Quantitative Metabolomics and Instationary 13C-Metabolic Flux Analysis Reveals Impact of Recombinant Protein Production on Trehalose and Energy Metabolism in Pichia pastoris
Abstract
:1. Introduction
2. Results and Discussion
2.1. Steady-State Chemostat Cultivations
| Strain | Glucose (mmol/gDCW·h) | Methanol (mmol/gDCW·h) | OUR (mmol/gDCW·h) | CER (mmol/gDCW·h) | Biomass (mCmol/gDCW·h) | RQ | Lipase Activity (UA/gDCW) |
|---|---|---|---|---|---|---|---|
| X-33 Control | −0.71 ± 0.01 | −0.94 ± 0.02 | −2.57 ± 0.03 | 2.03 ± 0.03 | 3.14 ± 0.04 | 0.79 ± 0.04 | 0 |
| X-33/ROL | −0.74 ± 0.03 | −1.05 ± 0.05 | −2.98 ± 0.04 | 2.39 ± 0.04 | 3.08 ± 0.05 | 0.81 ± 0.06 | 2417.05 ± 35.4 |
2.2. Intracellular and Extracellular Metabolite Pools
2.2.1. Energy and Redox Cofactors
| Metabolite | X-33 Control | X-33/ROL | ||
|---|---|---|---|---|
| Value | Sd | Value | Sd | |
| cAMP | 0.01 | 0.00 | 0.002 | 0.00 |
| AMP | 0.56 | 0.37 | 0.21 | 0.05 |
| ADP | 1.19 | 0.39 | 0.86 | 0.12 |
| ATP | 10.10 | 4.99 | 8.09 | 0.79 |
| GMP | 0.72 | 0.11 | 2.55 | 2.10 |
| GDP | 0.19 | 0.01 | 0.17 | 0.01 |
| GTP | 0.99 | 0.04 | 1.12 | 0.03 |
| Acetyl CoA | 0.18 | 0.03 | 0.21 | 0.05 |
| FAD | 0.73 | 0.12 | 1.04 | 0.06 |
| NAD+ + NADH | 64.29 | 13.16 | 60.07 | 24.32 |
| NADP+ + NADPH | 7.47 | 0.18 | 8.98 | 9.14 |
2.2.2. Central Carbon Metabolism and Storage Metabolites
| Metabolite | X-33 Control | X-33/ROL | ||||||
|---|---|---|---|---|---|---|---|---|
| Intra. (μmol/gDCW) | Extra. (μmol/L) | Intra. (μmol/gDCW) | Extra. (μmol/L) | |||||
| Value | Sd | Value | Sd | Value | Sd | Value | Sd | |
| Treh | 24.50 | 0.71 | 1.69 | 0.06 | 49.48 | 4.69 | 2.72 | 1.77 |
| Glc6P | 14.44 | 0.52 | 0.52 | 0.05 | 19.04 | 1.12 | 0.27 | 0.04 |
| Citrate | 7.17 | 0.25 | n.d | n.d. | 6.51 | 1.60 | n.d. | n.d. |
| Sed7P | 5.39 | 0.20 | 0.27 | 0.02 | 7.96 | 0.79 | 0.08 | 0.12 |
| Fru6P | 3.15 | 0.15 | 0.17 | 0.02 | 4.70 | 0.39 | 0.18 | 0.10 |
| MAL | 2.84 | 2.24 | 0.09 | 0.11 | 4.80 | 0.23 | 0.30 | 0.27 |
| SUCC | 1.97 | 0.15 | 0.27 | 0.04 | 1.29 | 0.15 | 0.27 | 0.26 |
| PG3 | 1.87 | 0.10 | 0.06 | 0.00 | 1.79 | 0.08 | 0.11 | 0.07 |
| αKG | 1.80 | 0.19 | 0.47 | 0.03 | 3.09 | 0.48 | 3.40 | 0.40 |
| Pyr | 1.47 | 0.20 | 43.19 | 6.28 | 1.57 | 0.35 | 36.02 | 4.29 |
| Man6P | 1.42 | 0.03 | 0.14 | 0.04 | 1.77 | 0.09 | 0.06 | 0.13 |
| FBP | 0.91 | 0.06 | 0.21 | 0.13 | 0.71 | 0.13 | 0.12 | 0.17 |
| Rib5P | 0.88 | 0.07 | 0.02 | 0.00 | 0.85 | 0.29 | 0.08 | 0.01 |
| Glc | 0.78 | 0.86 | 35.77 | 3.99 | 0.48 | 0.57 | 15.86 | 5.54 |
| FUM | 0.77 | 0.04 | 0.36 | 0.02 | 0.88 | 0.07 | 0.73 | 0.16 |
| Pep | 0.76 | 0.05 | 0.03 | 0.05 | 0.88 | 0.17 | 0.18 | 0.10 |
| DHAP | 0.71 | 0.02 | n.d. | n.d. | 0.49 | 0.26 | 0.02 | 0.03 |
| Rul5P | 0.23 | 0.03 | n.d. | n.d. | 0.29 | 0.03 | 0.13 | 0.14 |
| Xul5P | 0.16 | 0.02 | n.d. | n.d. | 0.34 | 0.04 | 0.05 | 0.03 |
| PG2 | 0.15 | 0.06 | 0.05 | 0.01 | 0.22 | 0.02 | 0.03 | 0.03 |
| T6P | 0.09 | 0.01 | 0.12 | 0.00 | 0.26 | 0.08 | 0.08 | 0.07 |
| E4P | 0.08 | 0.00 | 0.38 | 0.01 | 0.19 | 0.14 | 0.31 | 0.23 |
| Isocitrate | 0.03 | 0.03 | n.d. | n.d. | 0.07 | 0.02 | 0.02 | 0.03 |
| GA3P | 0.00 | 0.00 | n.d. | n.d. | 0.01 | 0.01 | 0.02 | 0.03 |
2.3. Intracellular Amino Acid Pools
- (1)
- The total free amino acid pool in the control strain was 11% lower (288.32 μmol/gDCW) compared to the Rol-expressing strain (324.58 μmol/gDCW).
- (2)
