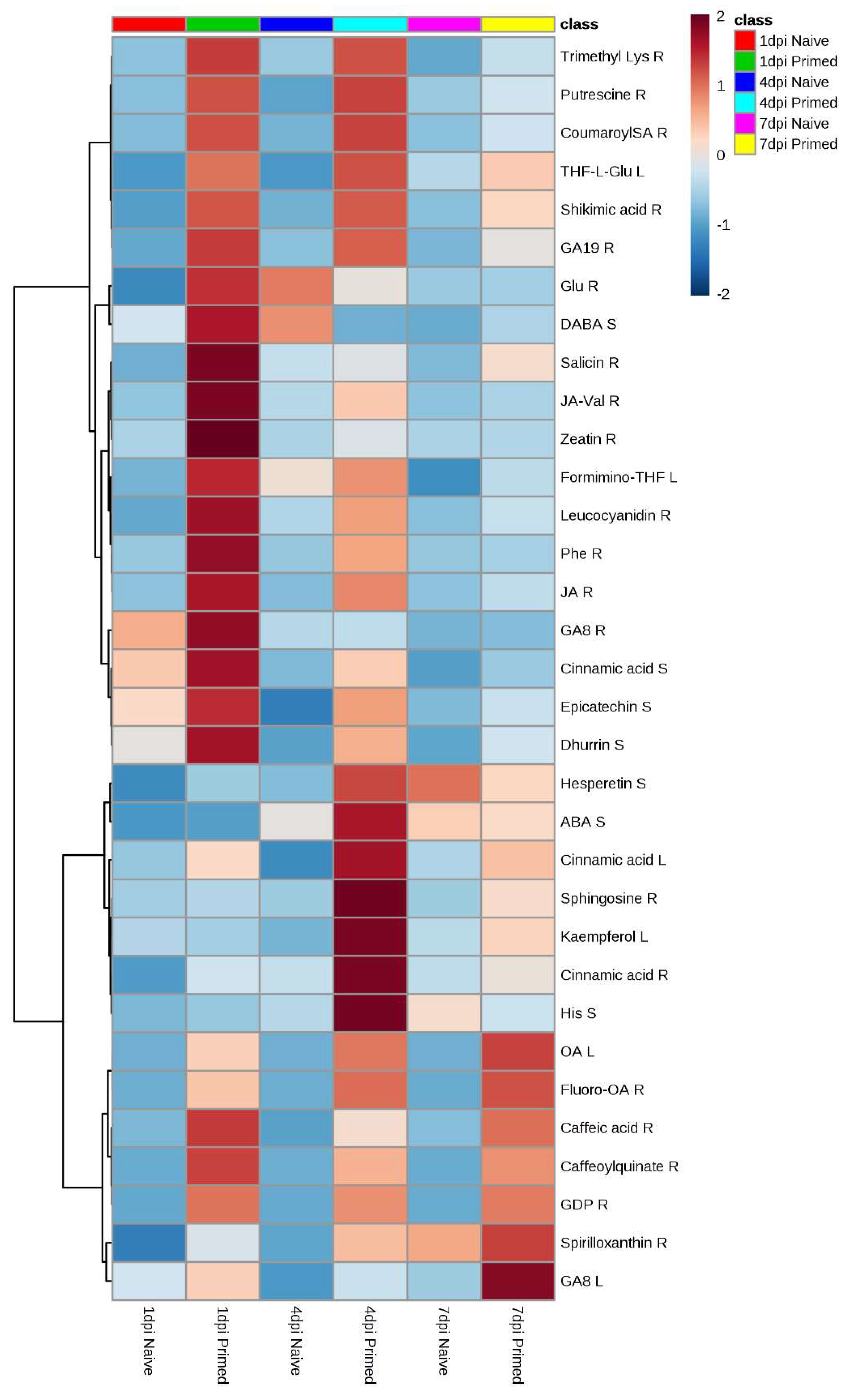

| Metabolite Fold Change (fc) and p-Value of Primed vs. Naïve Seedlings | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Heatmap Key | m/z | Rt (min) | ESI Mode | Molecular Formula/KEGG ID | Metabolite Class | 1 d.p.i. | 4 d.p.i. | 7 d.p.i. | ||||
| fc | p | fc | p | fc | p | ||||||||
| 1 | N-Carbamoylputrescine | Putrescine R | 176.08 | 4.66 | Pos | C5H13N3O/C00436 | Amino acid | 3.481 | 1.67 × 10−5 | 5.401 | 2.00 × 10−7 | 1.426 | 1.79 × 10−2 |
| 2 | L-2,4-Diaminobutanoate | DABA S | 163.05 | 5.59 | Pos | C4H10N2O2/C03283 | Amino acid | 2.373 | 3.97 × 10−2 | 0.287 | 4.30 × 10−2 | 1.660 | 5.44 × 10−1 |
| 3 | THF-L-Glutamate | THF-L-Glu L | 595.19 | 1.65 | Neg | C24H30N8O9/C09332 | Amino acid | 2.623 | 5.39 × 10−2 | 2.770 | 1.12 × 10−3 | 1.401 | 3.45 × 10−1 |
| 4 | Glutathione | Glu R | 326.12 | 2.93 | Pos | C10H17N3O6S/C00051 | Amino acid | 5.562 | 4.22 × 10−2 | 0.649 | 4.74 × 10−1 | 1.045 | 9.54 × 10−1 |
| 5 | 2-(3-Carboxy-3-aminopropyl)-L-histidine | His R | 339.07 | 5.76 | Neg | C10H16N4O4/C04441 | Amino acid | 2.845 | 1.21 × 10−3 | 0.694 | 7.96 × 10−2 | 0.643 | 8.00 × 10−3 |
| 6 | 2-(3-Carboxy-3-aminopropyl)-L-histidine | His S | 301.09 | 8.89 | Pos | C10H16N4O4/C04441 | Amino acid | 2.362 | 2.08 × 10−1 | 6.096 | 1.31 × 10−2 | 0.600 | 3.27 × 10−1 |
| 7 | L-Lysine | Lys R | 213.09 | 2.14 | Neg | C6H14N2O2/C00047 | Amino acid | 0.762 | 4.98 × 10−1 | 4.140 | 1.05 × 10−2 | 1.256 | 4.78 × 10−1 |
| 8 | N6,N6,N6-Trimethyl-L-lysine | Trimethyl Lys R | 313.09 | 3.60 | Neg | C9H20N2O2/C05546 | Amino acid | 5.498 | 1.60 × 10−3 | 4.479 | 1.70 × 10−4 | 4.019 | 1.2 × 10−2 |
| 9 | L-phenylalanine | Phe R | 373.11 | 5.49 | Pos | C12H22N4O7/C00079 | Amino acid | 32.594 | 3.39 × 10−2 | 20.705 | 1.92 × 10−2 | 3.180 | 1.54 × 10−1 |
| 10 | Guanosine 5’-diphosphate | GDP R | 675.88 | 1.15 | Neg | C10H17N5O17P4/C00035 | Purine nucleoside | 129.214 | 1.56 × 10−3 | 80.688 | 1.36 × 10−4 | 48.241 | 6.52 × 10−14 |
| 11 | 8’-Hydroxyabscisate | ABA S | 301.11 | 9.67 | Neg | C15H20O5/C15514 | Phytohormone | 2.403 | 6.41 × 10−2 | 2.502 | 7.68 × 10−3 | 0.913 | 1.98 × 10−2 |
