Pasture Feeding Changes the Bovine Rumen and Milk Metabolome
Abstract
1. Introduction
2. Results and Discussion
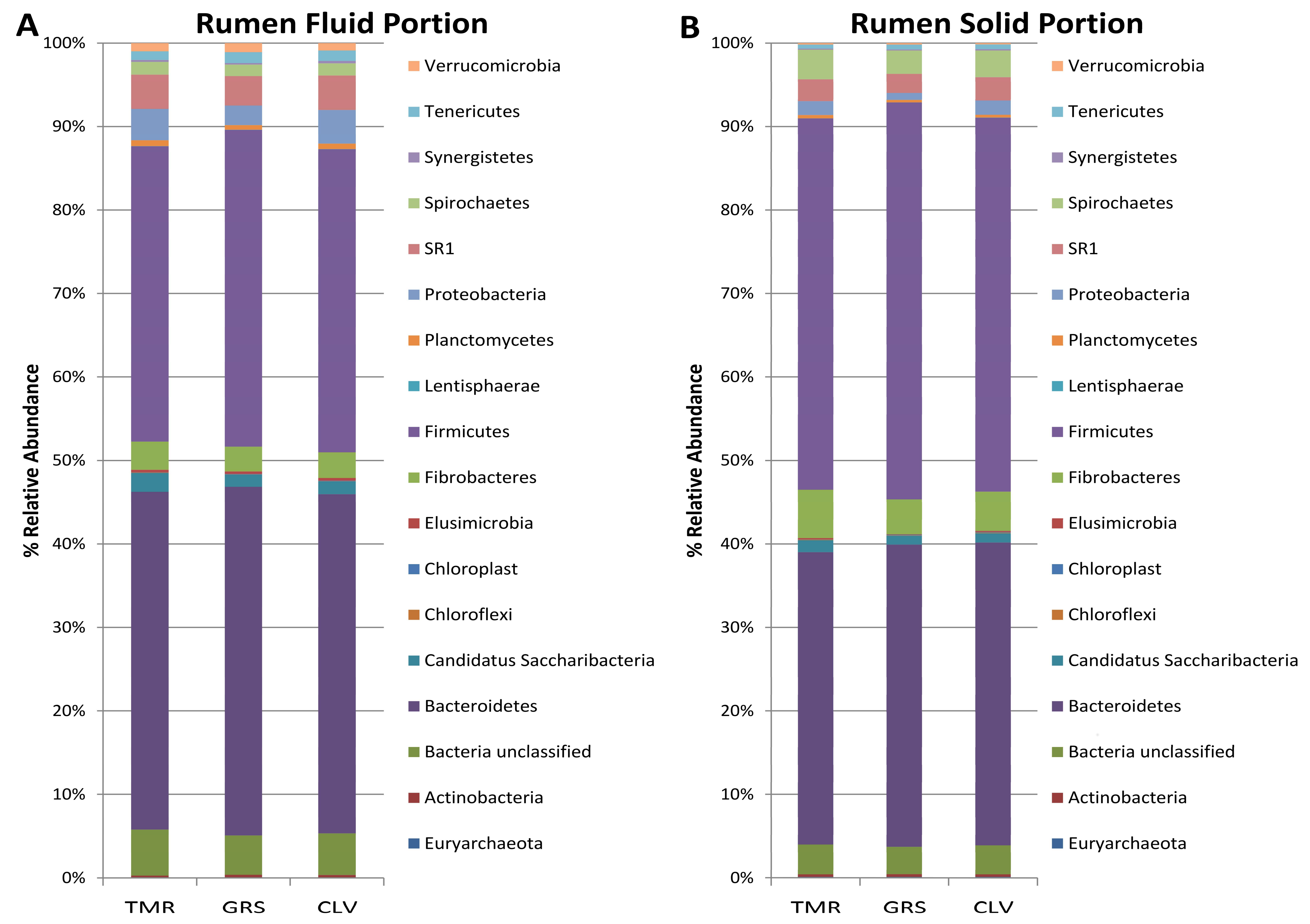
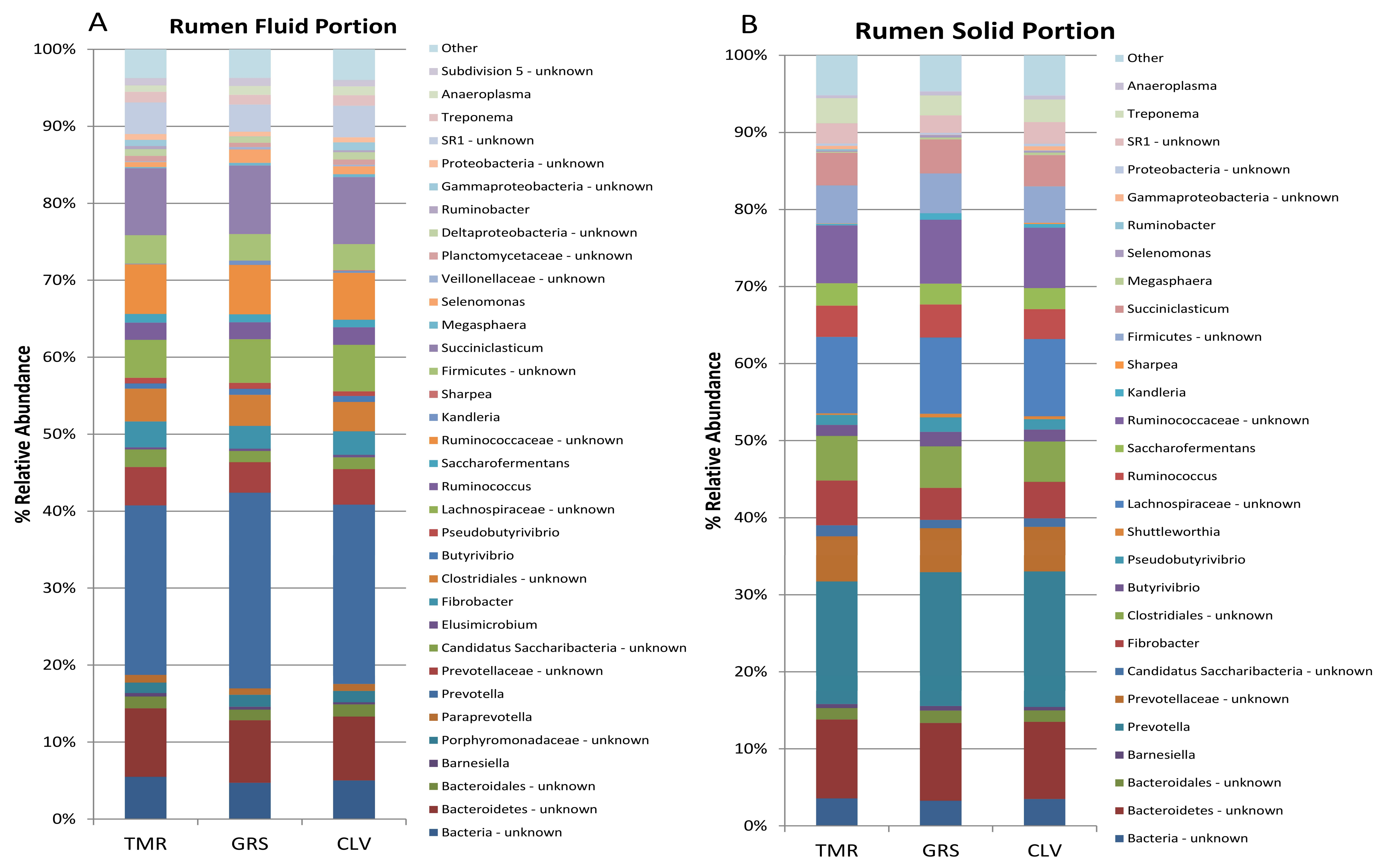
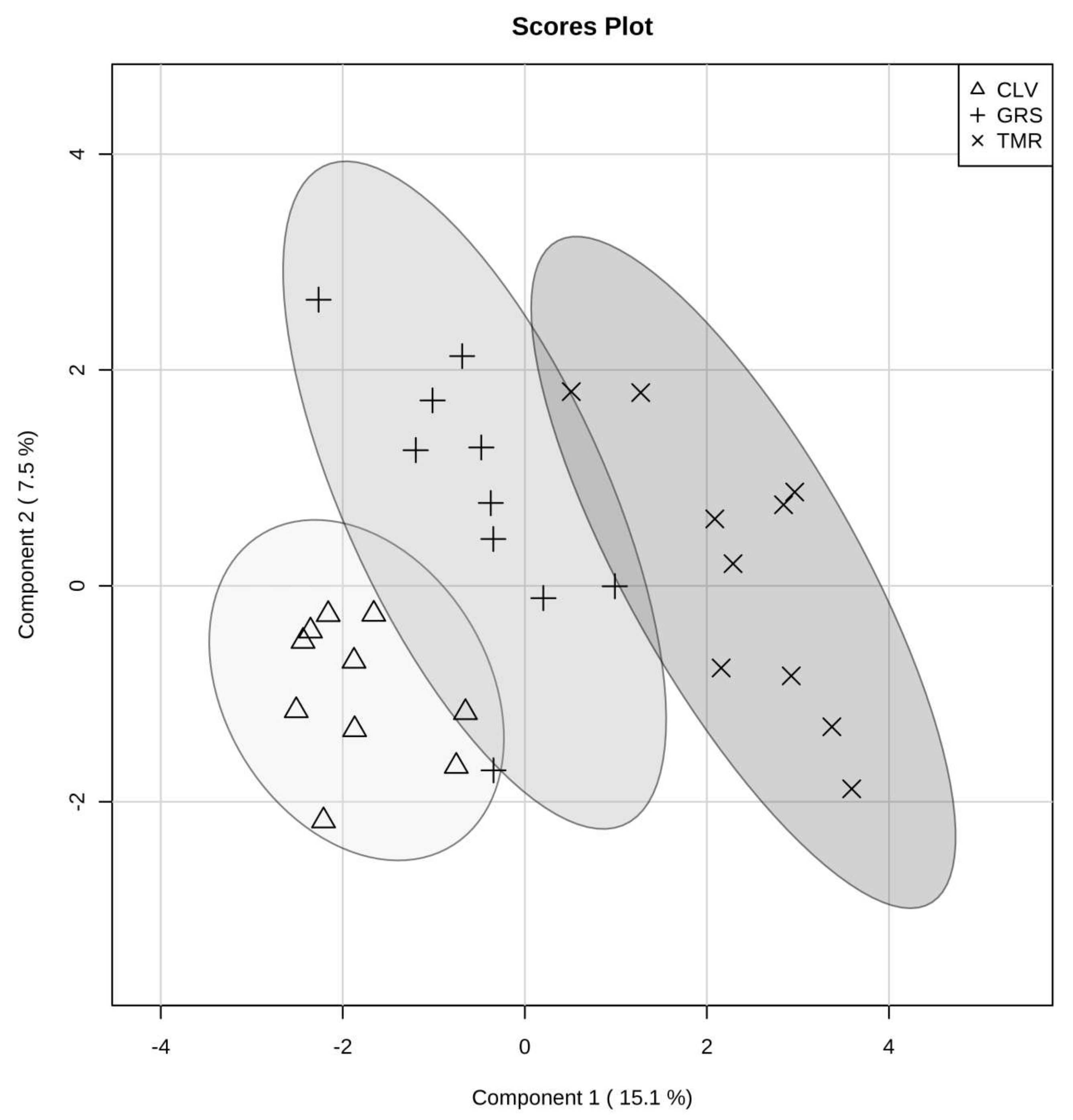
2.1. Effect of Cow Diet on Rumen Microbiota
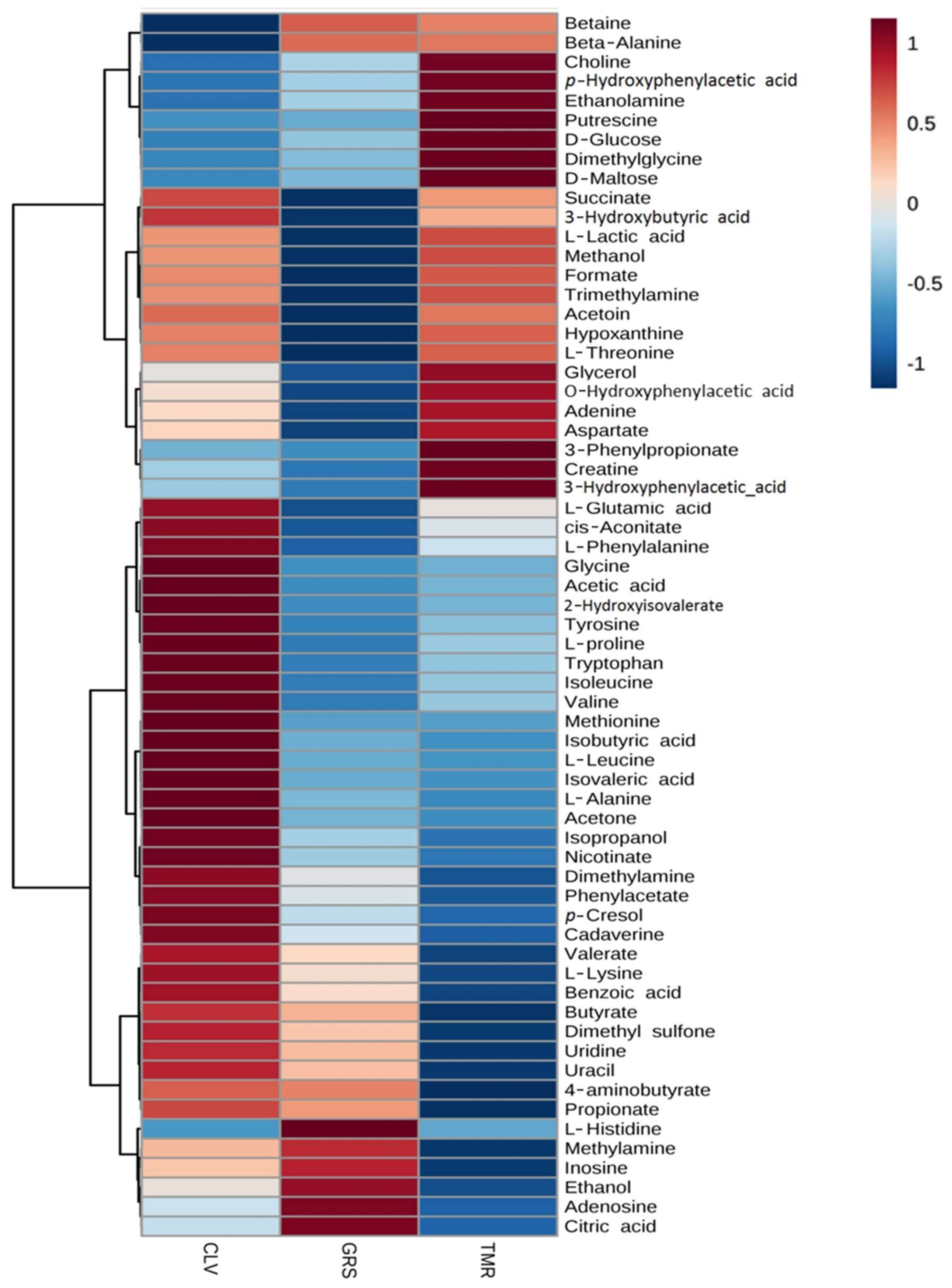
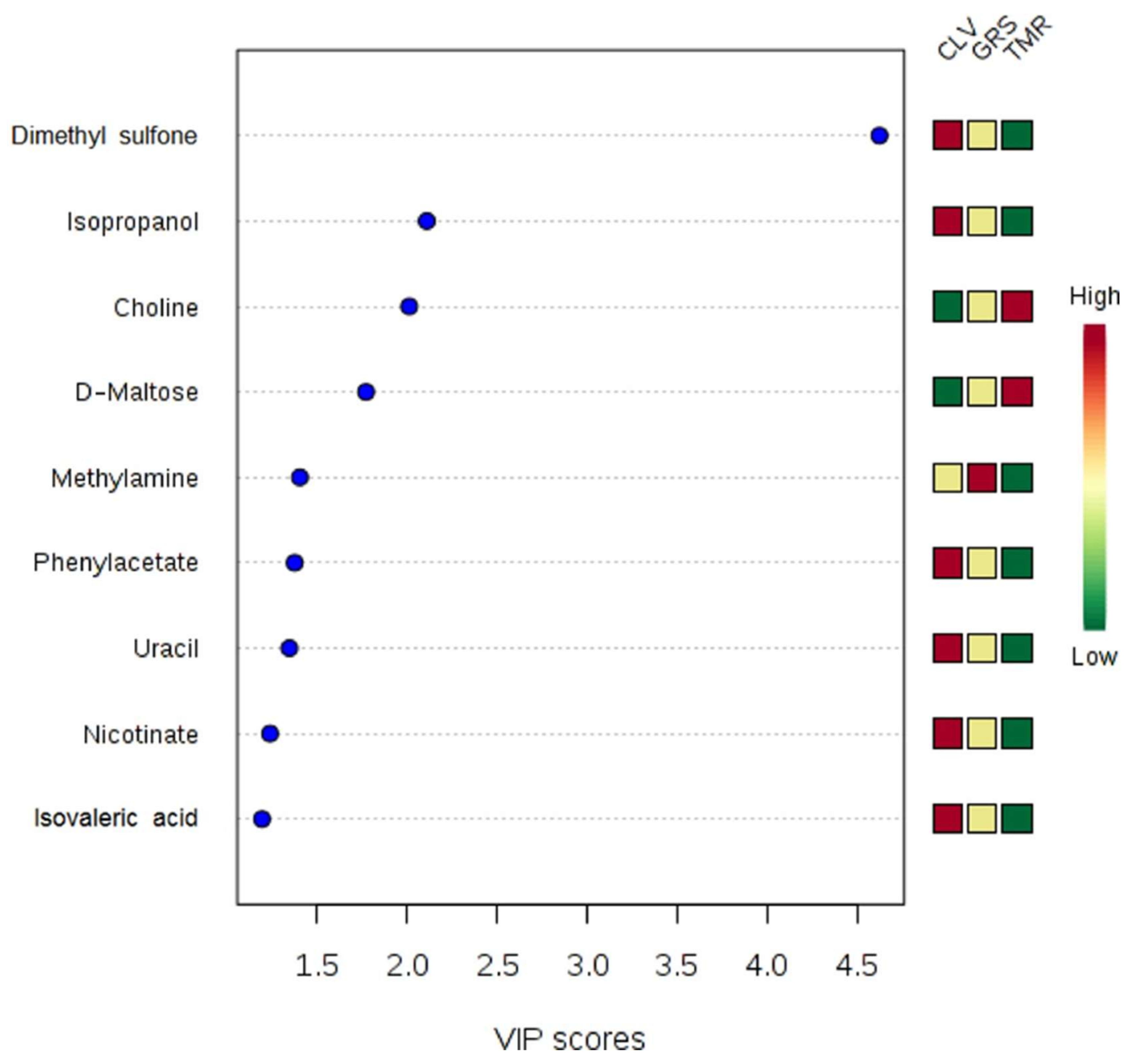
2.2. Rumen Fluid Metabolome
2.3. Milk Metabolome
3. Materials and Methods
3.1. Experimental Design and Sample Collection
3.2. Feed Compositional Analysis
3.3. Ethical Approval
3.4. NMR Sample Preparation
3.5. NMR Compound Identification and Quantification
3.6. DNA Extraction and MiSeq Sequencing
3.7. Bioinformatic (DNA) Analysis
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- United States Department of Agriculture (USDA). Dairy 2014: Dairy Cattle Management Practices in the United States, 2014; USDA: Fort Collins, CO, USA, 2016.
- Legrand, A.L.; von Keyserlingk, M.A.G.; Weary, D.M. Preference and usage of pasture versus free-stall housing by lactating dairy cattle. J. Dairy Sci. 2009, 92, 3651–3658. [Google Scholar] [CrossRef] [PubMed]
- Von Keyserlingk, M.A.G.; Rushen, J.; de Passillé, A.M.; Weary, D.M. Invited review: The welfare of dairy cattle—Key concepts and the role of science. J. Dairy Sci. 2009, 92, 4101–4111. [Google Scholar] [CrossRef] [PubMed]
- Getter, K.L.; Behe, B.K.; Howard, P.H.; Conner, D.S.; Spaniolo, L.M. Increasing demand for pasture-based dairy: What attributes and images do consumers want? In Re-Thinking Organic Food and Farming in a Changing World; Springer: Berlin, Germany, 2015; pp. 125–140. [Google Scholar]
- O’Callaghan, T.F.; Hennessy, D.; McAuliffe, S.; Kilcawley, K.N.; O’Donovan, M.; Dillon, P.; Ross, R.P.; Stanton, C. Effect of pasture versus indoor feeding systems on raw milk composition and quality over an entire lactation. J. Dairy Sci. 2016, 99, 9424–9440. [Google Scholar] [CrossRef] [PubMed]
- Saleem, F.; Bouatra, S.; Guo, A.C.; Psychogios, N.; Mandal, R.; Dunn, S.M.; Ametaj, B.N.; Wishart, D.S. The bovine ruminal fluid metabolome. Metabolomics 2013, 9, 360–378. [Google Scholar] [CrossRef]
- Sundekilde, U.; Larsen, L.; Bertram, H. NMR-based milk metabolomics. Metabolites 2013, 3, 204. [Google Scholar] [CrossRef] [PubMed]
- Sundekilde, U.K.; Larsen, L.B.; Bertram, H.C. NMR-based milk metabolomics. Metabolites 2013, 3, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Ametaj, B.N.; Zebeli, Q.; Saleem, F.; Psychogios, N.; Lewis, M.J.; Dunn, S.M.; Xia, J.; Wishart, D.S. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 2010, 6, 583–594. [Google Scholar] [CrossRef]
- Saleem, F.; Ametaj, B.; Bouatra, S.; Mandal, R.; Zebeli, Q.; Dunn, S.; Wishart, D. A metabolomics approach to uncover the effects of grain diets on rumen health in dairy cows. J. Dairy Sci. 2012, 95, 6606–6623. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Teather, R.; Forster, R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol. Ecol. 2010, 74, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Noel, S.J.; Attwood, G.T.; Rakonjac, J.; Moon, C.D.; Waghorn, G.C.; Janssen, P.H. Seasonal changes in the digesta-adherent rumen bacterial communities of dairy cattle grazing pasture. PLoS ONE 2017, 12, e0173819. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, M.L.; Cersosimo, L.M.; Wright, A.-D.G.; Kraft, J. Rumen bacterial communities shift across a lactation in holstein, jersey and holstein × jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 2016, 92, fiw059. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J. Effects of dilution rate and ph on the ruminal cellulolytic bacterium fibrobacter succinogenes s85 in cellulose-fed continuous culture. Arch. Microbiol. 1993, 160, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Pan, J.; Kobayashi, Y.; Tanaka, K. Kinetics of in sacco fiber-attachment of representative ruminal cellulolytic bacteria monitored by competitive pcr. J. Dairy Sci. 2003, 86, 1429–1435. [Google Scholar] [CrossRef]