- In particular, Asp, Orn, Ser, Asn, His, Thr, Pro, Val, Leu, Tyr, Phe pool sizes were statistically significantly higher in the Rol-expressing strain (Table 4), even though the biomass protein production demand of these amino acids was similar for both strains.
| Amino acid | X-33 Control | X-33/ROL | ||||||
|---|---|---|---|---|---|---|---|---|
| Intra. (μmol/gDCW) | Extra. (μmol/L) | Intra. (μmol/gDCW) | Extra. (μmol/L) | |||||
| Value | Sd | Value | Sd | Value | Sd | Value | Sd | |
| Glu | 84.85 | 2.97 | n.d. | n.d. | 91.98 | 4.21 | 0.03 | 0.04 |
| Gln | 84.97 | 2.40 | 0.01 | 0.00 | 88.40 | 3.54 | 26.05 | 36.68 |
| Asp | 39.56 | 0.56 | 0.08 | 0.00 | 45.58 | 3.58 | 0.03 | 0.04 |
| Orn | 22.59 | 1.87 | 1.53 | 1.10 | 28.28 | 1.75 | 0.16 | 0.32 |
| Ala | 15.01 | 1.21 | 1.19 | 0.22 | 13.56 | 0.71 | 2.77 | 3.50 |
| Lys | 10.22 | 0.18 | 0.27 | 0.22 | 12.27 | 0.44 | 1.15 | 1.77 |
| Ser | 5.94 | 1.20 | n.d | n.d. | 9.30 | 0.38 | 0.53 | 0.38 |
| Asn | 4.66 | 0.10 | 0.11 | 0.08 | 9.64 | 0.39 | 0.39 | 0.48 |
| His | 4.79 | 0.15 | 0.01 | 0.02 | 8.13 | 0.32 | 0.30 | 0.43 |
| Gly | 1.33 | 2.44 | 0.44 | 0.33 | 2.35 | 0.07 | 2.02 | 0.87 |
| Thr | 2.49 | 0.21 | 0.04 | 0.02 | 4.24 | 0.16 | 0.25 | 0.04 |
| Pro | 2.61 | 0.07 | 0.03 | 0.03 | 3.85 | 0.15 | 0.25 | 0.16 |
| Val | 1.30 | 0.14 | n.d. | n.d. | 2.41 | 0.17 | 0.24 | 0.36 |
| Leu | 0.69 | 0.23 | 0.06 | 0.04 | 1.55 | 0.22 | 0.39 | 0.52 |
| Ile | 0.33 | 0.14 | 0.04 | 0.02 | 0.53 | 0.11 | 0.22 | 0.25 |
| Tyr | 0.20 | 0.10 | 0.03 | 0.02 | 1.02 | 0.04 | 0.14 | 0.20 |
| Phe | 0.20 | 0.13 | 0.02 | 0.01 | 0.65 | 0.09 | 0.14 | 0.20 |
| Met | 0.48 | 0.05 | n.d. | n.d. | 0.53 | 0.04 | 0.04 | 0.07 |
| Trp | 0.09 | 0.03 | n.d. | n.d. | 0.33 | 0.02 | 0.08 | 0.14 |

2.4. Instationary 13C-MFA
- No significant differences have been observed in the amount of Glc6P entering the oxidative branch of the PPP: 77% and 69% of the Glc6P, in the control and Rol-expressing strains, respectively. However, the Rol-producing strain showed a slightly reduced biomass yield. Since the PPP is the main pathway for cytosolic NADPH formation, the flux through the oxidative branch of the PPP is generally directly correlated to the biosynthetic demand for NADPH [38]. The present study further supports the hypothesis of increased NADPH supply through the oxidative branch of the PPP in the Rol-producing strain. To confirm this hypothesis, the NADPH balances were reconstructed taking into account the stoichiometric model and the 13C flux estimations (which only balance carbon and labeling). In fact, calculation of NADPH biosynthetic demand for both strains was 0.85 ± 0.03 and 0.97 ± 0.07 mmol/gDCW/h for expressing and control strain cells. These values were lower than the total generated NADPH in both cases (1.04 ± 0.16 and 1.10 ± 0.10 mmol/gDCW/h, respectively). However, for both strains no statistically significant difference was observed between the generated NADPH and the demanded for biosynthesis. This could reflect the observation that some Crabtree negative yeast appear to have alternative mechanisms involved in the re-oxidation of the NADPH produced in the PPP, e.g. by mitochondrial external alternative dehydrogenases that use NADPH [39]. Alternatively, such effect on the oxidative branch of the PPP could be the indirect consequence of methanol assimilation, which requires Xul5P.
