Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma
Abstract
1. Introduction
2. Results
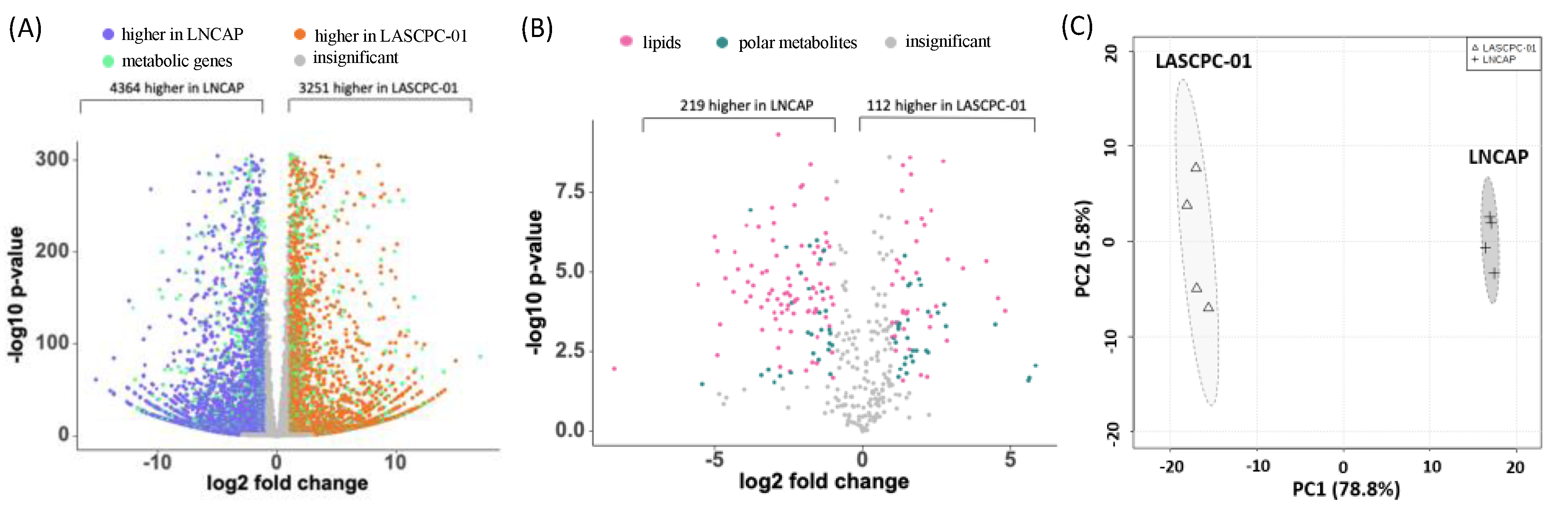
2.1. Transcriptome and Metabolome Profiles Were Different between LASCPC-01 and LNCAP Cell Lines
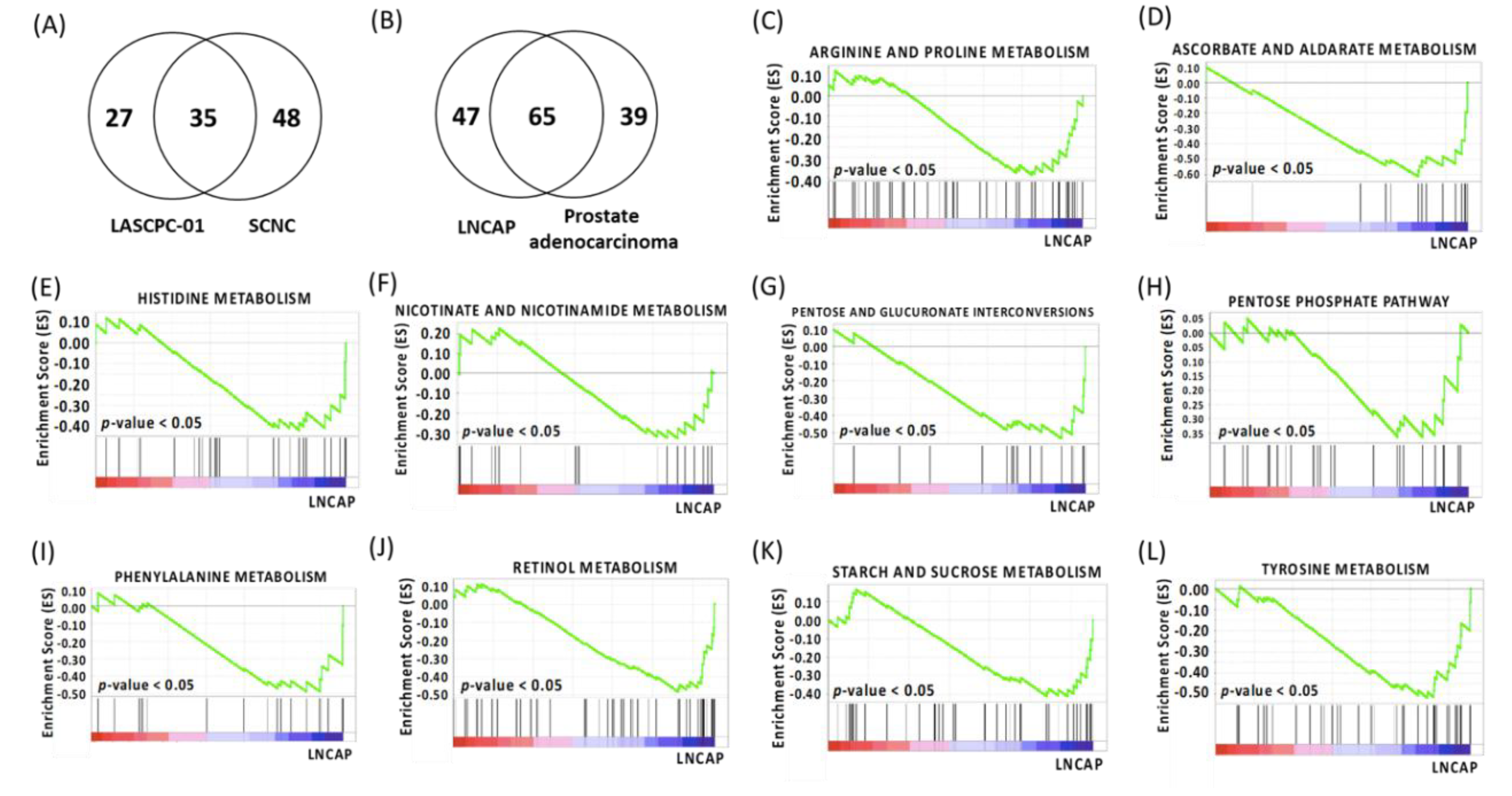
2.2. Gene Set Enrichment Analysis
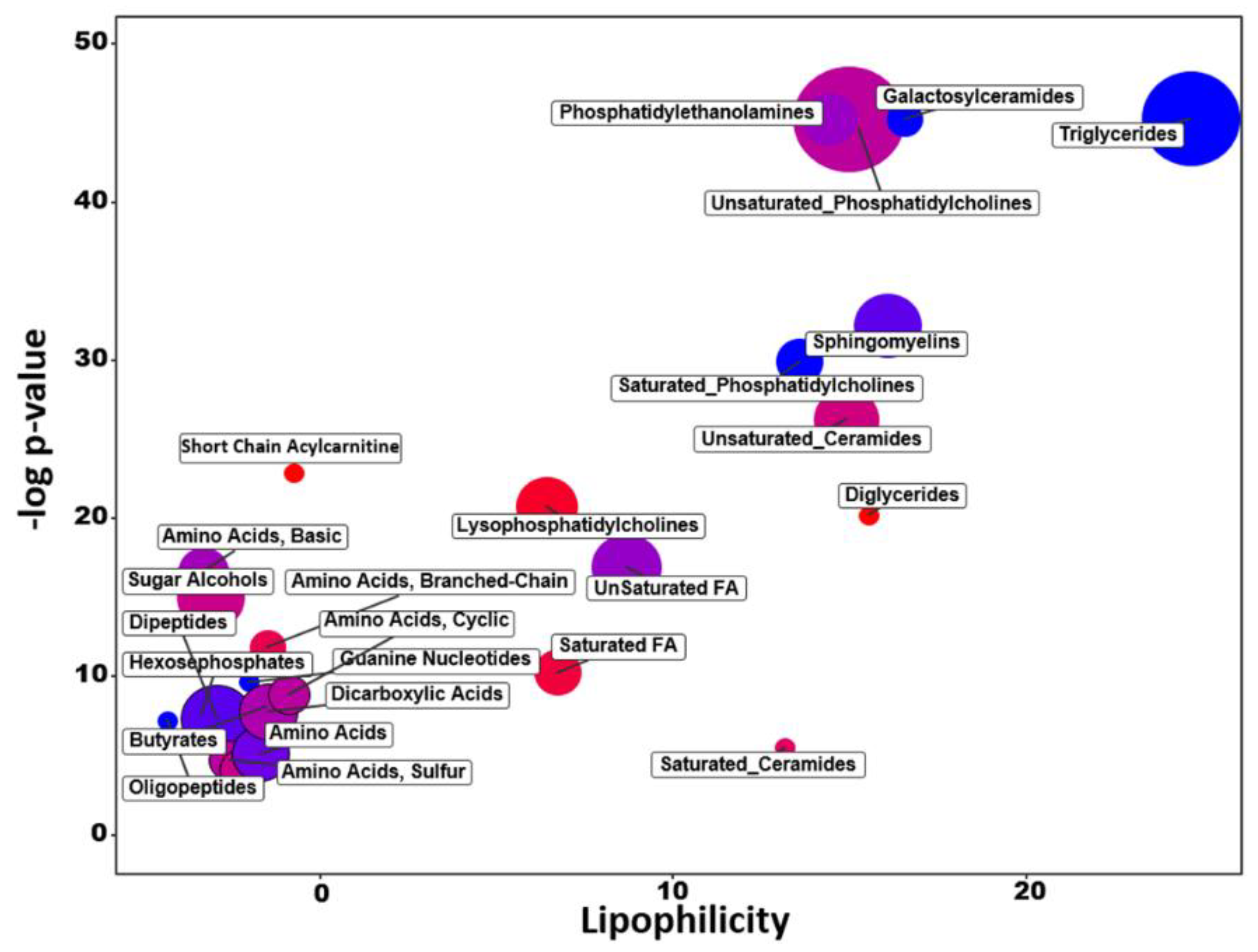
2.3. Chemical Similarity Enrichment Analysis
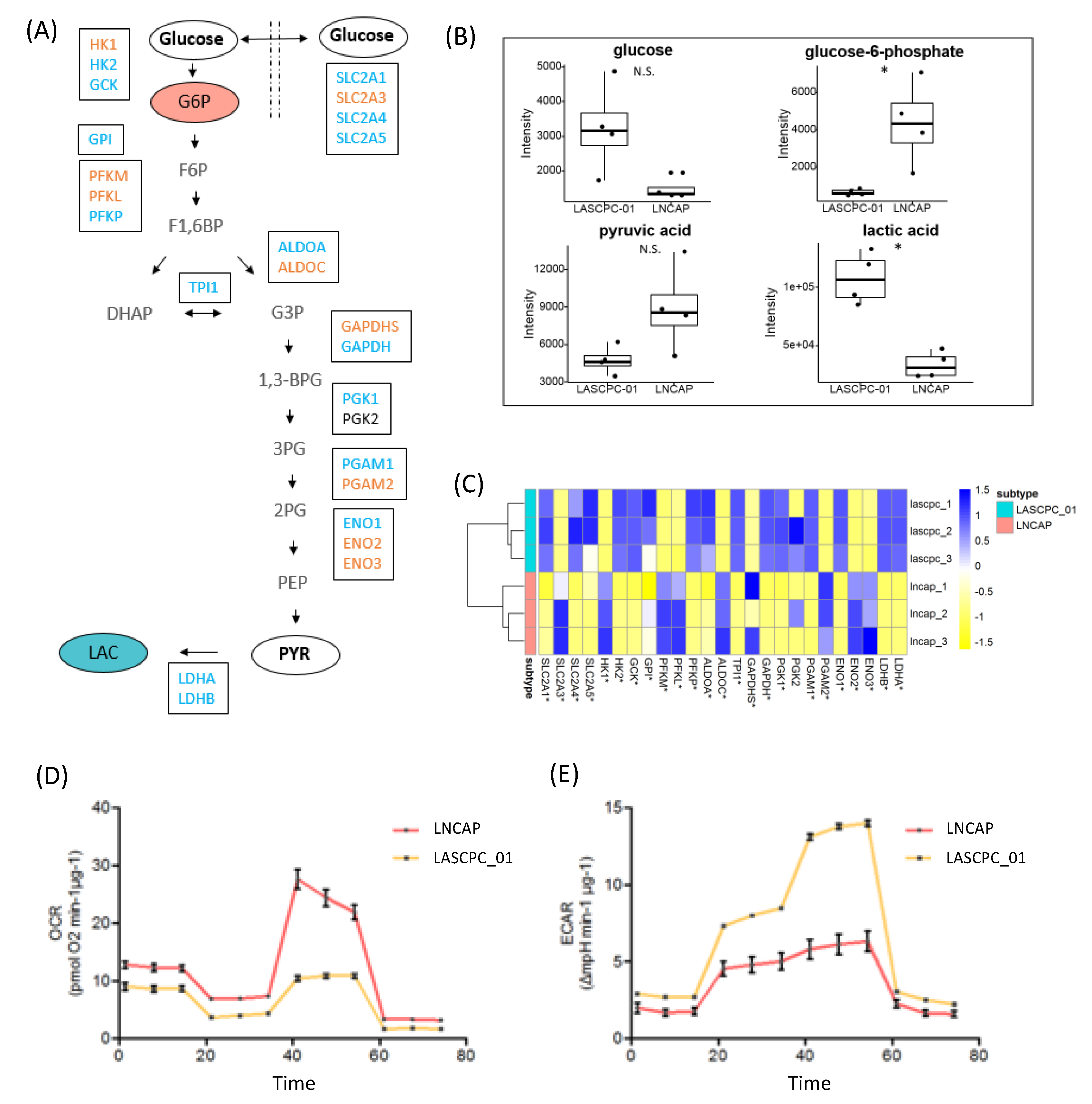
2.4. LASCPC-01 Exhibited a Higher Glycolytic Activity
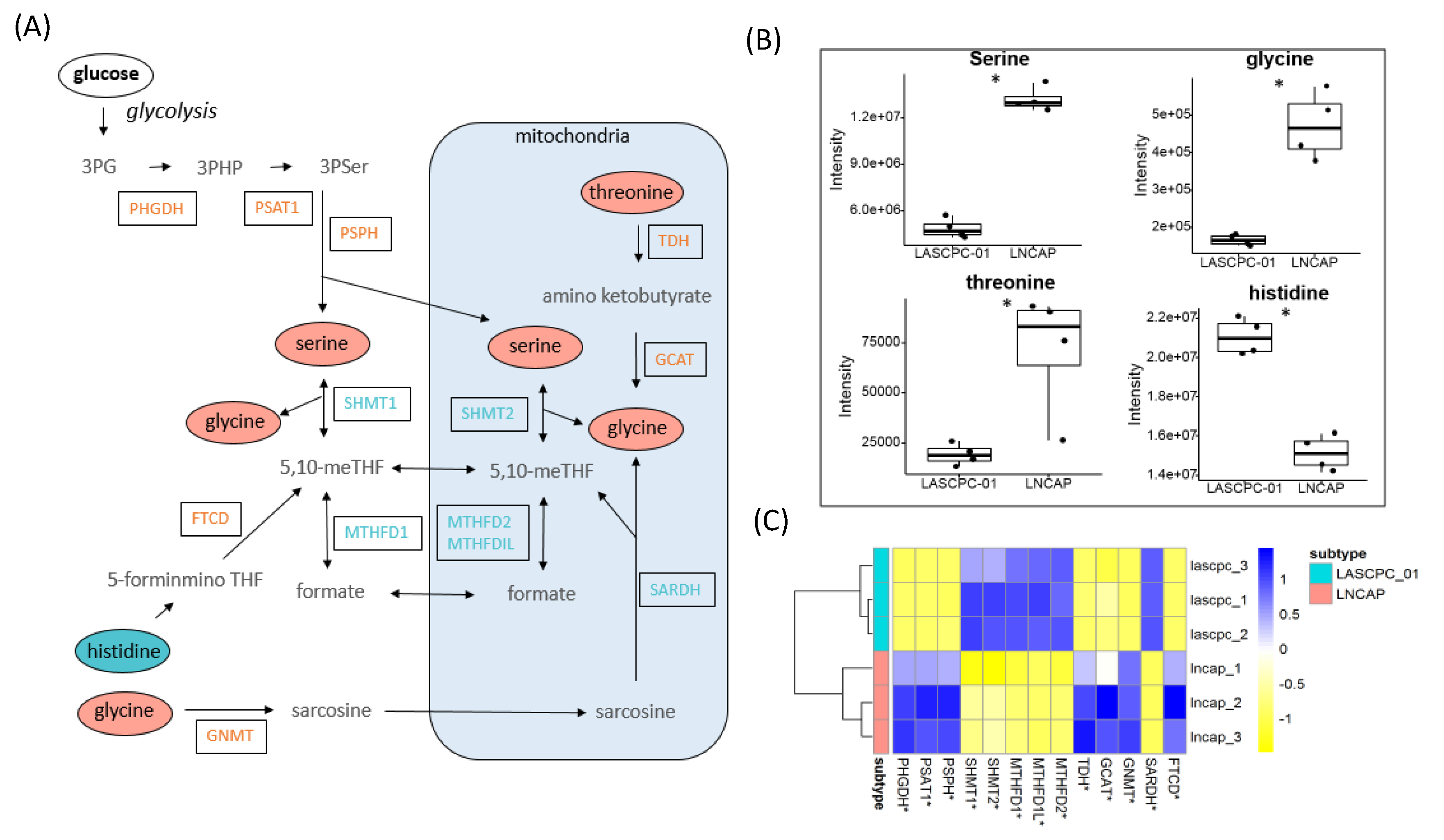
2.5. Elevated Levels of Serine and Glycine in LNCAP
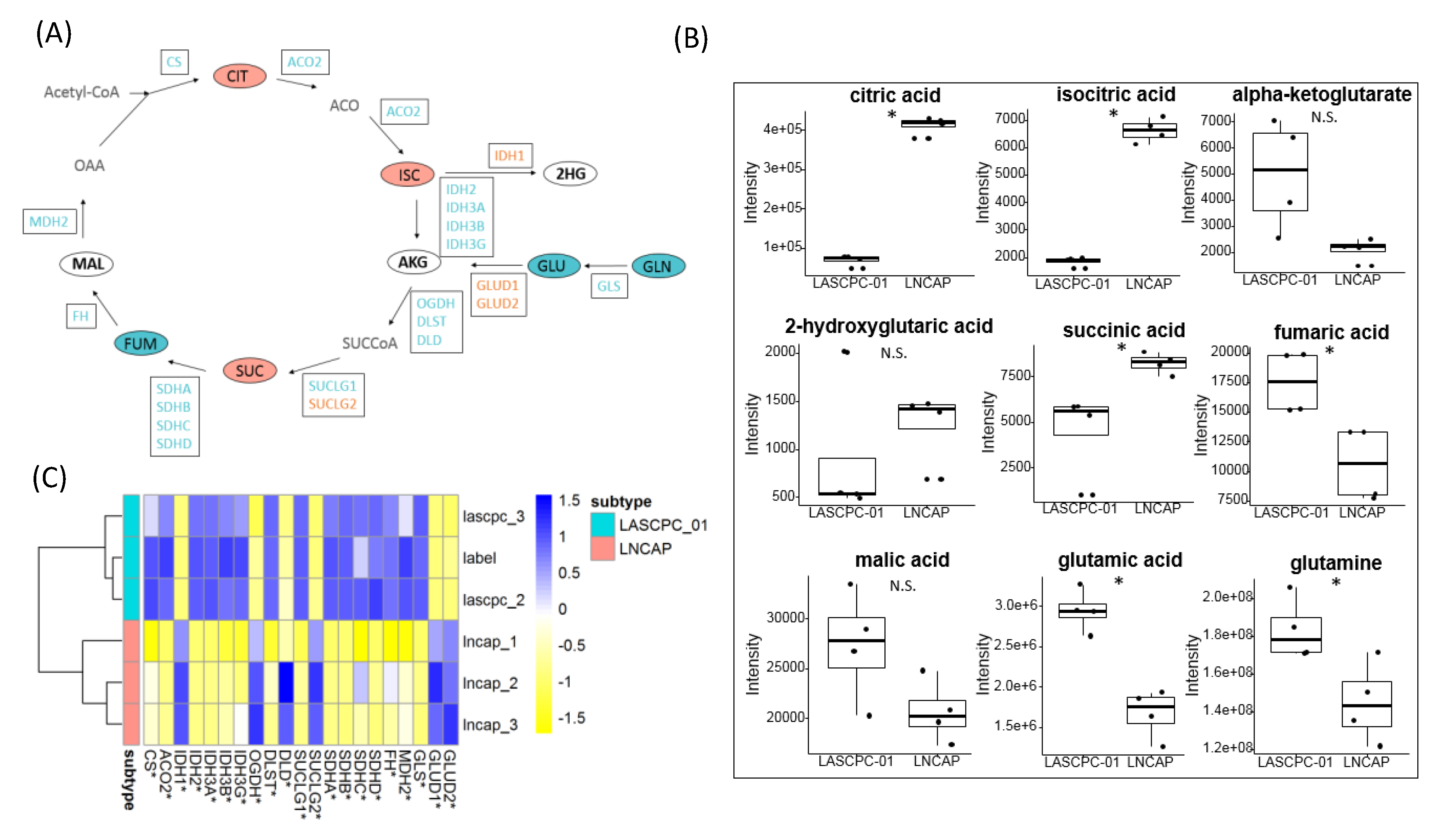
2.6. Citrate Accumulated in LNCAP Cells
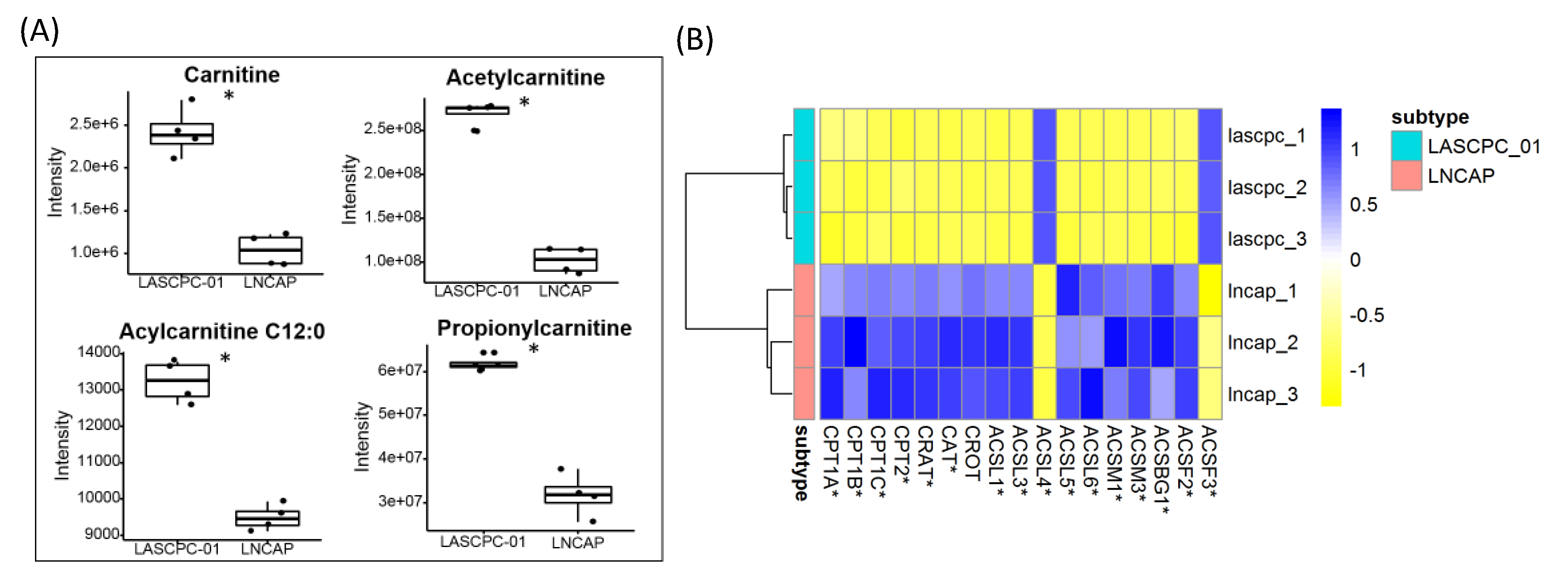
2.7. Short-Chain Acylcarnitines Were Lower in LNCAP
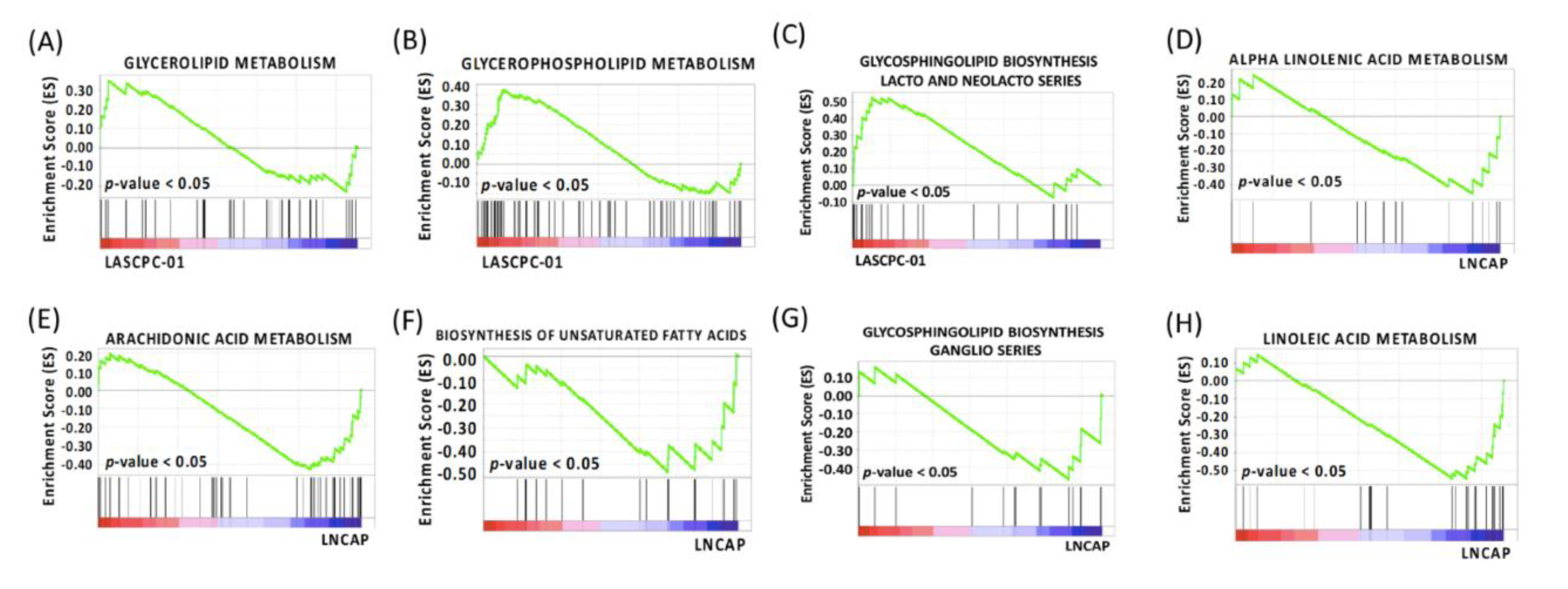
2.8. Different Lipid Metabolism
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Transcriptomics Library Construction and Data Analysis
4.3. Public Transcriptomics Data
4.4. Cellular Respiration
4.5. Profiling Primary Metabolism
4.6. Profiling Biogenic Amines
4.7. Profiling Complex Lipids
4.8. Liquid chromatography–mass spectrometry (LC-MS) Data Processing
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, C.D.; Welsbie, D.S.; Tran, C.; Baek, S.H.; Chen, R.; Vessella, R.; Rosenfeld, M.G.; Sawyers, C.L. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2003, 10, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.E.; Scher, H.I. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin. Cancer Res. 2009, 15, 4792–4798. [Google Scholar] [CrossRef] [PubMed]
