Metabolomic Profiling of Submaximal Exercise at a Standardised Relative Intensity in Healthy Adults
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Solvents
2.2. Subjects and Experimental Design
2.3. VO2max Test
2.4. Submaximal Test
2.5. Sample Collection and Preparation
2.6. LC-MS Method
2.7. LC-MS Data Processing and Statistical Analysis
2.8. Multivariate Analysis
3. Results
3.1. Physiological Responses to Submaximal Exercise
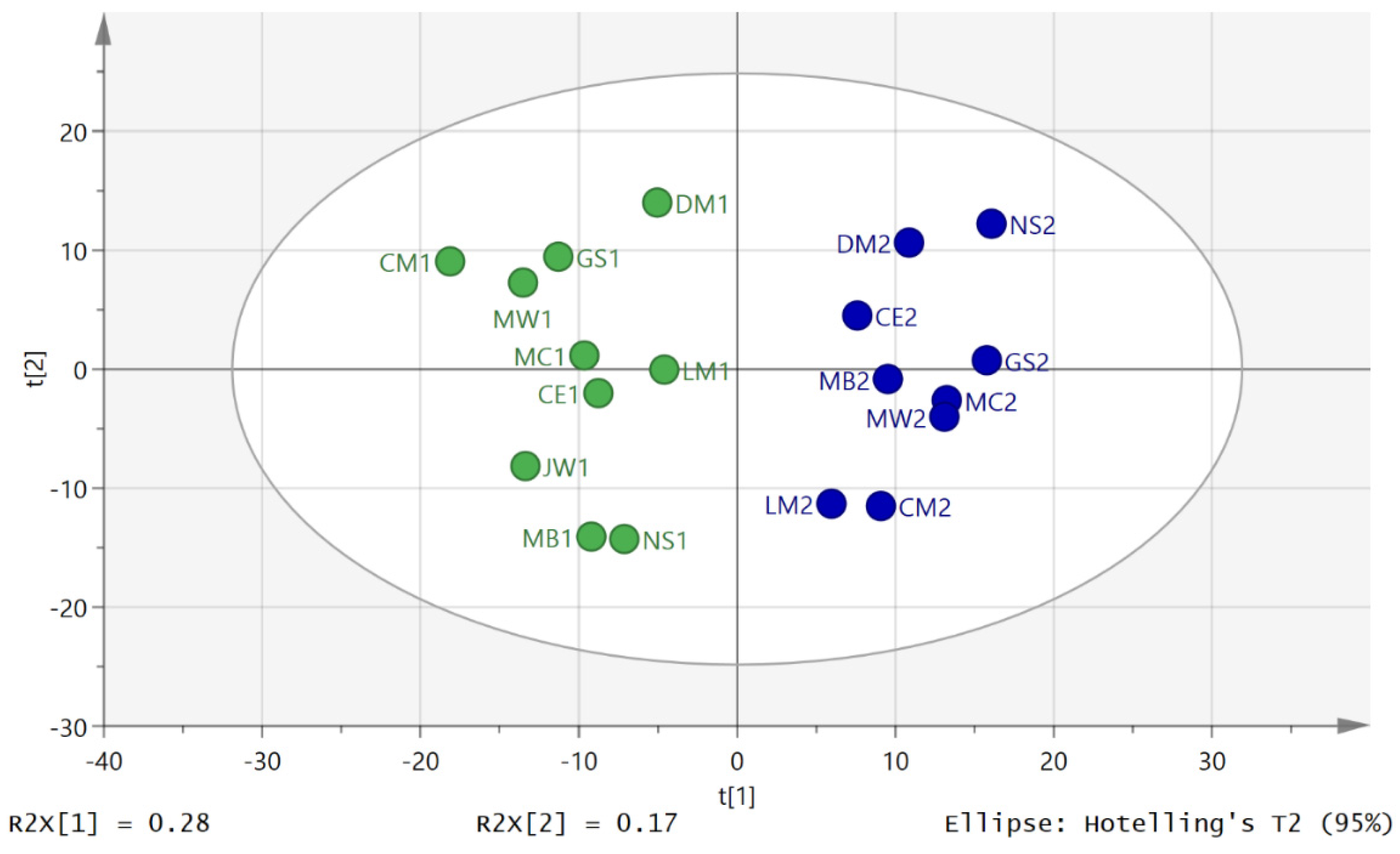
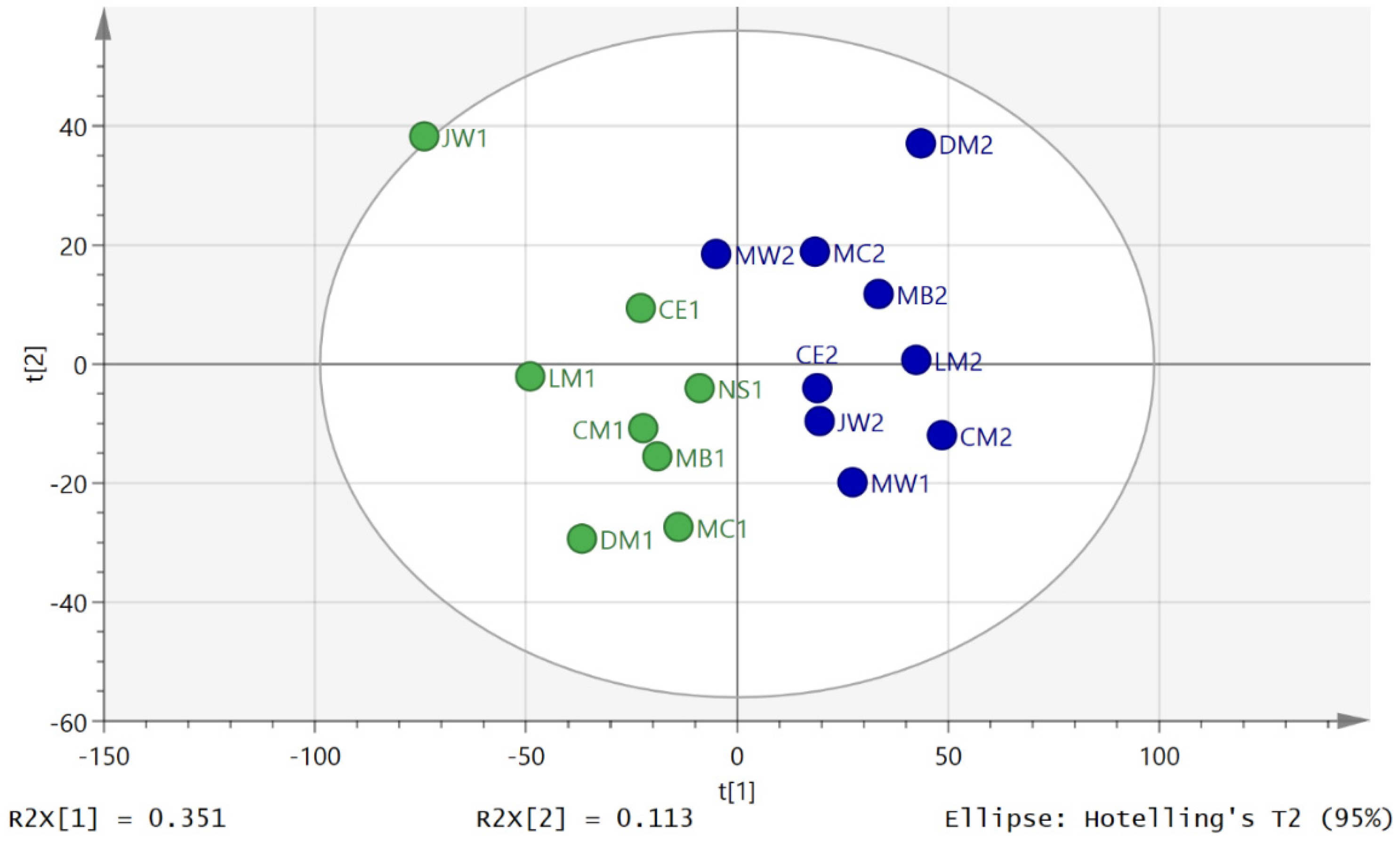
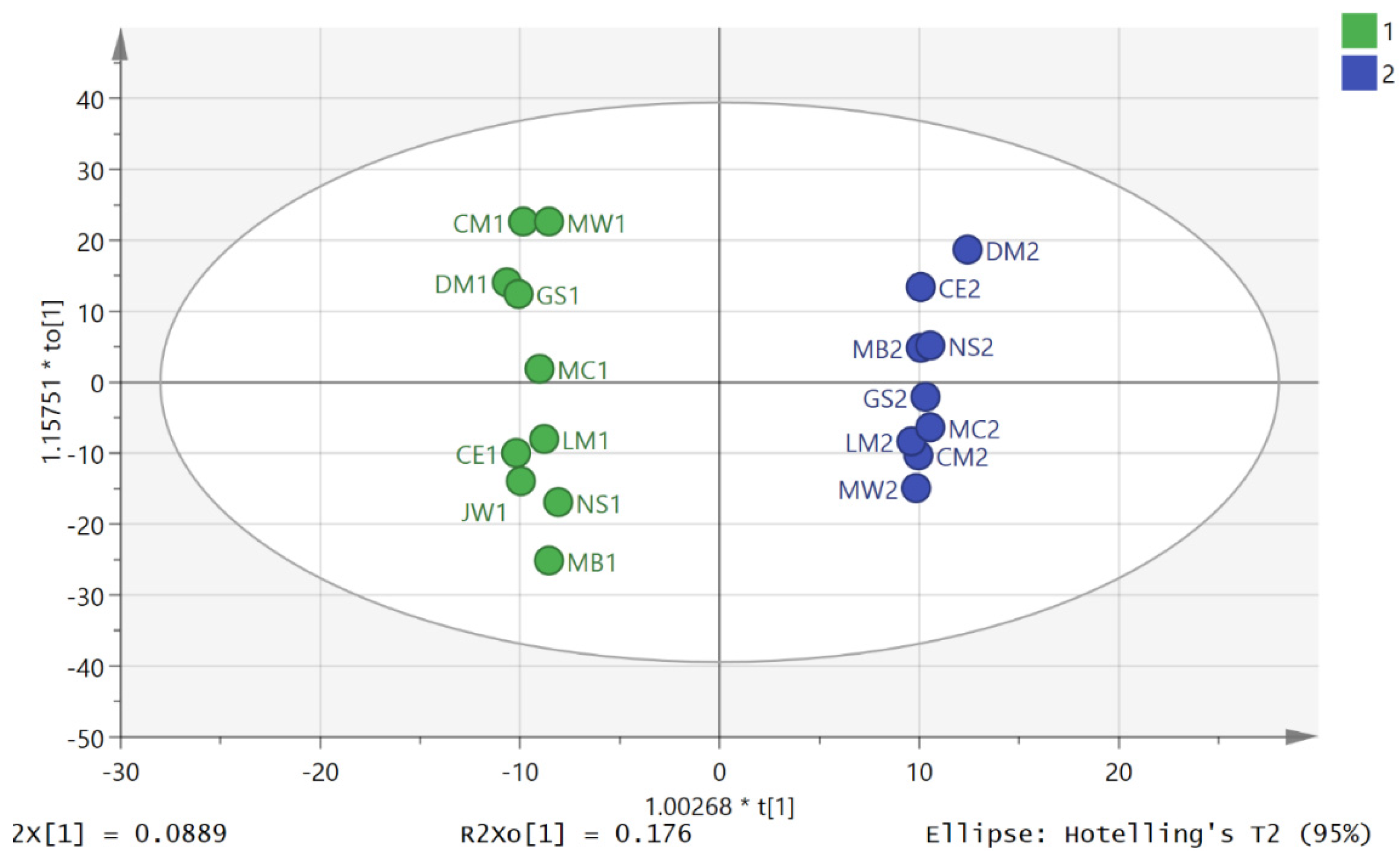
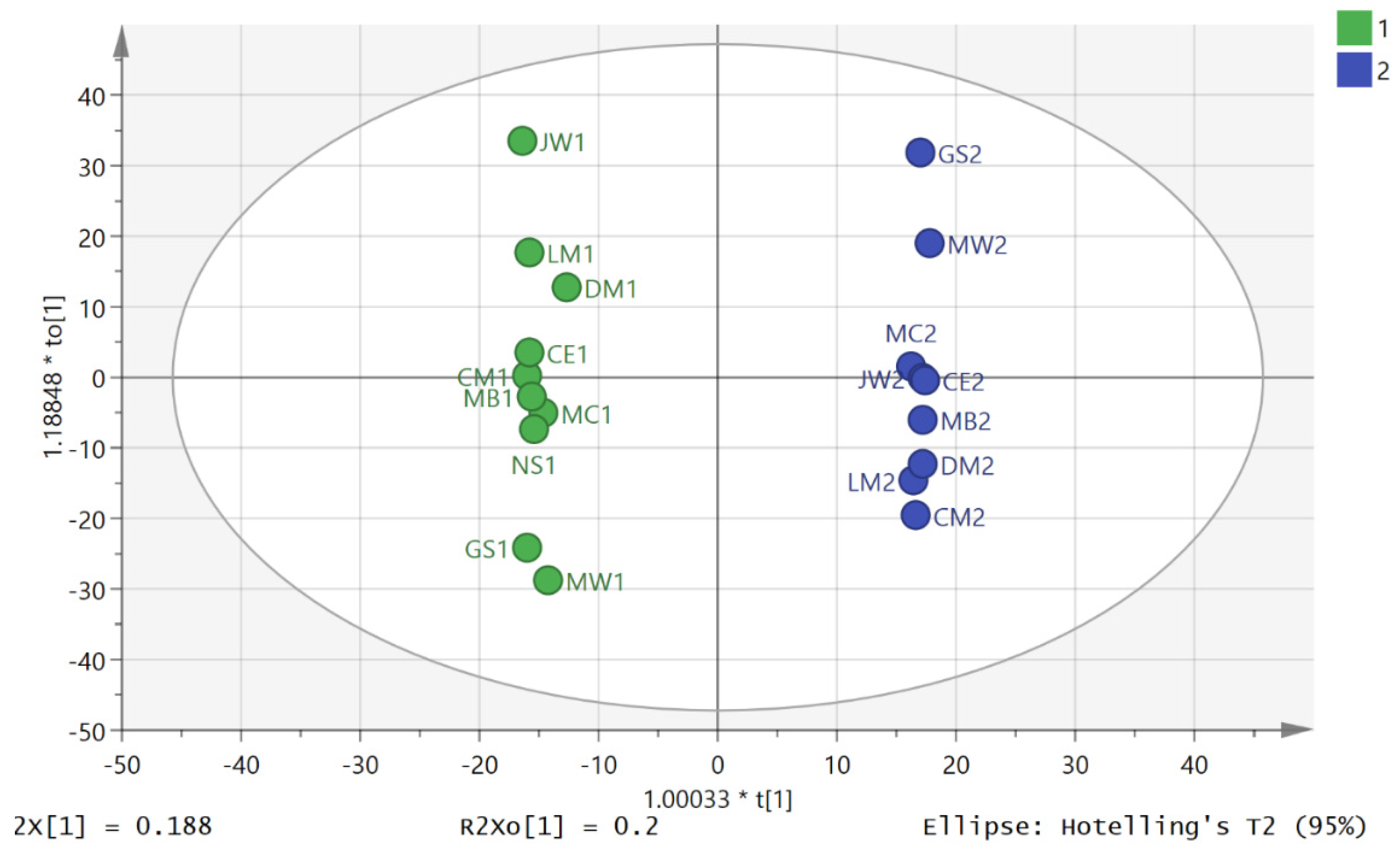
3.2. Metabolite Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Sarris, J.; O’Neil, A.; Coulson, C.E.; Schweitzer, I.; Berk, M. Lifestyle Medicine for Depression. BMC Psychiatry 2014, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Unhealthy Diets & Physical Inactivity; World Health Organization: Geneva, Switzerland, 2009; pp. 1–2. [Google Scholar]
- World Health Organization. Deaths from Cardiovascular Diseases and Diabetes. Available online: http://www.who.int/mediacentre/factsheets/fs317/en/ (accessed on 23 February 2016).