- The Rol-producing strain shows a tendency for a higher fraction of the assimilated methanol being directly oxidized to CO2 (60% vs. 50%, respectively). This trend can be seen in the enrichment of F6P and DHAP, which is higher (resp. less diluted from unlabelled carbon entering from methanol) in the producing strain. The increased methanol direct oxidation has also been observed using steady-state measurements [18]. The origin could be an increased energy requirement for Rol synthesis and secretion (2 mol NADH per mol of methanol are directly generated in methanol oxidation). Although the increased flux through the methanol oxidative pathway cannot be discriminated statistically, the trend is in agreement with the increased NADH production (increased oxygen uptake) in the Rol producer strain, as well as with our previous study, where such difference was assessed as statistically significant using the 13C-NMR based MFA approach [18].
- The flux through the trehalose cycle seems to be altered by Rol production. As it can be seen from Figure 2 and Figure 3, the absolute flux through the trehalose cycle tends to be higher (two-fold) in the Rol-expressing strain. Although such trend is not statistically significant, it is consistent with the statistically significantly higher trehalose concentration observed in this strain, further supporting increased recycling of this molecule building an ATP-futile cycle [32].
- The TCA cycle shows a trend towards higher flux for the producing strain. The flux through this part of the network cannot be discriminated statistically, but the trend is in agreement with the increased NADH production (increased oxygen uptake) in the Rol producer, as also observed in our previous study, where such differences were assessed as statistically significant using the 13C-NMR based MFA approach [18].
- As previously reported [25], the INST-13C approach provides additional insights regarding bidirectional reactions. Notably, the high exchange fluxes for oxaloacetate, malate, and Asp, which indicate amino acid pool buffering and the activity of Malate/Aspartate shuttle [42], are significantly reduced in the Rol-producing strain. In relation to this observation, the turnover time of succinate, fumarate and malate pools is drastically reduced in the producing strain, and the calculated mass action ratio (MAR, based on intracellular concentration measurements) of the fumarase reaction seems to be higher in this strain, although differences are not significant (Supplementary File 4). Also, the Rol-producing strain showed reduced exchange fluxes in some of the reactions of the non-oxidative branch of the PPP in relation to the reference strain, probably reflecting a reduced flux of methanol through its assimilatory pathway.
- The impact of Rol production on the methanol assimilation pathway results in altered behavior of the exchange fluxes in PPP. For instance, the exchange flux of the aldolase reaction was significantly increased in the Rol-producing strain, whereas the exchange flux between DHAP and GA3P was reduced (Figure 2 and Figure 3). As stated above, this trend can be directly observed in the enrichment dynamics of F6P, DHAP, 3PG and 2PG (Supplementary File 9). This may also result in slightly different MAR for the enolase reaction (Supplementary File 4), although this difference was not significant.