- Di Zazzo, E.; Galasso, G.; Giovannelli, P.; Di Donato, M.; Castoria, G. Estrogens and Their Receptors in Prostate Cancer: Therapeutic Implications. Front. Oncol. 2018, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- El Etreby, M.F.; Liang, Y.; Lewis, R.W. Induction of apoptosis by mifepristone and tamoxifen in human LNCaP prostate cancer cells in culture. Prostate 2000, 43, 31–42. [Google Scholar] [CrossRef]
- Kim, I.Y.; Seong, D.H.; Kim, B.-C.; Lee, D.K.; Remaley, A.T.; Leach, F.; Morton, R.A.; Kim, S.-J. Raloxifene, a Selective Estrogen Receptor Modulator, Induces Apoptosis in Androgen-responsive Human Prostate Cancer Cell Line LNCaP through an Androgen-independent Pathway. Cancer Res. 2002, 62, 3649. [Google Scholar] [PubMed]
- Aggarwal, R.; Huang, J.; Alumkal, J.J.; Zhang, L.; Feng, F.Y.; Thomas, G.V.; Weinstein, A.S.; Friedl, V.; Zhang, C.; Witte, O.N.; et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J. Clin. Oncol. 2018, 36, 2492–2503. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Asgari, Y.; Zabihinpour, Z.; Salehzadeh-Yazdi, A.; Schreiber, F.; Masoudi-Nejad, A. Alterations in cancer cell metabolism: The Warburg effect and metabolic adaptation. Genomics 2015, 105, 275–281. [Google Scholar] [CrossRef]
- Eidelman, E.; Twum-Ampofo, J.; Ansari, J.; Siddiqui, M.M. The Metabolic Phenotype of Prostate Cancer. Front. Oncol. 2017, 7, 131. [Google Scholar] [CrossRef]