- Robert, W. The bacterial community composition of the bovine rumen detected using pyrosequencing of 16S rRNA genes. Metagenomics 2012, 2012, 235571. [Google Scholar]
- Jami, E.; Mizrahi, I. Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- McCann, J.C.; Wickersham, T.A.; Loor, J.J. High-throughput methods redefine the rumen microbiome and its relationship with nutrition and metabolism. Bioinform. Biol. Insights 2014, 8, 109–125. [Google Scholar] [CrossRef] [PubMed]
- De Menezes, A.B.; Lewis, E.; O’Donovan, M.; O’Neill, B.F.; Clipson, N.; Doyle, E.M. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 2011, 78, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Lima, F.S.; Oikonomou, G.; Lima, S.F.; Bicalho, M.L.S.; Ganda, E.K.; de Oliveira Filho, J.C.; Lorenzo, G.; Trojacanec, P.; Bicalho, R.C. Prepartum and postpartum rumen fluid microbiomes: Characterization and correlation with production traits in dairy cows. Appl. Environ. Microbiol. 2015, 81, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Xue, F.; Nan, X.; Tang, Z.; Wang, K.; Beckers, Y.; Jiang, L.; Xiong, B. Illumina sequencing approach to characterize thiamine metabolism related bacteria and the impacts of thiamine supplementation on ruminal microbiota in dairy cows fed high-grain diets. Front. Microbiol. 2017, 8, 1818. [Google Scholar] [CrossRef] [PubMed]
- Van Gylswyk, N. Succiniclasticum ruminis gen. Nov., sp. Nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Evolut. Microbiol. 1995, 45, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; Rooke, J.A.; McKain, N.; Duthie, C.-A.; Hyslop, J.J.; Ross, D.W.; Waterhouse, A.; Watson, M.; Roehe, R. The rumen microbial metagenome associated with high methane production in cattle. BMC Genom. 2015, 16, 839. [Google Scholar] [CrossRef] [PubMed]
- Henning, P.H.; Horn, C.H.; Steyn, D.G.; Meissner, H.H.; Hagg, F.M. The potential of megasphaera elsdenii isolates to control ruminal acidosis. Anim. Feed Sci. Technol. 2010, 157, 13–19. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Krause, D.O.; Gozho, G.N.; McBride, B.W. Subacute ruminal acidosis in dairy cows: The physiological causes, incidence and consequences. Vet. J. 2008, 176, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Jewell, K.A.; McCormick, C.A.; Odt, C.L.; Weimer, P.J.; Suen, G. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl. Environ. Microbiol. 2015, 81, 4697–4710. [Google Scholar] [CrossRef] [PubMed]
- France, J.; Dijkstra, J. Volatile fatty acid production. Quant. Asp. Rumin. Dig. Metab. 2005, 2, 157–175. [Google Scholar]
- Lüscher, A.; Mueller-Harvey, I.; Soussana, J.F.; Rees, R.M.; Peyraud, J.L. Potential of legume-based grassland–livestock systems in europe: A review. Grass Forage Sci. 2014, 69, 206–228. [Google Scholar] [CrossRef] [PubMed]
- Nafikov, R.A.; Beitz, D.C. Carbohydrate and lipid metabolism in farm animals. J. Nutr. 2007, 137, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Andries, J.I.; Buysse, F.X.; De Brabander, D.L.; Cottyn, B.G. Isoacids in ruminant nutrition: Their role in ruminal and intermediary metabolism and possible influences on performances—A review. Anim. Feed Sci. Technol. 1987, 18, 169–180. [Google Scholar] [CrossRef]
- Taylor, B.F.; Kiene, R.P. Microbial metabolism of dimethyl sulfide. In Biogenic Sulfur in the Environment; American Chemical Society: Washington, DC, USA, 1989; Volume 393, pp. 202–221. [Google Scholar]
- Villeneuve, M.P.; Lebeuf, Y.; Gervais, R.; Tremblay, G.F.; Vuillemard, J.C.; Fortin, J.; Chouinard, P.Y. Milk volatile organic compounds and fatty acid profile in cows fed timothy as hay, pasture, or silage. J. Dairy Sci. 2013, 96, 7181–7194. [Google Scholar] [CrossRef] [PubMed]
- Chesson, A.; Provan, G.J.; Russell, W.R.; Scobbie, L.; Richardson, A.J.; Stewart, C. Hydroxycinnamic acids in the digestive tract of livestock and humans. J. Sci. Food Agric. 1999, 79, 373–378. [Google Scholar] [CrossRef]