- Allender, S.; Foster, C.; Scarborough, P.; Rayner, M. The Burden of Physical Activity-Related Ill Health in the UK. J. Epidemiol. Community Health 2007, 61, 344–348. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Health Risks: Mortality and Burden of Disease Attributable to Selected Major Risks; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Scarborough, P.; Bhatnagar, P.; Wickramasinghe, K.K.; Allender, S.; Foster, C.; Rayner, M. The Economic Burden of Ill Health due to Diet, Physical Inactivity, Smoking, Alcohol and Obesity in the UK: An Update to 2006–07 NHS Costs. J. Public Health 2011, 33, 527–535. [Google Scholar] [CrossRef] [PubMed]
- British Heart Foundation National Centre. Economic Costs of Physical Inactivity; British Heart Foundation National Centre: Loughborough, UK, 2013; pp. 1–8. [Google Scholar]
- Reilly, J.J.; Dick, S.; McNeill, G.; Tremblay, M.S. Results From Scotland’s 2013 Report Card on Physical Activity for Children and Youth. J. Phys. Act. Health 2014, 11, S93–S97. [Google Scholar] [CrossRef] [PubMed]
- Daskalaki, E.; Easton, C.; Watson, D. The Application of Metabolomic Profiling to the Effects of Physical Activity. Curr. Metabolomics 2015, 2, 233–263. [Google Scholar] [CrossRef]
- Carnethon, M.R.; Gulati, M.; Greenland, P. Prevalence and cardiovascular disease correlates of low cardiorespiratory fitness in adolescents and adults. J. Am. Med. Assoc. 2005, 294, 2981–2988. [Google Scholar] [CrossRef] [PubMed]
- Joyner, M.J.; Coyle, E. Endurance exercise performance: The physiology of champions. J. Physiol. 2008, 586, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Daskalaki, E.; Blackburn, G.; Kalna, G.; Zhang, T.; Anthony, N.; Watson, D.G. A Study of the Effects of Exercise on the Urinary Metabolome Using Normalisation to Individual Metabolic Output. Metabolites 2015, 5, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Creek, D.J.; Jankevics, A.; Breitling, R.; Watson, D.G.; Barrett, M.P.; Burgess, K.E.V. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: Improved metabolite identification by retention time prediction. Anal. Chem. 2011, 83, 8703–8710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Watson, D.G.; Wang, L.; Abbas, M.; Murdoch, L.; Bashford, L.; Ahmad, I.; Lam, N.Y.; Ng, A.C.F.; Leung, H.Y. Application of Holistic Liquid Chromatography-High Resolution Mass Spectrometry Based Urinary Metabolomics for Prostate Cancer Detection and Biomarker Discovery. PLoS ONE 2013, 8, e65880. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Watson, D.G.; Wang, L.; Westrop, G.D.; Coombs, G.H.; Zhang, T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A 2014, 1362, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.L.; Buskirk, E.; Henschel, A. Maximal oxygen uptake as an objective measure of cardiorespiratory performance. J. Appl. Physiol. 1955, 8, 73–80. [Google Scholar] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- MetaboLights. Available online: http://www.ebi.ac.uk/metabolights/ (accessed on 22 February 2016).