- Li, W.; Cohen, A.; Sun, Y.; Squires, J.; Braas, D.; Graeber, T.G.; Du, L.; Li, G.; Li, Z.; Xu, X.; et al. The Role of CD44 in Glucose Metabolism in Prostatic Small Cell Neuroendocrine Carcinoma. Mol. Cancer Res. Mcr 2016, 14, 344–353. [Google Scholar] [CrossRef]
- Cutruzzolà, F.; Giardina, G.; Marani, M.; Macone, A.; Paiardini, A.; Rinaldo, S.; Paone, A. Glucose Metabolism in the Progression of Prostate Cancer. Front. Physiol. 2017, 8, 97. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. Concepts of citrate production and secretion by prostate 1. Metabolic relationships. Prostate 1991, 18, 25–46. [Google Scholar] [CrossRef]
- Lima, A.R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. Biomarker Discovery in Human Prostate Cancer: an Update in Metabolomics Studies. Transl. Oncol. 2016, 9, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Franklin, R.B. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol. Cancer 2006, 5, 17. [Google Scholar] [CrossRef][Green Version]
- Costello, L.C.; Franklin, R.B. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology 2000, 59, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Phillips, J.W.; Smith, B.A.; Park, J.W.; Stoyanova, T.; McCaffrey, E.F.; Baertsch, R.; Sokolov, A.; Meyerowitz, J.G.; Mathis, C.; et al. N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell 2016, 29, 536–547. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef] [PubMed]