- Faulkner, H.; O’Callaghan, T.F.; McAuliffe, S.; Hennessy, D.; Stanton, C.; O’Sullivan, M.G.; Kerry, J.P.; Kilcawley, K.N. Effect of different forage types on the volatile and sensory properties of bovine milk. J. Dairy Sci. 2018, 101, 1034–1047. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.K. The origin of urinary aromatic compounds excreted by ruminants: 3. The metabolism of phenolic compounds to simple phenols. Br. J. Nutr. 1982, 48, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, W.; Jiang, L.; Mao, S. Comparative metabolome analysis of ruminal changes in holstein dairy cows fed low- or high-concentrate diets. Metabolomics 2017, 13, 74. [Google Scholar] [CrossRef]
- McAllan, A.B.; Smith, R.H. Degradation of nucleic acids in the rumen. Br. J. Nutr. 1973, 29, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Bruss, M.L.; Lopez, M.J. Mixed ruminal microbes of cattle produce isopropanol in the presence of acetone but not 3-d-hydroxybutyrate. J. Dairy Sci. 2000, 83, 2580–2584. [Google Scholar] [CrossRef]
- Martin, C.; Mirande, C.; Morgavi, D.P.; Forano, E.; Devillard, E.; Mosoni, P. Methionine analogues hmb and hmbi increase the abundance of cellulolytic bacterial representatives in the rumen of cattle with no direct effects on fibre degradation. Anim. Feed Sci. Technol. 2013, 182, 16–24. [Google Scholar] [CrossRef]
- Carlson, J.R.; Breeze, R.G. Ruminal metabolism of plant toxins with emphasis on indolic compounds. J. Anim. Sci. 1984, 58, 1040–1049. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zepka, L.Q.; Garruti, D.S.; Sampaio, K.L.; Mercadante, A.Z.; Da Silva, M.A.A.P. Aroma compounds derived from the thermal degradation of carotenoids in a cashew apple juice model. Food Res. Int. 2014, 56, 108–114. [Google Scholar] [CrossRef]
- Noziere, P.; Graulet, B.; Lucas, A.; Martin, B.; Grolier, P.; Doreau, M. Carotenoids for ruminants: From forages to dairy products. Anim. Feed Sci. Technol. 2006, 131, 418–450. [Google Scholar] [CrossRef]
- Prache, S.; Priolo, A.; Tournadre, H.; Jailler, R.; Dubroeucq, H.; Micol, D.; Martin, B.; Durand, J.; Emile, J.; Huyghe, C. Traceability of Grass-Feeding by Quantifying the Signature of Carotenoid Pigments in Herbivores Meat, Milk and Cheese, Multi-function grasslands: Quality forages, animal products and landscapes. In Proceedings of the 19th General Meeting of the European Grassland Federation, La Rochelle, France, 27–30 May 2002; pp. 592–593. [Google Scholar]
- Kilic, M.; Lindsay, R.C. Distribution of conjugates of alkylphenols in milk from different ruminant species. J. Dairy Sci. 2005, 88, 7–12. [Google Scholar] [CrossRef]
- Leiva, T.; Cooke, R.F.; Brandão, A.P.; Marques, R.S.; Vasconcelos, J.L.M. Effects of rumen-protected choline supplementation on metabolic and performance responses of transition dairy cows 1. J. Anim. Sci. 2015, 93, 1896–1904. [Google Scholar] [CrossRef] [PubMed]
- Neill, A.R.; Grime, D.W.; Snoswell, A.M.; Northrop, A.J.; Lindsay, D.B.; Dawson, R.M. The low availability of dietary choline for the nutrition of the sheep. Biochem. J. 1979, 180, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Artegoitia, V.M.; Middleton, J.L.; Harte, F.M.; Campagna, S.R.; de Veth, M.J. Choline and choline metabolite patterns and associations in blood and milk during lactation in dairy cows. PLoS ONE 2014, 9, e103412. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H.; Da Costa, K.A.; Franklin, P.D.; Alexander, E.A.; Lamont, J.T.; Sheard, N.F.; Beiser, A. Choline, an essential nutrient for humans. FASEB J. 1991, 5, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Baldi, A.; Pinotti, L. Choline metabolism in high-producing dairy cows: Metabolic and nutritional basis. Can. J. Anim. Sci. 2006, 86, 207–212. [Google Scholar] [CrossRef]