| 12 | Gibberellin A19 | GA19 R | 429.15 | 2.85 | Neg | C20H26O6/C02034 | Phytohormone | 2.055 | 2.94 × 10−5 | 1.777 | 4.46 × 10−2 | 1.345 | 3.92 × 10−2 |
| 13 | Gibberellin A8-catabolite | GA8 L | 383.11 | 8.87 | Neg | C19H22O7/C11870 | Phytohormone | 0.819 | 3.96 × 10−1 | 1.822 | 4.08 × 10−6 | 1.626 | 2.11 × 10−3 |
| 14 | Gibberellin A8-catabolite | GA8 R | 383.11 | 8.91 | Neg | C19H22O7/C11870 | Phytohormone | 1.781 | 1.62 × 10−8 | 1.135 | 6.42 × 10−1 | 2.585 | 2.33 × 10−2 |
| 15 | Salicin | Salicin R | 353.08 | 3.65 | Neg | C13H18O7/C01451 | Phytohormone | 14.920 | 2.96 × 10−2 | 1.278 | 6.61 × 10−1 | 4.174 | 1.14 × 10−1 |
| 16 | (-)-11-Hydroxy-9,10-dihydrojasmonic acid 11-beta-D-glucoside | JA R | 435.19 | 9.22 | Neg | C18H28O9/C21385 | Phytohormone | 27.667 | 1.05 × 10−3 | 78.994 | 9.42 × 10−3 | 4.373 | 2.32 × 10−2 |
| 17 | (-)-Jasmonoyl-L-valine | JA-Val R | 368.16 | 5.49 | Pos | C17H27NO4/C21509 | Phytohormone | 25.519 | 2.37 × 10−2 | 3.025 | 2.40 × 10−1 | 3.127 | 2.64 × 10−1 |
| 18 | Dihydrozeatin | Zeatin R | 242.10 | 6.14 | Neg | C10H15N5O/C02029 | Phytohormone | 96.477 | 2.10 × 10−3 | 15.249 | 1.08 × 10−3 | 2.199 | 2.04 × 10−1 |
| 19 | (-)-Epicatechin | Epicatechin S | 289.07 | 3.50 | Neg | C15H14O6/C09727 | Phenylpropanoid | 1.455 | 7.08 × 10−3 | 2.749 | 1.50 × 10−2 | 1.299 | 4.98 × 10−1 |
| 20 | (-)-Hesperetin | Hesperetin S | 301.07 | 8.28 | Neg | C16H14O6/C01709 | Phenylpropanoid | 3.301 | 1.81 × 10−2 | 3.799 | 4.39 × 10−4 | 0.697 | 4.01 × 10−1 |
| 21 | 4-Coumaroylshikimate | CoumaroylSA R | 336.11 | 4.70 | Neg | C16H16O7/C02947 | Phenylpropanoid | 4.580 | 6.11 × 10−6 | 5.575 | 5.88 × 10−6 | 1.760 | 5.16 × 10−6 |
| 22 | 7-O-D-Glucosyl-apigenin | Apigenin S | 477.10 | 5.95 | Neg | C21H20O10/C04608 | Phenylpropanoid | 0.804 | 5.84 × 10−1 | 1.935 | 1.77 × 10−1 | 9.563 | 3.73 × 10−4 |
| 23 | 8-C-Glucosylnaringenin | Naringenin R | 433.11 | 6.00 | Neg | C21H22O10/C16492 | Phenylpropanoid | 0.631 | 8.01 × 10−2 | 2.588 | 7.39 × 10−3 | 1.683 | 6.31 × 10−2 |
| 24 | Kaempferol 3-O-D-Glucosylgalactoside | Kaempferol L | 609.15 | 5.44 | Neg | C27H30O16/C16490 | Phenylpropanoid | 0.877 | 8.41 × 10−1 | 5.543 | 3.2 × 10−3 | 1.650 | 3.54 × 10−1 |