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; Robards, K.; Prenzler, P.; Kendall, M. Recent and potential developments in the analysis of urine: A review. Anal. Chim. Acta 2011, 684, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate a Practical and powerful Approach to Multiple Testing. J. Roy. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]
- McDonald, J.H. Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MD, USA, 2014. [Google Scholar]
- Scripps Center for Metabolomics. METLIN. Available online: https://metlin.scripps.edu/index.php (accessed on 22 February 2016).
- Zieliński, J.; Krasińska, B.; Kusy, K. Hypoxanthine as a predictor of performance in highly trained athletes. Int. J. Sports Med. 2013, 34, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, J.; Kusy, K. Training-induced adaptation in purine metabolism in high-level sprinters vs. triathletes. J. Appl. Physiol. 2012, 112, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Stathis, C.G.; Carey, M.F.; Hayes, A.; Garnham, A.P.; Snow, R.J. Sprint training reduces urinary purine loss following intense exercise in humans. Appl. Physiol. Nutr. Metab. 2006, 31, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Sahlin, K.; Tonkonogi, M.; Söderlund, K. Plasma hypoxanthine and ammonia in humans during prolonged exercise. Eur. J. Appl. Physiol. Occup. Physiol. 1999, 80, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Dudzinska, W.; Lubkowska, A.; Dolegowska, B.; Safranow, K.; Jakubowska, K. Adenine, guanine and pyridine nucleotides in blood during physical exercise and restitution in healthy subjects. Eur. J. Appl. Physiol. 2010, 110, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, J.; Kusy, K.; Rychlewski, T. Effect of training load structure on purine metabolism in middle-distance runners. Med. Sci. Sports Exerc. 2011, 43, 1798–1807. [Google Scholar] [CrossRef] [PubMed]
- Dansie, L.E.; Reeves, S.; Miller, K.; Zano, S.P.; Frank, M.; Pate, C.; Wang, J.; Jackowski, S. Physiological Roles of the Pantothenate Kinases. Biochem. Soc. Trans. 2014, 42, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Bracken, R.M.; Linnane, D.M.; Brooks, S. Plasma catecholamine and nephrine responses to brief intermittent maximal intensity exercise. Amino Acids. 2009, 36, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Lustgarten, M.S.; Price, L.L.; Chalé, A.; Fielding, R.A. Metabolites Related to Gut Bacterial Metabolism, Peroxisome Proliferator-Activated Receptor-Alpha Activation, and Insulin Sensitivity Are Associated with Physical Function in Functionally-Limited Older Adults. Aging Cell 2014, 13, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Dovio, A.; Roveda, E.; Sciolla, C.; Montaruli, A.; Raffaelli, A.; Saba, A.; Calogiuri, G.; de Francia, S.; Borrione, P.; Salvadori, P. Intense physical exercise increases systemic 11β-hydroxysteroid dehydrogenase type 1 activity in healthy adult subjects. Eur. J. Appl. Physiol. 2010, 108, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Hafezi-Moghadam, A.; Simoncini, T.; Yang, Z.; Limbourg, F.P.; Plumier, J.-C.; Rebsamen, M.C.; Hsieh, C.-M.; Chui, D.