- Shahsavari, A.; D’Occhio, M.J.; Al Jassim, R. The role of rumen-protected choline in hepatic function and performance of transition dairy cows. Br. J. Nutr. 2016, 116, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Wolin, M.J.; Miller, T.L.; Stewart, C.S. Microbe-microbe interactions. In The Rumen Microbial Ecosystem; Hobson, P.N., Stewart, C.S., Eds.; Springer: Dordrecht, The Netherlands, 1997; pp. 467–491. [Google Scholar]
- Wolin, M.J. The rumen fermentation: A model for microbial interactions in anaerobic ecosystems. In Advances in Microbial Ecology: Volume 3; Alexander, M., Ed.; Springer: Boston, MA, USA, 1979; pp. 49–77. [Google Scholar]
- Hungate, R.E.; Smith, W.; Bauchop, T.; Yu, I.; Rabinowitz, J.C. Formate as an intermediate in the bovine rumen fermentation. J. Bacteriol. 1970, 102, 389–397. [Google Scholar] [PubMed]
- Lovley, D.R.; Greening, R.C.; Ferry, J.G. Rapidly growing rumen methanogenic organism that synthesizes coenzyme m and has a high affinity for formate. Appl. Environ. Microbiol. 1984, 48, 81–87. [Google Scholar] [PubMed]
- Collard, B.L.; Boettcher, P.J.; Dekkers, J.C.M.; Petitclerc, D.; Schaeffer, L.R. Relationships between energy balance and health traits of dairy cattle in early lactation. J. Dairy Sci. 2000, 83, 2683–2690. [Google Scholar] [CrossRef]
- Contarini, G.; Povolo, M.; Leardi, R.; Toppino, P.M. Influence of heat treatment on the volatile compounds of milk. J. Agric. Food Chem. 1997, 45, 3171–3177. [Google Scholar] [CrossRef]
- Heuer, C.; Wangler, A.; Schukken, Y.H.; Noordhuizen, J.P.T.M. Variability of acetone in milk in a large low-production dairy herd: A longitudinal case study. Vet. J. 2001, 161, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, A.H.; Emanuelson, U. Milk acetone concentration as an indicator of hyperketonaemia in dairy cows: The critical value revised. Anim. Sci. 1996, 63, 183–188. [Google Scholar] [CrossRef]
- Coppa, M.; Ferlay, A.; Monsallier, F.; Verdier-Metz, I.; Pradel, P.; Didienne, R.; Farruggia, A.; Montel, M.; Martin, B. Milk fatty acid composition and cheese texture and appearance from cows fed hay or different grazing systems on upland pastures. J. Dairy Sci. 2011, 94, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Butler, W.R. Effect of protein nutrition on ovarian and uterine physiology in dairy cattle. J. Dairy Sci. 1998, 81, 2533–2539. [Google Scholar] [CrossRef]
- Harris, S.L.; Auldist, M.J.; Clark, D.A.; Jansen, E.B. Effects of white clover content in the diet on herbage intake, milk production and milk composition of new zealand dairy cows housed indoors. J. Dairy Res. 1998, 65, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Spek, J.; Dijkstra, J.; Van Duinkerken, G.; Bannink, A. A review of factors influencing milk urea concentration and its relationship with urinary urea excretion in lactating dairy cattle. J. Agric. Sci. 2013, 151, 407–423. [Google Scholar] [CrossRef]
- Huhtanen, P.; Cabezas-Garcia, E.H.; Krizsan, S.J.; Shingfield, K.J. Evaluation of between-cow variation in milk urea and rumen ammonia nitrogen concentrations and the association with nitrogen utilization and diet digestibility in lactating cows. J. Dairy Sci. 2015, 98, 3182–3196. [Google Scholar] [CrossRef] [PubMed]
- Patton, S. The presence of hippuric acid in milk. J. Dairy Sci. 1953, 36, 943–947. [Google Scholar] [CrossRef]
- Besle, J.M.; Viala, D.; Martin, B.; Pradel, P.; Meunier, B.; Berdagué, J.L.; Fraisse, D.; Lamaison, J.L.; Coulon, J.B. Ultraviolet-absorbing compounds in milk are related to forage polyphenols. J. Dairy Sci. 2010, 93, 2846–2856. [Google Scholar] [CrossRef] [PubMed]
- Carpio, A.; Rodríguez-Estévez, V.; Sánchez-Rodríguez, M.; Arce, L.; Valcárcel, M. Differentiation of organic goat’s milk based on its hippuric acid content as determined by capillary electrophoresis. Electrophoresis 2010, 31, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Boudonck, K.J.; Mitchell, M.W.; Wulff, J.; Ryals, J.A. Characterization of the biochemical variability of bovine milk using metabolomics. Metabolomics 2009, 5, 375–386. [Google Scholar] [CrossRef]