- Flavin, R.; Zadra, G.; Loda, M. Metabolic alterations and targeted therapies in prostate cancer. J. Pathol. 2011, 223, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Chiara, C.; Matteo, S.; Francesco, M.; Alessandra, M.; Francesco, P.; Alessandro, C.; Roberta, M.; Liang, C.; Antonio, L.-B.; Marina, S.; et al. Metabolic Alterations in Renal and Prostate Cancer. Curr. Drug Metab. 2016, 17, 150–155. [Google Scholar] [CrossRef]
- Trock, B.J. Application of metabolomics to prostate cancer. Urol. Oncol. 2011, 29, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Poulose, N.; Amoroso, F.; Steele, R.E.; Singh, R.; Ong, C.W.; Mills, I.G. Genetics of lipid metabolism in prostate cancer. Nat. Genet. 2018, 50, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Feng, P.; Milon, B.; Tan, M.; Franklin, R.B. Role of zinc in the pathogenesis and treatment of prostate cancer: Critical issues to resolve. Prostate Cancer Prostatic Dis. 2004, 7, 111–117. [Google Scholar] [CrossRef]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Schlaepfer, I.R.; Rider, L.; Rodrigues, L.U.; Gijón, M.A.; Pac, C.T.; Romero, L.; Cimic, A.; Sirintrapun, S.J.; Glodé, L.M.; Eckel, R.H.; et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol. Cancer Ther. 2014, 13, 2361–2371. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.C.; Maddocks, O.D.K. Serine and Functional Metabolites in Cancer. Trends Cell Biol. 2017, 27, 645–657. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Nikiforov, M.A.; Chandriani, S.; O’Connell, B.; Petrenko, O.; Kotenko, I.; Beavis, A.; Sedivy, J.M.; Cole, M.D. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 2002, 22, 5793–5800. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography–Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.34.31–30.34.32. [Google Scholar] [CrossRef]
- Skogerson, K.; Wohlgemuth, G.; Barupal, D.K.; Fiehn, O. The volatile compound BinBase mass spectral database. BMC Bioinform. 2011, 12, 321. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib – mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- DeFelice, B.C.; Mehta, S.S.; Samra, S.; Čajka, T.; Wancewicz, B.; Fahrmann, J.F.; Fiehn, O. Mass Spectral Feature List Optimizer (MS-FLO): A Tool To Minimize False Positive Peak Reports in Untargeted Liquid Chromatography–Mass Spectroscopy (LC-MS) Data Processing. Anal. Chem. 2017, 89, 3250–3255. [Google Scholar] [CrossRef]
- Yoav, B.; Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
| Gene Sets | LASCPC-01 | SCNC | ||||||
|---|---|---|---|---|---|---|---|---|
| Number | ES | p-value | FDR | Number | ES | p-value | FDR | |
| Glyoxylate and Dicarboxylate Metabolism | 7 (16) | 0.45 | 0.08 | 0.31 | 4 (16) | 0.21 | 0.93 | 0.99 |
| Cysteine and Methionine Metabolism | 17 (30) | 0.42 | 0.08 | 0.28 | 10 (34) | 0.23 | 0.82 | 1.00 |
| Pyruvate Metabolism | 8 (36) | 0.33 | 0.16 | 0.31 | 8 (39) | 0.30 | 0.50 | 1.00 |
| Pyrimidine Metabolism | 40 (94) | 0.31 | 0.19 | 0.30 | 16 (95) | 0.20 | 0.77 | 1.00 |
| Inositol Phosphate Metabolism | 13 (53) | 0.26 | 0.20 | 0.28 | 3 (54) | 0.14 | 0.99 | 0.98 |