-S.; Thomas, K.L.; Prorock, A.J.; et al. Acute cardiovascular protective effects of corticosteroids are mediated by non-transcriptional activation of endothelial nitric oxide synthase. Nat. Med. 2002, 8, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Gatti, R.; Cappellin, E.; Zecchin, B.; Antonelli, G.; Spinella, P.; Mantero, F.; de Palo, E.F. Urinary high performance reverse phase chromatography cortisol and cortisone analyses before and at the end of a race in elite cyclists. J. Chromatogr. B 2005, 824, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.S.; Copeland, J.L.; van Helder, W. Effect of training status and exercise mode on endogenous steroid hormones in men. J. Appl. Physiol. 2004, 96, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Balderas, C.; Rupérez, F.J.; Ibañez, E.; Señorans, J.; Guerrero-Fernández, J.; Casado, I.G.; Gracia-Bouthelier, R.; García, A.; Barbas, C. Plasma and urine metabolic fingerprinting of type 1 diabetic children. Electrophoresis 2013, 34, 2882–2890. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-L.; Aoyama, S.; Nagai, R.; Miyoshi, N.; Ohshima, H. Inhibition of L-arginine metabolizing enzymes by L-arginine-derived advanced glycation end products. J. Clin. Biochem. Nutr. 2010, 46, 177. [Google Scholar] [CrossRef] [PubMed]
- Lustgarten, M.S.; Price, L.L.; Logvinenko, T.; Hatzis, C.; Padukone, N.; Reo, N.V.; Phillips, E.M.; Kirn, D.; Mills, J.; Fielding, R.A. Identification of serum analytes and metabolites associated with aerobic capacity. Eur. J. Appl. Physiol. 2013, 113, 1311–1320. [Google Scholar] [CrossRef] [PubMed]

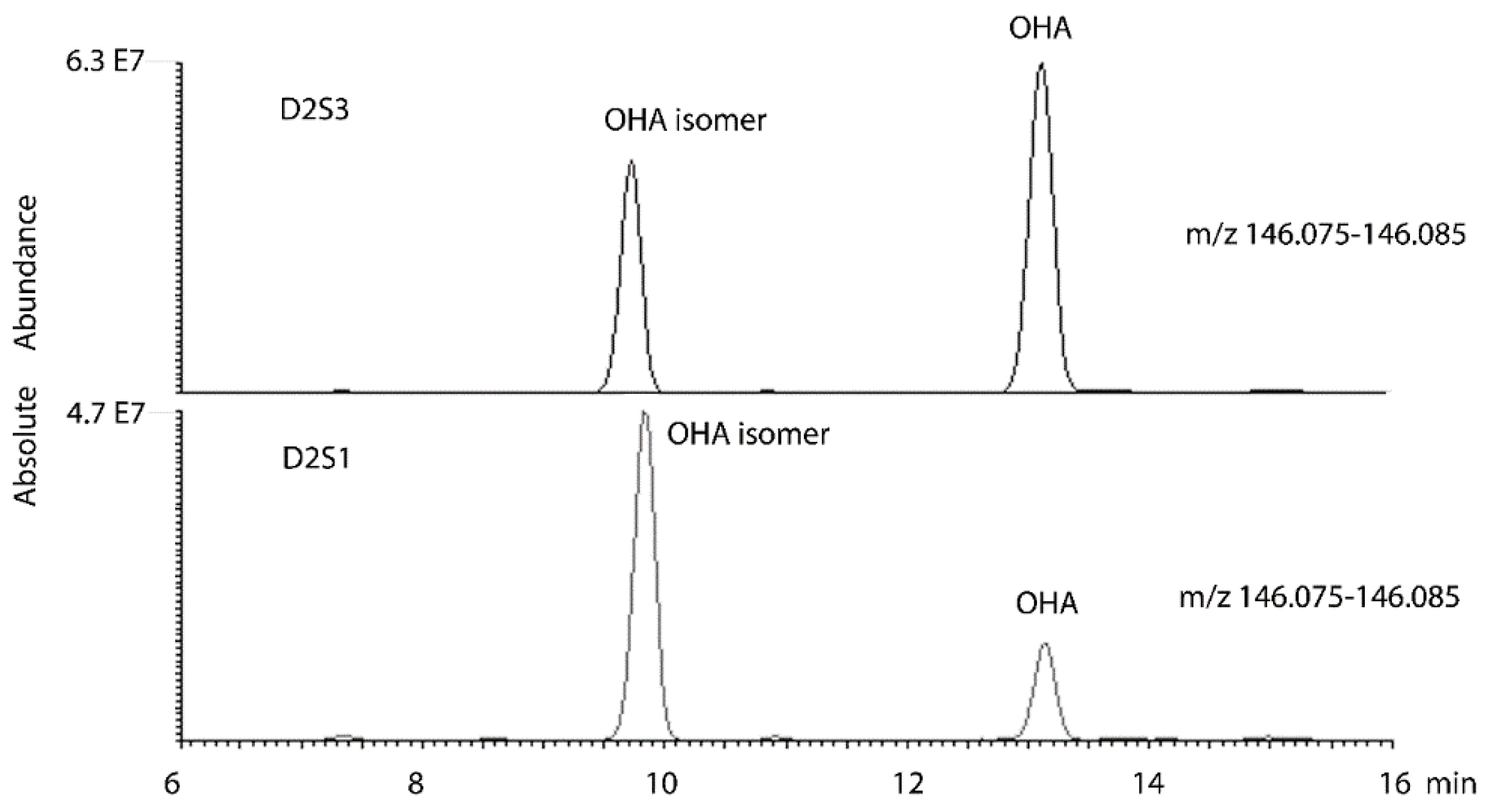
| Polarity | m/z | Rt | Metabolite | p-Value*** D2S1D2S3 | Ratio D2S3D2S1 | p-Value*** D1S1D1S3 | Ratio D1S3D1S1 |
|---|---|---|---|---|---|---|---|
| Purine Metabolism | |||||||
| + | 137.046 | 10.8 | * Hypoxanthine | <0.001 | 3.75 | 0.516 | 0.832 |
| + | 152.057 | 13.1 | * Guanine | 0.02 | 2.56 | na | na |
| − | 167.021 | 13.5 | * Urate | 0.015 | 0.748 | <0.001 | 0.534 |
| − | 181.037 | 10.8 | 1-Methyluric acid | 0.01 | 0.59 | 0.012 | 0.561 |
| + | 253.093 | 9.2 | Deoxyinosine | 0.01 | 4.97 | na | na |
| + | 269.088 | 11.5 | * Inosine | 0.02 | 16.27 | 0.947 | 0.967 |
| + | 285.083 | 13.1 | * Xanthosine | 0.01 | 2.33 | 0.012 | 0.506 |
| + | 282.12 | 13.9 | Methyladenosine | 0.24 | 0.856 | <0.01 | 0.516 |
| + | 296.136 | 17.8 | Dimethyladenosine | 0.20 | 2.390 | <0.01 | 0.491 |
| + | 312.13 | 9.2 | Dimethylguanosine | 0.90 | 0.983 | <0.003 | 0.562 |
| Neurotransmitter Metabolism | |||||||
| − | 101.024 | 12.7 | Oxobutanoate | 0.030 | 1.212 | 0.690 | 0.953 |
| + | 104.071 | 14.9 | * 4-Aminobutanoate | < 0.001 | 0.737 | 0.003 | 0.801 |
| − | 182.997 | 17.0 | Hydroxybutyric acid sulfate | <0.001 | 2.172 | 0.519 | 1.133 |
| + | 198.113 | 16.8 | * Metanephrine | 0.082 | 3.000 | 0.074 | 0.762 |
| − | 246.992 | 12.8 | Dihydroxy phenyl acetic acid sulfate or isomer | 0.020 | 1.442 | 0.513 | 1.212 |
| − | 261.007 | 10.5 | Homovanillicacid sulfate | 0.142 | 1.397 | 0.857 | 0.932 |
| − | 263.023 | 8.5 | Methoxyhydroxy phenylethyleneglycol sulfate | 0.071 | 1.385 | 0.405 | 0.833 |
| Collagen Metabolism | |||||||