- Carpio, A.; Bonilla-Valverde, D.; Arce, C.; Rodriguez-Estevez, V.; Sanchez-Rodriguez, M.; Arce, L.; Valcarcel, M. Evaluation of hippuric acid content in goat milk as a marker of feeding regimen. J. Dairy Sci. 2013, 96, 5426–5434. [Google Scholar] [CrossRef] [PubMed]
- Gulati, A.; Galvin, N.; Lewis, E.; Hennessy, D.; O’Donovan, M.; McManus, J.J.; Fenelon, M.A.; Guinee, T.P. Outdoor grazing of dairy cows on pasture versus indoor feeding on total mixed ration: Effects on gross composition and mineral content milk during lactation. J. Dairy Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.; Lynch, M.B.; Hennessy, D. Including white clover in nitrogen fertilized perennial ryegrass swards: Effects on dry matter intake and milk production of spring calving dairy cows. J. Agric. Sci. 2017, 155, 657–668. [Google Scholar] [CrossRef]
- O’Connor, P.J.; Hennessy, D.; Lynch, M.B.; Slattery, H.; Lewis, E. The effect of dicyandiamide on rumen and blood metabolites, diet digestibility and urinary excretion. Lives. Sci. 2013, 155, 30–37. [Google Scholar] [CrossRef]
- Institut National de la Recherche Agronomique (INRA). Alimentation des Bovins, Ovins et Caprins, Éditions Quæ; INRA: Paris, France, 2007. [Google Scholar]
- Saude, E.; Slupsky, C.; Sykes, B. Optimization of NMR analysis of biological fluids for quantitative accuracy. Metabolomics 2006, 2, 113–123. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of pcr-quality community DNA from digesta and fecal samples. Biotechniques 2004, 36, 808–813. [Google Scholar] [PubMed]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboke, NJ, USA, 2002. [Google Scholar]
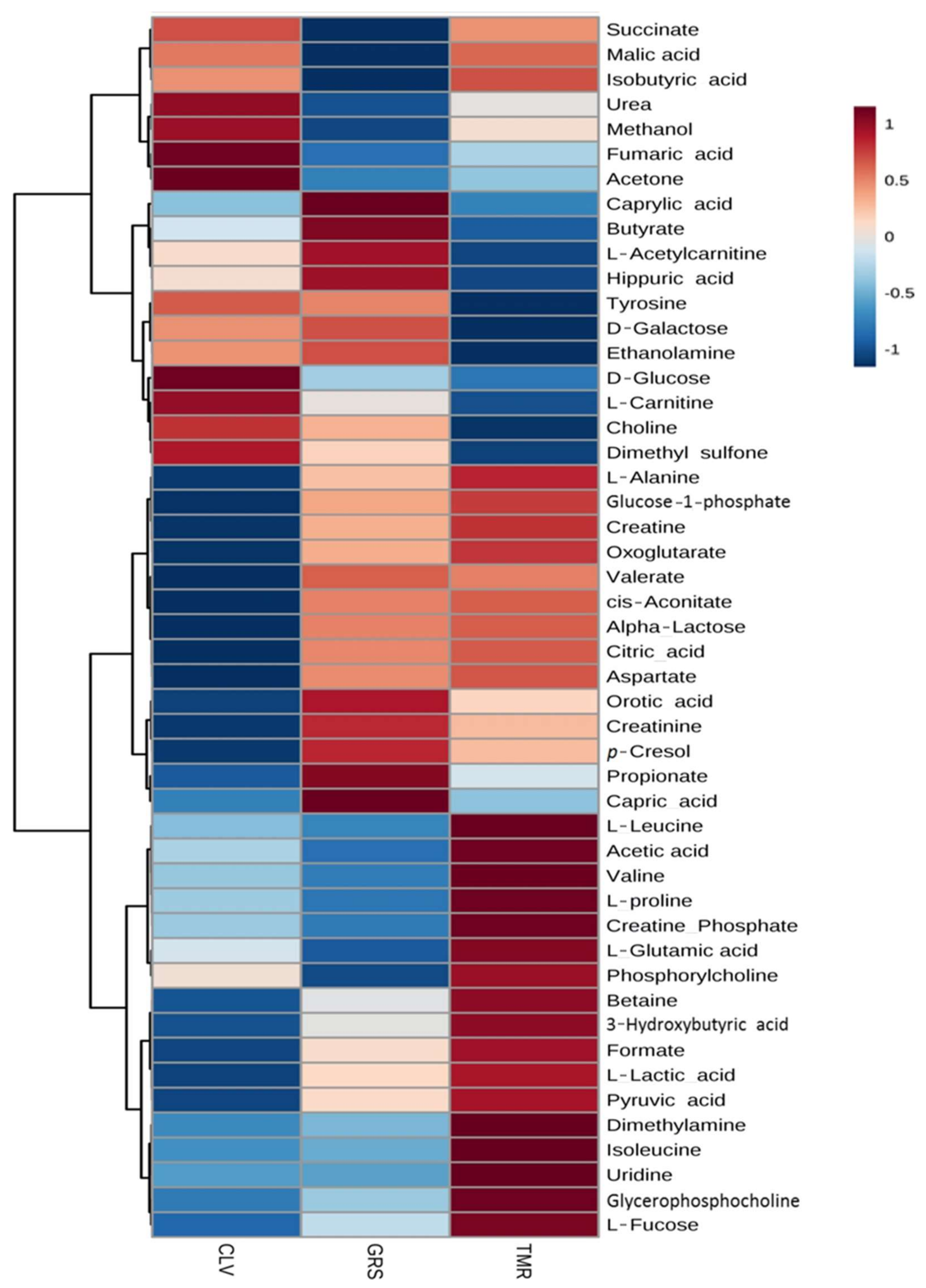
| Metabolite (μM) | Diet | p-Value | ||||
|---|---|---|---|---|---|---|
| TMR | GRS | CLV | Diet | Time | Diet × Time | |
| 2-hydroxyisovalerate | 7.06 (±6.35) | 6.17 (±4.99) | 7.49 (±5.32) | 0.78 | 0.81 | 0.21 |
| 3-Hydroxybutyric acid | 12.78 (±14.29) | 8.68 (±5.09) | 12.63 (±13.27) | 0.51 | 0.13 | 0.28 |
| 3-Hydroxyphenylacetic acid | 26.71 (±9.76) | 22.41 (±9.00) | 22.85 (±7.60) | 0.25 | <0.01 | 0.16 |
| 3-Phenylpropionate | 745.91 (±112.81) b | 632.94 (±170.49) a | 634.19 (±124.84) a | <0.01 | <0.01 | <0.01 |
| 4-Aminobutyrate | 36.39 (±18.73) | 52.15 (±35.68) | 48.77 (±20.07) | 0.36 | 0.26 | 0.86 |
| Acetic acid | 55,295.87 (±7149.21) b | 54,836.35 (±6828.43) a | 58,322.59 (±3754.77) b | 0.01 | <0.01 | 0.03 |
| Acetoin | 25.41 (±9.68) | 21.36 (±6.28) | 25.29 (±8.02) | 0.26 | 0.1 | 0.68 |
| Acetone | 8.67 (±3.16) a | 8.53 (±2.01) a | 12.03 (±6.44) b | 0.01 | <0.01 | 0.07 |
| Adenine | 26.43 (±7.53) | 26.12 (±12.30) | 30.54 (±22.58) | 0.51 | <0.01 | 0.22 |
| Adenosine | 4.55 (±3.34) | 6.78 (±5.51) | 5.08 (±4.26) | 0.35 | 0.16 | 0.49 |
| Aspartate | 153.24 (±54.47) b | 115.83 (±45.41) a | 148.01 (±79.63) b | 0.03 | <0.01 | <0.01 |
| Benzoic acid | 23.88 (±4.87) | 26.31 (±7.47) | 27.46 (±5.34) | 0.02 d | <0.01 | 0.04 |
| Beta Alanine | 18.43 (±14.98) | 18.24 (±16.25) | 17.44 (±14.00) | 0.94 | <0.01 | 0.01 |
| Betaine | 6.91 (±10.81) | 5.08 (±5.66) | 3.80 (±1.87) | 0.51 | 0.36 | 0.79 |
| Butyrate | 12,585.29 (±2066.81) | 14,245.39 (±2839.45) | 14,728.79 (±2514.33) | 0.02 d | 0.05 | 0.21 |
| Cadaverine | 82.27 (±39.12) | 96.51 (±76.25) | 107.09 (±27.17) | 0.43 | 0.97 | 0.16 |
| Choline | 25.89 (±14.19) b | 14.68 (±8.23) a | 12.59 (±8.65) a | <0.01 | 0.19 | 0.03 |
| cis-Aconitate | 5.58 (±3.40) | 8.46 (±9.56) | 7.59 (±5.55) | 0.50 | 0.30 | 0.44 |
| Citric acid | 7.46 (±6.93) | 7.53 (±4.36) | 6.62 (±2.72) | 0.83 | 0.02 | 0.67 |
| Creatine | 8.69 (±5.98) | 6.93 (±5.02) | 6.93 (±3.56) | 0.51 | 0.71 | 0.96 |
| d-Glucose | 520.63 (±287.13) | 544.86 (±383.52) | 602.03 (±536.85) | 0.63 | <0.01 | 0.03 |
| d-Maltose | 71.21 (±66.10) b | 34.40 (±27.63) a | 33.35 (±26.23) a | 0.02 | <0.01 | 0.18 |