| 25 | Leucocyanidin | Leucocyanidin R | 322.09 | 4.67 | Neg | C15H14O7/C05906 | Phenylpropanoid | 6.203 | 3.03 × 10−10 | 2.208 | 6.51 × 10−3 | 1.622 | 8.90E × 10−2 |
| 26 | Neohesperidin | Neohesperidin R | 609.18 | 4.91 | Neg | C28H34O15/C09806 | Phenylpropanoid | 0.968 | 6.11 × 10−1 | 2.025 | 6.52 × 10−5 | 2.579 | 3.58 × 10−4 |
| 27 | 5-O-Caffeoylshikimic acid | Shikimic acid R | 352.10 | 4.01 | Neg | C16H16O8/C10434 | Phenylpropanoid | 743.006 | 9.33 × 10−8 | 11.814 | 9.87 × 10−5 | 4.179 | 4.54 × 10−5 |
| 28 | Caffeic acid 3-glucoside | Caffeic acid R | 341.09 | 6.25 | Neg | C15H18O9/C10431 | Phenylpropanoid | 6.106 | 5.83 × 10−2 | 6.521 | 5.20 × 10−2 | 4.845 | 2.63 × 10−2 |
| 29 | Caffeoylquinate | Caffeoylquinate R | 353.09 | 2.62 | Neg | C16H17KO9/C00852 | Phenylpropanoid | 7.071 | 2.65 × 10−2 | 4.685 | 1.35 × 10−2 | 5.660 | 8.72 × 10−2 |
| 30 | Trans-D-Glucosyl-2-hydroxycinnamate | Cinnamic acid L | 325.09 | 3.71 | Neg | C15H18O8/C05158 | Phenylpropanoid | 1.517 | 3.37 × 10−1 | 3.486 | 4.97 × 10−2 | 1.485 | 3.20 × 10−1 |
| 31 | Trans-D-Glucosyl-2-hydroxycinnamate | Cinnamic acid R | 325.09 | 3.75 | Neg | C15H18O8/C05158 | Phenylpropanoid | 1.907 | 1.33 × 10−1 | 2.338 | 2.18 × 10−2 | 1.224 | 4.75 × 10−1 |
| 32 | Trans-D-Glucosyl-2-hydroxycinnamate | Cinnamic acid S | 325.09 | 6.94 | Neg | C15H18O8/C05158 | Phenylpropanoid | 1.644 | 9.05 × 10−3 | 2.238 | 4.44 × 10−5 | 1.634 | 1.73 × 10−1 |
| 33 | (6Z,9Z,12Z)-Octadecatrienoic acid | γ-linolenic acid R | 345.20 | 1.92 | Neg | C18H30O2/C06426 | Lipid | 3.122 | 1.18 × 10−8 | 3.157 | 4.37 × 10−8 | 2.392 | 2.61 × 10−9 |
| 34 | 9-Hydroperoxy-12,13-epoxy-10-octadecenoic acid | OA L | 371.18 | 5.27 | Neg | C18H32O4/C08368 | Lipid | 445.872 | 2.44 × 10−14 | 691.790 | 2.44 × 10−14 | 818.658 | 2.44 × 10−14 |
| 35 | 12,13-Epoxy-9-hydroxy-10-octadecenoate | Oleic acid R | 361.20 | 1.14 | Neg | C18H32O4/C14832 | Lipid | 2.041 | 2.15 × 10−8 | 1.459 | 1.17 × 10−5 | 1.322 | 1.04 × 10−3 |
| 36 | Methyl 9-hydroperoxy-10,12,13,15-bisepidioxy-16E-octadecenoate | Oleic acid L | 461.14 | 1.60 | Neg | C19H32O8/C14832 | Lipid | 3.756 | 4.42 × 10−3 | 0.419 | 2.13 × 10−1 | 0.968 | 9.51 × 10−1 |