| Nitrogen Metabolism | 7 (23) | 0.38 | 0.20 | 0.31 | 9 (23) | 0.42 | 0.34 | 1.00 |
| Galactose Metabolism | 7 (24) | 0.33 | 0.30 | 0.33 | 7 (26) | 0.29 | 0.65 | 1.00 |
| One Carbon Pool By Folate | 4 (17) | 0.29 | 0.49 | 0.47 | 5 (17) | 0.28 | 0.73 | 1.00 |
| Gene Sets | LNCAP | Prostate Adenocarcinoma | ||||||
|---|---|---|---|---|---|---|---|---|
| Number | ES | p-value | FDR | Number | ES | p-value | FDR | |
| Arginine and Proline Metabolism | 17 (49) | −0.38 | 0 | 0.11 | 14 (52) | −0.31 | 0.53 | 0.94 |
| Ascorbate and Aldarate Metabolism | 11 (18) | −0.61 | 0 | 0.18 | 8 (16) | −0.61 | 0.21 | 1.00 |
| Histidine Metabolism | 12 (27) | −0.42 | 0 | 0.13 | 5 (28) | −0.24 | 0.88 | 0.97 |
| Nicotinate and Nicotinamide Metabolism | 8 (21) | −0.33 | 0 | 0.15 | 7 (22) | −0.28 | 0.83 | 0.99 |
| Pentose and Glucuronate Interconversions | 7 (20) | −0.54 | 0 | 0.17 | 8 (18) | −0.63 | 0.14 | 1.00 |
| Pentose Phosphate Pathway | 9 (25) | −0.36 | 0 | 0.10 | 18 (26) | −0.20 | 0.97 | 0.98 |
| Phenylalanine Metabolism | 10 (18) | −0.49 | 0 | 0.12 | 6 (18) | −0.40 | 0.50 | 0.99 |
| Retinol Metabolism | 17 (48) | −0.48 | 0 | 0.10 | 20 (54) | −0.42 | 0.59 | 0.98 |
| Starch and Sucrose Metabolism | 13 (41) | −0.41 | 0 | 0.10 | 8 (43) | −0.38 | 0.47 | 0.97 |
| Tyrosine Metabolism | 14 (38) | −0.52 | 0 | 0.10 | 13 (42) | −0.40 | 0.40 | 0.99 |
| Butanoate Metabolism | 10 (30) | −0.41 | 0.09 | 0.13 | 8 (34) | −0.39 | 0.29 | 0.95 |