| Dimethyl sulfone | 3.83 (±2.43) a | 16.81 (±7.70) b | 33.38 (±12.49) c | <0.01 | 0.03 | 0.21 |
| Dimethylamine | 4.25 (±7.97) | 2.31 (±1.32) | 4.80 (±9.51) | 0.64 | 0.26 | 0.69 |
| Dimethylglycine | 14.28 (±21.99) | 4.55 (±4.93) | 7.24 (±13.14) | 0.31 | 0.81 | 0.79 |
| Ethanol | 25.63 (±22.49) | 59.29 (±109.16) | 31.70 (±20.26) | 0.37 | 0.66 | 0.28 |
| Ethanolamine | 29.38 (±13.94) | 28.26 (±13.02) | 29.48 (±19.30) | 0.94 | <0.01 | 0.44 |
| Formate | 118.49 (±4.16) a,b | 114.91 (±5.35) a | 118.09 (±2.89) b | 0.01 | 0.01 | 0.06 |
| Glycerol | 254.92 (±55.91) b | 242.33 (±34.69) a | 248.91 (±48.42) a,b | 0.51 d | <0.01 | 0.28 |
| Glycine | 121.49 (±37.66) a | 119.16 (±40.23) a | 158.48 (±60.01) b | 0.01 | <0.01 | 0.03 |
| Hypoxanthine | 171.16 (±32.82) b | 159.21 (±56.02) a | 179.41 (±70.06) b | 0.27 d | <0.01 | 0.03 |
| Inosine | 11.53 (±6.29) a | 27.24 (±26.18) b | 22.14 (±18.14) b | 0.01 | <0.01 | 0.03 |
| Isobutyric acid | 796.76 (±88.10) a | 811.95 (±103.96) a | 997.57 (±191.24) b | <0.01 | <0.01 | <0.01 |
| Isoleucine | 84.44 (±23.65) a,b | 83.46 (±35.14) a | 105.76 (±38.25) b | 0.07 d | <0.01 | 0.27 |
| Isopropanol | 18.16 (±3.86) a | 22.25 (±6.37) a | 42.18 (±27.01) b | <0.01 | <0.01 | <0.01 |
| Isovaleric acid | 683.44 (±112.48) a | 701.31 (±135.43) a | 951.09 (±283.61) b | <0.01 | <0.01 | <0.01 |
| l-Alanine | 180.40 (±45.02) a | 189.58 (±55.46) a | 238.88 (±76.37) b | <0.01 | <0.01 | 0.04 |
| l-Glutamic acid | 293.98 (±71.48) b | 252.06 (±72.63) a | 350.58 (±97.44) b | <0.01 | 0.02 | 0.06 |
| l-Histidine | 32.96 (±11.46) | 36.68 (±14.08) | 34.49 (±15.58) | 0.65 | <0.01 | 0.32 |
| l-Lactic acid | 30.66 (±22.16) | 29.41 (±44.10) | 27.49 (±18.07) | 0.94 | 0.06 | 0.54 |
| l-Leucine | 89.73 (±19.51) a,b | 93.85 (±31.78) a | 119.58 (±38.95) b | 0.01 | <0.01 | 0.21 |
| l-Lysine | 149.54 (±64.36) | 173.82 (±77.85) | 205.94 (±123.11) | 0.14 | <0.01 | 0.45 |
| l-Phenylalanine | 51.48 (±13.84) | 50.64 (±19.03) | 57.76 (±22.60) | 0.32 | <0.01 | 0.17 |
| l-Proline | 84.33 (±34.94) | 80.50 (±37.46) | 108.33 (±63.16) | 0.09 | <0.01 | 0.09 |
| l-Threonine | 108.67 (±41.15) b | 85.44 (±32.09) a | 106.61 (±42.64) a,b | 0.08 d | <0.01 | 0.04 |
| Methanol | 13.74 (±14.69) | 9.69 (±1.28) | 11.44 (±3.02) | 0.51 | 0.81 | 0.44 |
| Methionine | 33.63 (±9.78) | 35.13 (±13.72) | 40.76 (±12.37) | 0.26 | 0.01 | 0.34 |
| Methylamine | 4.64 (±11.01) | 11.39 (±16.79) | 16.25 (±33.42) | 0.25 | 0.15 | 0.04 |
| Nicotinate | 25.77 (±7.67) a | 28.56 (±9.80) a | 35.93 (±9.57) b | <0.01 | <0.01 | 0.03 |
| o-Hydroxyphenylacetic acid | 17.38 (±3.47) c | 13.08 (±3.78) a | 15.66 (±5.49) b | <0.01 | <0.01 | 0.03 |
| p-Cresol | 58.38 (±15.58) a | 65.95 (±24.67) a | 85.18 (±38.62) b | <0.01 | <0.01 | <0.01 |
| p-Hydroxyphenylacetic acid | 17.86 (±4.95) | 14.62 (±3.72) | 13.77 (±4.27) | 0.07 | 0.87 | 0.72 |
| Phenylacetate | 199.58 (±56.74) a | 262.11 (±109.84) a | 381.02 (±191.25) b | <0.01 | <0.01 | <0.01 |
| Propionate | 17,119.26 (±3923.21) | 18,084.74 (±3647.63) | 18,140.78 (±2724.51) | 0.43 | 0.01 | 0.04 |
| Putrescine | 58.93 (±21.87) | 56.48 (±55.10) | 45.56 (±16.64) | 0.51 | 0.02 | 0.56 |
| Succinate | 123.57 (±122.66) b | 74.56 (±51.26) a | 90.93 (±54.20) a,b | 0.01 | <0.01 | <0.01 |
| Trimethylamine | 5.80 (±14.48) | 1.85 (±1.38) | 5.40 (±9.03) | 0.51 | 0.37 | 0.13 |
| Tryptophan | 7.17 (±1.65) | 7.39 (±3.02) | 8.19 (±3.41) | 0.36 | <0.01 | 0.11 |
| Tyrosine | 44.27 (±12.59) a,b | 43.55 (±16.83) a | 56.05 (±20.71) b | 0.03 | <0.01 | 0.21 |
| Uracil | 200.49 (±91.29) a | 264.18 (±106.92) b | 297.61 (±107.14) b | <0.01 | <0.01 | 0.55 |
| Uridine | 7.40 (±8.28) | 7.49 (±5.34) | 8.81 (±8.03) | 0.83 | 0.42 | 0.28 |
| Valerate | 1030.42 (±266.50) | 1256.89 (±602.06) | 1328.14 (±439.27) | 0.08 | 0.01 | 0.04 |
| Valine | 93.59 (±23.59) a,b | 90.46 (±35.70) a | 128.79 (±65.36) b | 0.01 | <0.01 | 0.11 |
| Early Lactation | Mid Lactation | Late Lactation | ||||
|---|---|---|---|---|---|---|
| GRS | CLV | GRS | CLV | GRS | CLV | |