| 37 | 18-Fluoro-octadecanoic acid | Fluoro-OA R | 347.23 | 1.57 | Pos | C18H35FO2/C01530 | Lipid | 76.144 | 1.20 × 10−2 | 176.354 | 7.82 × 10−9 | 474.046 | 6.61 × 10−4 |
| 38 | (4E,8E,10E-d18:3) Sphingosine | Sphingosine R | 318.24 | 1.02 | Pos | C18H33NO2/C00319 | Lipid | 9.621 | 2.96 × 10−1 | 288.427 | 2.68 × 10−2 | 111.911 | 2.14 × 10−1 |
| 39 | 3,4,3′,4′-tetrahydrospirilloxanthin | Spirilloxanthin R | 673.42 | 1.21 | Neg | C42H64O2/C15888 | Lipid | 9.271 | 2.34 × 10−2 | 3.857 | 5.20 × 10−3 | 1.329 | 9.69 × 10−2 |
| 40 | 5-Formiminotetrahydrofolate | Formimino-THF L | 516.14 | 5.12 | Neg | C20H24N8O6/C00664 | Vitamin | 3.140 | 3.34 × 10−3 | 1.363 | 3.79 × 10−1 | 2.074 | 6.33 × 10−2 |
| 41 | Dhurrin | Dhurrin S | 310.09 | 2.82 | Neg | C14H17NO7/C05143 | Cyanogenic glucoside | 2.186 | 5.67 × 10−4 | 4.514 | 1.96 × 10−3 | 2.545 | 1.04 × 10−1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Labuschagne, N. Differential Metabolic Reprogramming in Paenibacillus alvei-Primed Sorghum bicolor Seedlings in Response to Fusarium pseudograminearum Infection. Metabolites 2019, 9, 150. https://doi.org/10.3390/metabo9070150
Carlson R, Tugizimana F, Steenkamp PA, Dubery IA, Labuschagne N. Differential Metabolic Reprogramming in Paenibacillus alvei-Primed Sorghum bicolor Seedlings in Response to Fusarium pseudograminearum Infection. Metabolites. 2019; 9(7):150. https://doi.org/10.3390/metabo9070150
Chicago/Turabian StyleCarlson, René, Fidele Tugizimana, Paul A. Steenkamp, Ian A. Dubery, and Nico Labuschagne. 2019. "Differential Metabolic Reprogramming in Paenibacillus alvei-Primed Sorghum bicolor Seedlings in Response to Fusarium pseudograminearum Infection" Metabolites 9, no. 7: 150. https://doi.org/10.3390/metabo9070150
APA StyleCarlson, R., Tugizimana, F., Steenkamp, P. A., Dubery, I. A., & Labuschagne, N. (2019). Differential Metabolic Reprogramming in Paenibacillus alvei-Primed Sorghum bicolor Seedlings in Response to Fusarium pseudograminearum Infection. Metabolites, 9(7), 150. https://doi.org/10.3390/metabo9070150