| + | 173.092 | 13.3 | Glycylproline | <0.001 | 0.54 | 0.820 | 1.064 |
| + | 189.123 | 13.6 | Glycylleucine | <0.001 | 0.72 | 0.001 | 0.370 |
| Sugar Metabolism | |||||||
| − | 87.0092 | 8.6 | * Pyruvate | 0.256 | 1.179 | 0.017 | 0.565 |
| − | 89.0242 | 10.2 | * Lactate | 0.02 | 2.19 | 0.867 | 1.04 |
| − | 151.061 | 13.6 | * Xylitol or isomer | 0.016 | 0.734 | 0.036 | 0.617 |
| − | 163.061 | 9.9 | * Rhamnose or isomer | 0.025 | 0.667 | <0.01 | 0.629 |
| 165.04 | 8.9 | Arabinonate | 0.023 | 0.648 | <0.01 | 0.520 | |
| − | 181.072 | 14.7 | * Sorbitol or isomer | 0.05 | 0.70 | 0.126 | 0.653 |
| − | 193.036 | 15.2 | * Glucuronate or isomer | 0.018 | 0.628 | 0.024 | 0.641 |
| Fatty Acid Conjugates | |||||||
| − | 202.109 | 5.8 | ǂ Hydroxyhepatonyl glycine | <0.01 | 1.76 | 0.238 | 1.276 |
| + | 218.139 | 6.0 | Propanoylcarnitine | 0.03 | 0.360 | 0.363 | 1.124 |
| + | 288.217 | 6.4 | Octanoylcarnitine | 0.5397 | 1.115 | 0.002 | 0.540 |
| + | 300.218 | 6.0 | Nonanoyl carnitine | 0.015 | 1.63 | 0.499 | 0.850 |
| + | 314.234 | 5.6 | Decanoyl carnitine | 0.01 | 1.56 | 0.391 | 0.825 |
| + | 330.227 | 6.6 | Keto-decanoylcarnitine | 0.032 | 1.61 | 0.499 | 0.850 |
| Steroids | |||||||
| − | 367.158 | 3.9 | Testosterone sulfate | 0.04 | 1.57 | 0.319 | 1.23 |
| − | 539.25 | 6.7 | Tetrahydroaldosterone glucuronide | <0.01 | 1.62 | 0.369 | 1.21 |
| Bile Acids | |||||||
| − | 405.265 | 5.6 | Dihydroxyoxocholanate | <0.01 | 0.44 | 0.078 | 0.512 |
| − | 407.281 | 6.6 | Cholic acid | <0.01 | 0.34 | <0.01 | 0.340 |
| − | 583.312 | 10.3 | Cholic acid glucuronide | <0.01 | 0.27 | < 0.01 | 0.170 |
| − | 583.312 | 11.5 | Cholic acid glucuronide | <0.01 | 0.23 | <0.01 | 0.170 |
| Amino Acids and Their Metabolites | |||||||
| − | 118.051 | 12.0 | * Threonine | <0.01 | 0.64 | 0.685 | 1.06 |
| − | 131.046 | 15.8 | * Asparagine | <0.01 | 0.76 | 0.879 | 1.05 |
| + | 132.102 | 11.3 | * Leucine | 0.030 | 0.593 | 0.061 | 0.675 |
| − | 148.043 | 14.0 | * Methionine | <0.01 | 0.58 | 0.061 | 0.675 |
| + | 161.128 | 23.0 | Methyllysine | 0.114 | 1.526 | 0.002 | 2.669 |
| − | 172.098 | 6.1 | N-Acetylleucine | <0.01 | 0.65 | 0.010 | 0.590 |
| + | 175.108 | 9.3 | ** N-Acetylornithine isomer | <0.01 | 0.70 | < 0.01 | 0.566 |
| − | 187.072 | 11.4 | * N-Acetylglutamine | <0.01 | 0.67 | < 0.01 | 0.628 |
| + | 221.092 | 6.8 | ** 5-Hydroxytryptophan isomer | 0.06 | 2.24 | 0.530 | 0.952 |
| − | 214.028 | 14.0 | Chlorotyrosine | 0.04 | 2.09 | <0.01 | 2.91 |
| − | 216.099 | 11.8 | N-Acetylcitrulline | <0.01 | 0.60 | <0.01 | 0.570 |
| + | 219.134 | 13.7 | Carboxyethyllysine | <0.01 | 0.65 | <0.01 | 0.467 |
| + | 247.14 | 15.0 | Carboxyethylarginine | <0.01 | 2.23 | 0.022 | 0.544 |
| Vitamins and Co-Factors | |||||||
| + | 132.077 | 15.4 | * Creatine | 0.011 | 0.538 | 0.018 | 0.440 |
| + | 220.118 | 9.4 | * Pantothenate | <0.01 | 1.40 | <0.01 | 0.529 |
| − | 375.13 | 8.1 | * Riboflavin | 0.013 | 0.66 | 0.032 | 0.500 |
| Microbial Metabolites | |||||||
| − | 153.02 | 14.0 | Dihydroxybenzoate | 0.078 | 1.324 | 0.884 | 0.964 |
| − | 192.067 | 6.0 | Phenylacetylglycine | <0.01 | 1.76 | 0.277 | 0.756 |