| Glycine Serine and Threonine Metabolism | 12 (29) | −0.40 | 0.10 | 0.18 | 4 (31) | −0.31 | 0.76 | 1.00 |
| Tryptophan Metabolism | 15 (37) | −0.34 | 0.10 | 0.30 | 8 (39) | −0.27 | 0.81 | 0.97 |
| Valine Leucine and Isoleucine Degradation | 20 (44) | −0.31 | 0.30 | 0.33 | 12 (44) | −0.35 | 0.31 | 0.99 |
| Amino Sugar and Nucleotide Sugar Metabolism | 14 (43) | −0.22 | 0.31 | 0.42 | 18 (44) | −0.33 | 0.40 | 0.99 |
| Gene Sets | LASCPC-01 | SCNC | ||||||
|---|---|---|---|---|---|---|---|---|
| Number | ES | p-value | FDR | Number | ES | p-value | FDR | |
| Glycerolipid Metabolism | 7 (39) | 0.34 | 0 | 0.30 | 5 (43) | 0.27 | 0.68 | 1.00 |
| Glycerophospholipid Metabolism | 20 (67) | 0.38 | 0 | 0.31 | 9 (70) | 0.25 | 0.73 | 1.00 |
| Glycosphingolipid Biosynthesis Lacto and Neolacto Series | 9 (24) | 0.53 | 0 | 0.19 | 11 (25) | 0.39 | 0.41 | 1.00 |
| Ether Lipid Metabolism | 7 (26) | 0.31 | 0.19 | 0.30 | 9 (29) | 0.37 | 0.39 | 1.00 |
| Gene Sets | LNCAP | Prostate Adenocarcinoma | ||||||
|---|---|---|---|---|---|---|---|---|
| Number | ES | p-value | FDR | Number | ES | p-value | FDR | |
| Alpha Linolenic Acid Metabolism | 5 (15) | −0.46 | 0 | 0.13 | 6 (18) | −0.34 | 0.70 | 0.96 |
| Arachidonic Acid Metabolism | 21 (47) | −0.43 | 0 | 0.10 | 24 (57) | −0.37 | 0.43 | 0.96 |
| Biosynthesis of Unsaturated Fatty Acids | 11 (20) | −0.48 | 0 | 0.10 | 2 (20) | −0.27 | 0.77 | 0.98 |
| Glycosphingolipid Biosynthesis Ganglio Series | 6 (15) | −0.47 | 0 | 0.25 | 9 (15) | −0.31 | 0.77 | 1.00 |