| OM (g/kg of DM) | 881.2 ± 22.3 | 883.7 ± 32.4 | 864.1 ± 42.8 | 867.2 ± 32.8 | 807.5 ± 49.9 | 814.8 ± 28.2 |
| CP (g/kg of DM) | 218.4 ± 30.7 | 228.1 ± 19.3 | 216.9 ± 30.0 | 239.9 ± 28 | 266.5 ± 17.2 | 279.7 ± 25.2 |
| ADF (g/kg of DM) | 284.6 ± 18.4 | 283.0 ± 23.1 | 294.1 ± 17.1 | 298.4 ± 21.1 | 321.7 ± 24.7 | 314.3 ± 23.2 |
| NDF (g/kg of DM) | 378.2 ± 21.5 | 350.4 ± 27.5 | 386.1 ± 31.0 | 366.3 ± 31.2 | 429.5 ± 35.9 | 385.4 ± 22.0 |
| Ash (g/kg of DM) | 61.8 ± 10.1 | 60.4 ± 9.2 | 68.7 ± 16.9 | 61.8 ± 11.4 | 75.1 ± 8.9 | 65.1 ± 10.5 |
| UFL (/kg of DM) | 0.96 ± 0.03 | 0.97 ± 0.04 | 0.94 ± 0.03 | 0.95 ± 0.04 | 0.91 ± 0.05 | 0.94 ± 0.04 |
| PDIA (/kg of DM) | 42.8 ± 4.0 | 44.0 ± 2.5 | 42.6 ± 3.9 | 45.6 ± 3.7 | 49.1 ± 2.5 | 51.1 ± 3.7 |
| PDIE (/kg of DM) | 101.2 ± 4.0 | 102.9 ± 1.9 | 99.7 ± 3.9 | 103.4 ± 3.5 | 104 ± 3.6 | 108.4 ± 3.2 |
| PDIN (/kg of DM) | 141.1 ± 20.6 | 147.4 ± 13.0 | 140.0 ± 20.0 | 155.4 ± 18.9 | 173.5 ± 11.9 | 182.8 ± 17.5 |
| Early Lactation | Mid Lactation | Late Lactation | ||||
|---|---|---|---|---|---|---|
| Grass Silage | Maize Silage | Grass Silage | Maize Silage | Grass Silage | Maize Silage | |
| DM (g/kg of DM) | 238.6 ± 12.1 | 279.5 ± 13.9 | 466.8 ± 83.3 | 280.0 ± 25.7 | 340.2 ± 155.6 | 238.7 ± 3.3 |
| CP (g/kg of DM) | 96.7 ± 6.4 | 66.3 ± 8.1 | 128.2 ± 10.5 | 56.7 ± 5.0 | 118.4 ± 7.8 | 64.7 ± 9.0 |
| Starch (g/kg of DM) | NA | 221.3 ± 14.9 | NA | 235.7 ± 33.7 | NA | 198.3 ± 6.3 |
| ADF (g/kg of DM) | 262.5 ± 12.2 | NA | 270.6 ± 6.1 | NA | 275.7 ± 17.7 | NA |
| NDF (g/kg of DM) | 386.5 ± 27.4 | 470.5 ± 32.2 | 407.3 ± 10.3 | 510.3 ± 9.0 | 409.6 ± 35.7 | 522.7 ± 4.7 |
| ASH (g/kg of DM) | 92.3 ± 5.9 | 27.0 ± 1.6 | 82.8 ± 2.5 | 28.3 ± 4.6 | 89.1 ± 4.6 | 29.7 ± 0.5 |
| UFL (/kg of DM) | 1.02 ± 0.03 | 0.90 ± 0.1 | 1.0 ± 0.02 | 0.90 ± 0.1 | 0.97 ± 0.05 | 0.9 ± 0.1 |
| PDIA (/kg of DM) | 17.0 ± 1.0 | 14.4 ± 1.8 | 31.0 ± 5.1 | 12.3 ± 1.1 | 24.6 ± 6.7 | 14.1 ± 2.0 |
| PDIE (/kg of DM) | 57.0 ± 4.1 | 64.5 ± 1.7 | 81.0 ± 8.2 | 60.9 ± 0.8 | 72.6 ± 7.2 | 62.5 ± 2.2 |
| PDIN (/kg of DM) | 70.6 ± 1.2 | 40.7 ± 5.0 | 77.5 ± 4.8 | 34.8 ± 3.1 | 70.6 ± 6.8 | 39.7 ± 5.5 |
| Concentrate Ingredient Composition | % as Fed |
| Maize | 13.00 |
| Beet pulp molassed | 15.50 |
| Soyabean meal 48% CP | 30.00 |
| Maize distillers | 12.00 |
| Acid buffer | 0.70 |
| Maize/Beet Min Balancer | 2.50 |
| Salt | 0.50 |
| Barley (rolled) | 15.00 |
| Rapeseed meal | 7.50 |
| Megalac | 3.30 |
| Chemical Analysis | /kg as fed |
| DM, g | 874.90 |
| UFL | 1.02 |
| UFV | 0.99 |
| Crude protein % | 24.28 |
| PDIN, g | 169.26 |
| PDIE, g | 133.91 |
| Starch % | 18.10 |
| Sugar % | 6.92 |
| Crude fibre % | 6.10 |
| Oil % | 5.12 |
| Ash % | 7.99 |
| Copper mg/kg DM | 94.66 |
| ME MJ/kg DM | 11.16 |
| Metabolite (μM) | TMR | GRS | CLV | p-Value z |
|---|---|---|---|---|
| 3-Hydroxybutyric acid | 34.06 (±10.63) b | 28.18 (±2.92) a | 24.63 (±4.75) a | 0.07 d |
| Acetic acid | 71.81 (±41.67) | 41.47 (±10.17) | 47.88 (±12.36) | 0.12 |
| Acetone | 13.20 (±3.88) a | 11.66 (±1.22) a | 18.85 (±3.54) b | <0.01 |
| Alpha-Lactose | 109,682.11 (±4835.98) | 109,179.54 (±4189.15) | 103,625.15 (±5267.57) | 0.07 |
| Aspartate | 28.86 (±11.37) b | 28.63 (±13.11) b | 20.35 (±7.08) a | 0.33 d |
| Betaine | 90.15 (±25.33) a | 72.22 (±16.17) a,b | 64.41 (±31.44) b | 0.23 d |
| Butyrate | 29.22 (±5.72) a | 43.93 (±16.09) b | 35.91 (±16.47) b | 0.23 d |
| Capric acid | 20.66 (±5.75) | 23.59 (±7.41) | 19.89 (±5.74) | 0.58 |
| Caprylic acid | 12.92 (±6.01) a | 23.01 (±8.03) b | 14.23 (±6.69) a | 0.04 |
| Choline | 214.22 (±58.37) a | 298.26 (±146.01) b | 310.54 (±77.69) b | 0.23 d |
| cis-Aconitate | 35.20 (±4.97) | 35.13 (±6.74) | 32.12 (±3.51) | 0.55 |
| Citric acid | 4595.41 (±448.18) | 4602.04 (±617.46) | 4453.36 (±225.49) | 0.77 |
| Creatine | 436.42 (±79.99) | 432.95 (±84.67) | 427.70 (±115.63) | 0.98 |
| Creatine Phosphate | 60.35 (±28.96) | 49.33 (±52.66) | 38.24 (±39.15) | 0.64 |