| − | 197.046 | 9.8 | Dihydroxyphenyllactate | 0.025 | 1.43 | 0.383 | 0.855 |
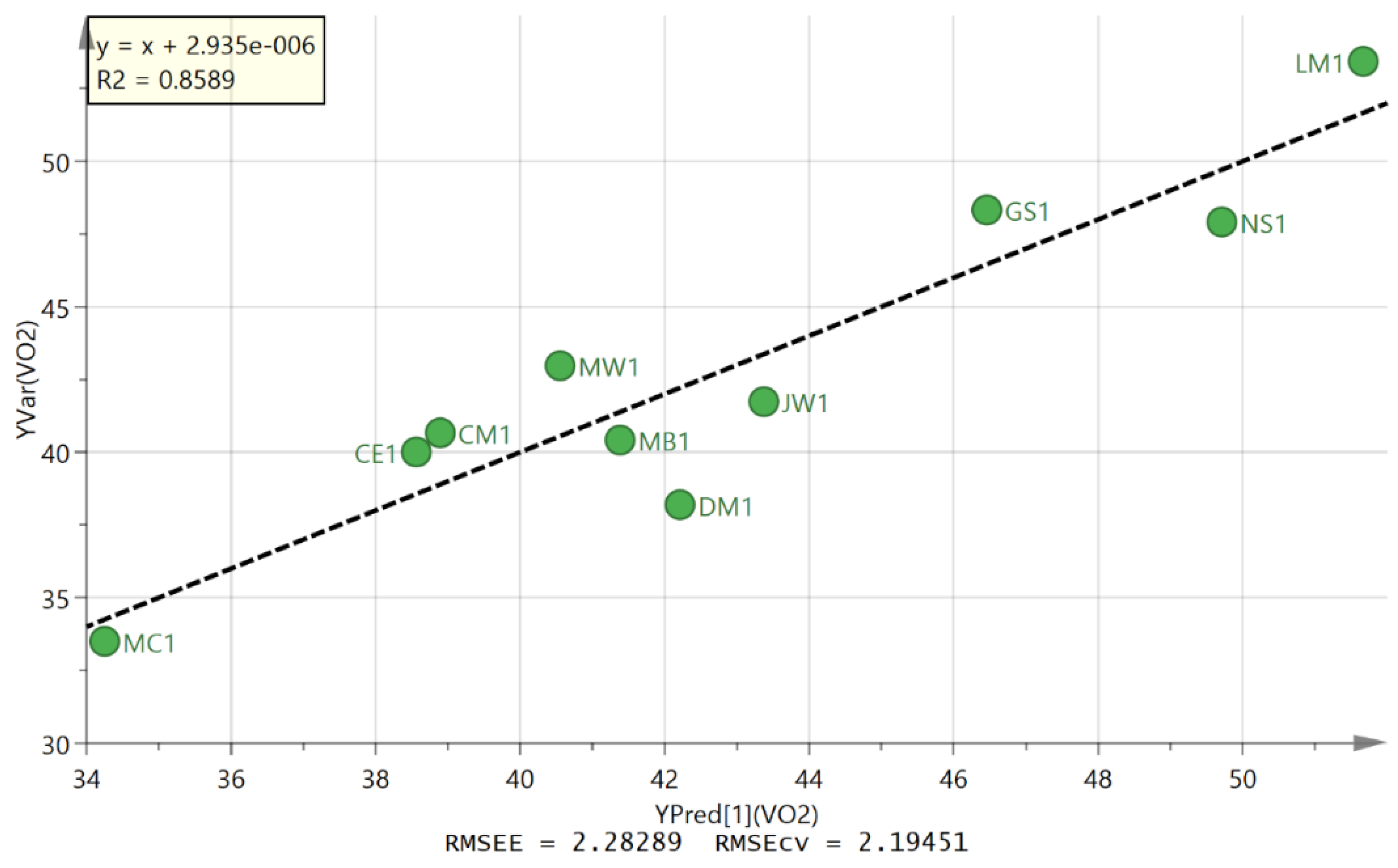
| Primary ID | Set | OAHA Normalised Level | VO2max | Predicted VO2max |
|---|---|---|---|---|
| 105 | D2S3 | 0.431 | 40 | 38.5 |
| 65 | D2S3 | 0.464 | 40.7 | 38.9 |
| 51 | D2S3 | 0.800 | 38.2 | 42.2 |
| 24 | D2S3 | 1.225 | 48.3 | 46.5 |
| 91 | D2S3 | 0.917 | 41.7 | 43.4 |
| 12 | D2S3 | 1.749 | 53.4 | 51.7 |
| 118 | D2S3 | 0.717 | 40.4 | 41.4 |
| 78 | D2S3 | 0.000 | 33.5 | 34.3 |
| 37 | D2S3 | 0.633 | 43 | 40.6 |
| 128 | D2S3 | 1.554 | 47.9 | 49.7 |
| 108 | D2S1 | 1.000 | 40 | 44.2 |
| 68 | D2S1 | 1.069 | 40.7 | 44.9 |
| 54 | D2S1 | 2.544 | 38.2 | 59.6 |
| 27 | D2S1 | 1.532 | 48.3 | 49.5 |
| 94 | D2S1 | 2.152 | 41.7 | 55.7 |
| 15 | D2S1 | 0.335 | 53.4 | 37.6 |
| 121 | D2S1 | 1.994 | 40.4 | 54.1 |
| 81 | D2S1 | 1.989 | 33.5 | 54.1 |
| 40 | D2S1 | 2.850 | 43 | 62.6 |
| 133 | D2S1 | 0.399 | 47.9 | 38.2 |
| 107 | D2S2 | 0.418 | 40 | 38.4 |
| 67 | D2S2 | 0.812 | 40.7 | 42.3 |
| 53 | D2S2 | 0.777 | 38.2 | 42.0 |
| 26 | D2S2 | 0.634 | 48.3 | 40.6 |
| 93 | D2S2 | 0.318 | 41.7 | 37.4 |
| 14 | D2S2 | 1.624 | 53.4 | 50.4 |
| 120 | D2S2 | 1.180 | 40.4 | 46.0 |
| 80 | D2S2 | 1.419 | 33.5 | 48.4 |
| 39 | D2S2 | 0.948 | 43 | 43.7 |
| 132 | D2S2 | 1.320 | 47.9 | 47.4 |
| 103 | D2S4 | 1.705 | 40 | 51.2 |
| 63 | D2S4 | 1.384 | 40.7 | 48.0 |
| 49 | D2S4 | 0.398 | 38.2 | 38.2 |
| 22 | D2S4 | 0.867 | 48.3 | 42.9 |
| 89 | D2S4 | 1.014 | 41.7 | 44.4 |
| 10 | D2S4 | 0.937 | 53.4 | 43.6 |
| 116 | D2S4 | 0.706 | 40.4 | 41.3 |
| 76 | D2S4 | 0.666 | 33.5 | 40.9 |
| 35 | D2S4 | 0.392 | 43 | 38.2 |
| 129 | D2S4 | 1.671 | 47.9 | 50.9 |
| 101 | D2S5 | 1.446 | 40 | 48.7 |
| 61 | D2S5 | 1.270 | 40.7 | 46.9 |
| 47 | D2S5 | 0.481 | 38.2 | 39.1 |
| 20 | D2S5 | 0.743 | 48.3 | 41.7 |
| 87 | D2S5 | 0.599 | 41.7 | 40.2 |
| 8 | D2S5 | 0.355 | 53.4 | 37.8 |
| 114 | D2S5 | 0.403 | 40.4 | 38.3 |
| 74 | D2S5 | 0.926 | 33.5 | 43.5 |
| 33 | D2S5 | 0.177 | 43 | 36.0 |
| 127 | D2S5 | 0.057 | 47.9 | 34.8 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhsen Ali, A.; Burleigh, M.; Daskalaki, E.; Zhang, T.; Easton, C.; Watson, D.G. Metabolomic Profiling of Submaximal Exercise at a Standardised Relative Intensity in Healthy Adults. Metabolites 2016, 6, 9. https://doi.org/10.3390/metabo6010009
Muhsen Ali A, Burleigh M, Daskalaki E, Zhang T, Easton C, Watson DG. Metabolomic Profiling of Submaximal Exercise at a Standardised Relative Intensity in Healthy Adults. Metabolites. 2016; 6(1):9. https://doi.org/10.3390/metabo6010009
Chicago/Turabian StyleMuhsen Ali, Ali, Mia Burleigh, Evangelia Daskalaki, Tong Zhang, Chris Easton, and David G. Watson. 2016. "Metabolomic Profiling of Submaximal Exercise at a Standardised Relative Intensity in Healthy Adults" Metabolites 6, no. 1: 9. https://doi.org/10.3390/metabo6010009
APA StyleMuhsen Ali, A., Burleigh, M., Daskalaki, E., Zhang, T., Easton, C., & Watson, D. G. (2016). Metabolomic Profiling of Submaximal Exercise at a Standardised Relative Intensity in Healthy Adults. Metabolites, 6(1), 9. https://doi.org/10.3390/metabo6010009