| Linoleic Acid Metabolism | 14 (25) | −0.54 | 0 | 0.10 | 8 (28) | −0.32 | 0.79 | 0.96 |
| Glycosylphosphatidylinositol Gpi Anchor Biosynthesis | 12 (24) | −0.26 | 0.29 | 0.46 | 13 (25) | −0.34 | 0.36 | 0.93 |
| Fatty Acid Metabolism | 18 (39) | −0.31 | 0.29 | 0.19 | 16 (42) | −0.45 | 0.20 | 1.00 |
| Sphingolipid Metabolism | 5 (33) | −0.25 | 0.48 | 0.51 | 10 (36) | −0.32 | 0.39 | 0.98 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, B.; Lue, H.-W.; Podolak, J.; Fan, S.; Zhang, Y.; Serawat, A.; Alumkal, J.J.; Fiehn, O.; Thomas, G.V. Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites 2019, 9, 82. https://doi.org/10.3390/metabo9050082
Gao B, Lue H-W, Podolak J, Fan S, Zhang Y, Serawat A, Alumkal JJ, Fiehn O, Thomas GV. Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites. 2019; 9(5):82. https://doi.org/10.3390/metabo9050082
Chicago/Turabian StyleGao, Bei, Hui-Wen Lue, Jennifer Podolak, Sili Fan, Ying Zhang, Archana Serawat, Joshi J. Alumkal, Oliver Fiehn, and George V. Thomas. 2019. "Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma" Metabolites 9, no. 5: 82. https://doi.org/10.3390/metabo9050082
APA StyleGao, B., Lue, H.-W., Podolak, J., Fan, S., Zhang, Y., Serawat, A., Alumkal, J. J., Fiehn, O., & Thomas, G. V. (2019). Multi-Omics Analyses Detail Metabolic Reprogramming in Lipids, Carnitines, and Use of Glycolytic Intermediates between Prostate Small Cell Neuroendocrine Carcinoma and Prostate Adenocarcinoma. Metabolites, 9(5), 82. https://doi.org/10.3390/metabo9050082