| Creatinine | 50.69 (±7.42) a,b | 53.41 (±9.12) b | 45.12 (±8.40) a | 0.23 |
| d-Galactose | 730.39 (±277.50) | 901.88 (±624.82) | 1023.29 (±936.59) | 0.74 |
| d-Glucose | 331.24 (±84.17) | 340.62 (±65.95) | 387.43 (±91.21) | 0.47 |
| Dimethyl sulfone | 11.67 (±1.59) a | 27.97 (±7.90) b | 46.41 (±12.72) c | <0.01 |
| Dimethylamine | 14.60 (±2.77) a | 9.95 (±1.96) b | 9.40 (±2.30) b | <0.01 |
| Ethanolamine | 84.09 (±27.64) a | 125.81 (±39.77) b | 116.79 (±23.05) b | 0.07 d |
| Formate | 117.40 (±1.66) | 116.44 (±1.93) | 115.20 (±2.75) | 0.23 |
| Fumaric acid | 11.63 (±3.65) | 11.20 (±4.36) | 12.54 (±2.98) | 0.77 |
| Glucose-1-phosphate | 52.60 (±27.92) | 86.70 (±128.89) | 28.31 (±27.81) | 0.47 |
| Glycerophosphocholine | 617.18 (±83.87) | 532.34 (±107.89) | 531.67 (±240.19) | 0.58 |
| Hippuric acid | 112.76 (±33.00) a | 227.93 (±38.90) c | 165.93 (±32.77) b | <0.01 |
| Isobutyric acid | 15.87 (±20.47) | 5.10 (±4.79) | 14.45 (±15.29) | 0.46 |
| Isoleucine | 5.15 (±1.64) | 4.33 (±1.74) | 4.20 (±1.53) | 0.58 |
| l-Acetylcarnitine | 45.97 (±7.39) | 51.71 (±12.86) | 49.66 (±12.65) | 0.64 |
| l-Alanine | 29.20 (±5.91) b | 27.45 (±5.67) b | 23.82 (±5.36) a | 0.27 d |
| l-Carnitine | 75.91 (±18.17) | 81.36 (±20.84) | 85.71 (±17.08) | 0.64 |
| l-Fucose | 27.79 (±13.14) b | 21.03 (±14.33) a,b | 15.36 (±7.37) a | 0.23 d |
| l-Glutamic acid | 196.16 (±64.07) | 188.62 (±77.08) | 190.87 (±67.63) | 0.98 |
| l-Lactic acid | 73.34 (±93.23) | 47.31 (±26.19) | 33.43 (±9.04) | 0.49 |
| l-Leucine | 4.64 (±1.55) | 3.80 (±1.31) | 3.81 (±0.83) | 0.47 |
| l-proline | 82.350 (±29.96) b | 55.55 (±12.68) a | 60.57 (±15.15) a | 0.07 d |
| Malic acid | 92.79 (±29.23) | 83.66 (±28.83) | 91.15 (±26.69) | 0.78 |
| Methanol | 27.43 (±15.84) | 23.64 (±7.16) | 29.85 (±19.30) | 0.74 |
| Orotic acid | 542.52 (±165.89) | 581.43 (±134.08) | 487.36 (±177.47) | 0.59 |
| Oxoglutarate | 117.93 (±17.22) | 116.48 (±18.59) | 111.39 (±15.62) | 0.75 |
| p-Cresol | 13.87 (±11.56) | 13.39 (±10.02) | 9.36 (±2.04) | 0.62 |
| Phosphorylcholine | 72.12 (±59.74) b | 30.83 (±34.97) a | 44.14 (±32.01) a,b | 0.27 d |
| Propionate | 5.71 (±7.02) | 7.58 (±6.92) | 3.84 (±2.27) | 0.58 |
| Pyruvic acid | 38.03 (±8.02) | 36.52 (±14.70) | 31.75 (±8.44) | 0.58 |
| Succinate | 23.20 (±6.60) | 19.73 (±3.18) | 23.06 (±3.75) | 0.41 |
| Tyrosine | 4.71 (±0.96) a | 8.11 (±1.97) b | 8.65 (±2.51) b | <0.01 |
| Urea | 282.72 (±45.21) a | 221.68 (±102.60) a | 389.71 (±57.62) b | <0.01 |
| Uridine | 16.07 (±4.91) b | 11.76 (±1.81) a | 11.78 (±2.37) a | 0.06 d |
| Valerate | 6.50 (±3.19) | 6.47 (±2.84) | 5.44 (±2.61) | 0.74 |
| Valine | 11.27 (±3.98) b | 7.92 (±1.59) a | 8.55 (±2.14) a | 0.1 d |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Callaghan, T.F.; Vázquez-Fresno, R.; Serra-Cayuela, A.; Dong, E.; Mandal, R.; Hennessy, D.; McAuliffe, S.; Dillon, P.; Wishart, D.S.; Stanton, C.; et al. Pasture Feeding Changes the Bovine Rumen and Milk Metabolome. Metabolites 2018, 8, 27. https://doi.org/10.3390/metabo8020027
O’Callaghan TF, Vázquez-Fresno R, Serra-Cayuela A, Dong E, Mandal R, Hennessy D, McAuliffe S, Dillon P, Wishart DS, Stanton C, et al. Pasture Feeding Changes the Bovine Rumen and Milk Metabolome. Metabolites. 2018; 8(2):27. https://doi.org/10.3390/metabo8020027
Chicago/Turabian StyleO’Callaghan, Tom F., Rosa Vázquez-Fresno, Arnau Serra-Cayuela, Edison Dong, Rupasri Mandal, Deirdre Hennessy, Stephen McAuliffe, Pat Dillon, David S. Wishart, Catherine Stanton, and et al. 2018. "Pasture Feeding Changes the Bovine Rumen and Milk Metabolome" Metabolites 8, no. 2: 27. https://doi.org/10.3390/metabo8020027
APA StyleO’Callaghan, T. F., Vázquez-Fresno, R., Serra-Cayuela, A., Dong, E., Mandal, R., Hennessy, D., McAuliffe, S., Dillon, P., Wishart, D. S., Stanton, C., & Ross, R. P. (2018). Pasture Feeding Changes the Bovine Rumen and Milk Metabolome. Metabolites, 8(2), 27. https://doi.org/10.3390/metabo8020027