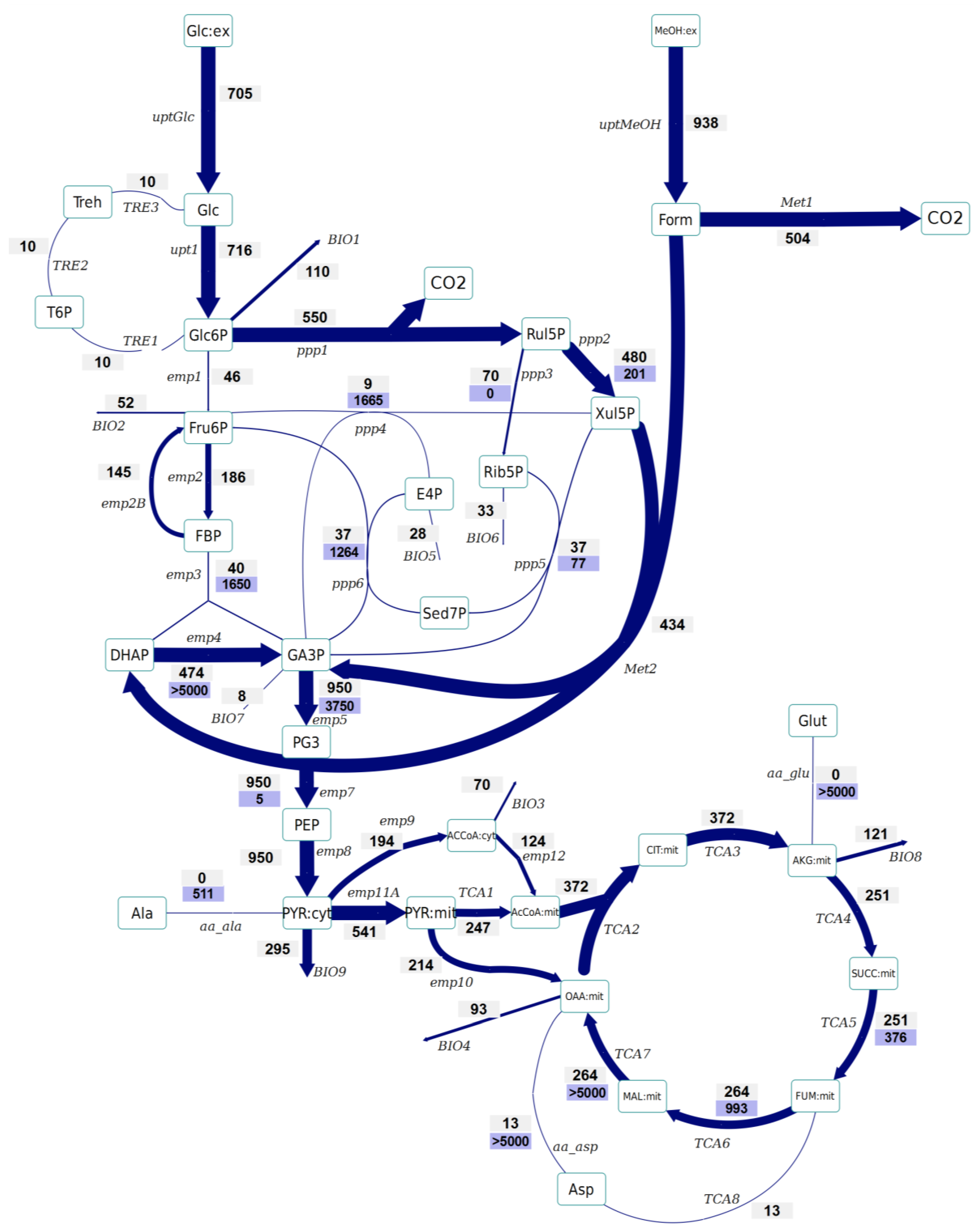
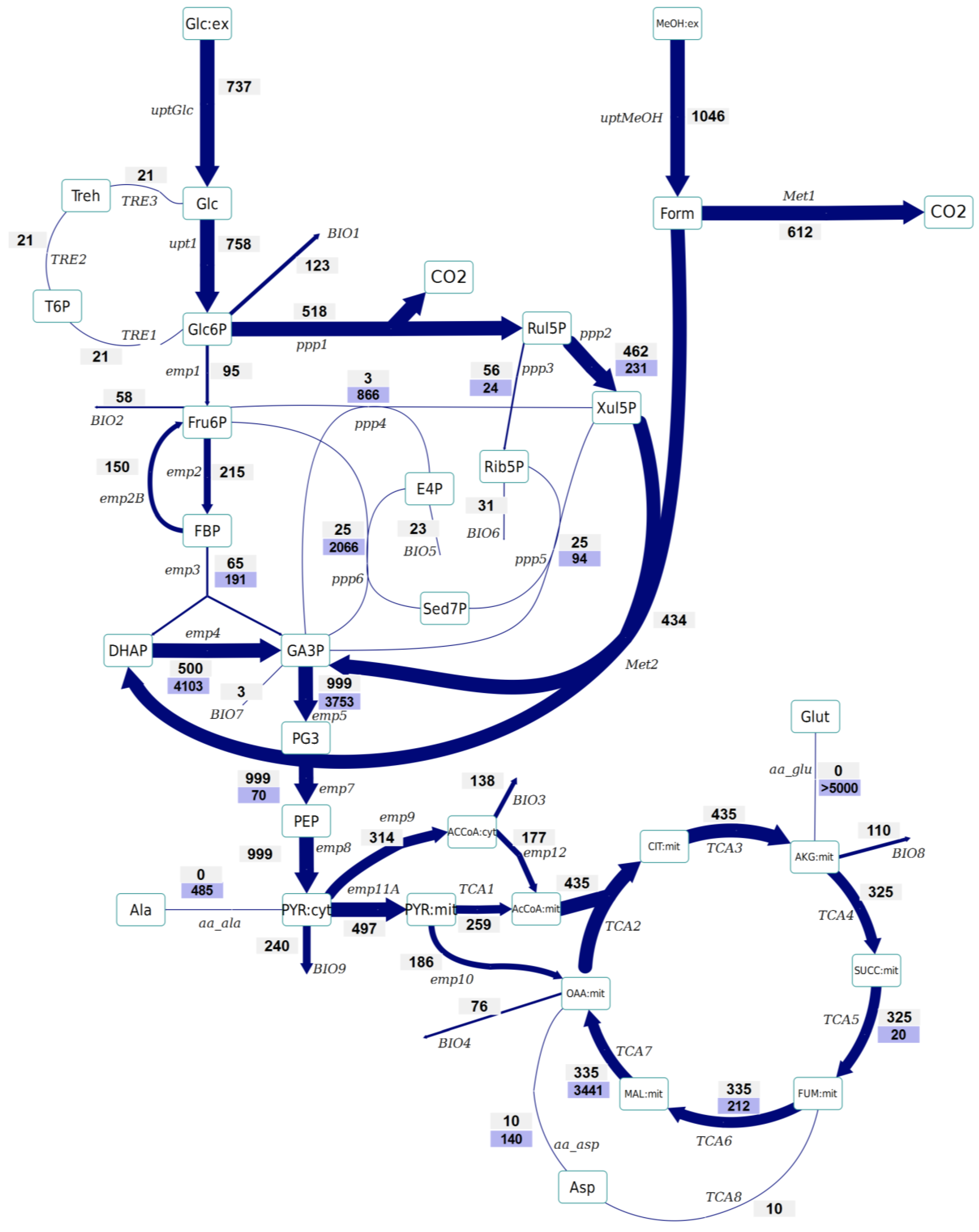
3. Experimental Section
3.1. Strain and Cultivation Conditions
3.2. Sampling and Experiment Design
3.3. 13C-Based Metabolic Flux Analysis (13C-MFA)
3.4. Analytical Procedures
4. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Corchero, J.L.; Gasser, B.; Resina, D.; Smith, W.; Parrilli, E.; Vázquez, F.; Abasolo, I.; Giuliani, M.; Jantti, J.; et al. Unconventional microbial systems for the cost-efficient production of high-quality protein therapeutics. Biotechnol. Adv. 2013, 31, 140–153. [Google Scholar] [CrossRef]
- Mattanovich, D.; Branduardi, P.; Dato, L.; Gasser, B.; Sauer, M.; Porro, D. Recombinant protein production in yeasts. Methods Mol. Biol. 2012, 824, 329–358. [Google Scholar] [CrossRef]
- Damasceno, L.M.; Huang, C.J.; Batt, C.A. Protein secretion in Pichia pastoris and advances in protein production. Appl. Microbiol. Biotechnol. 2012, 93, 31–39. [Google Scholar] [CrossRef]
- Bollok, M.; Resina, D.; Valero, F.; Ferrer, P. Recent patents on the Pichia pastoris expression system: Expanding the toolbox for recombinant protein production. Recent Pat. Biotechnol. 2009, 3, 192–201. [Google Scholar]
- Cos, O.; Serrano, A.; Montesinos, J.L.; Ferrer, P.; Cregg, J.M.; Valero, F. Combined effect of the methanol utilization (Mut) phenotype and gene dosage on recombinant protein production in Pichia pastoris fed-batch cultures. J. Biotechnol. 2005, 116, 321–335. [Google Scholar] [CrossRef]
- Hohenblum, H.; Gasser, B.; Maurer, M.; Borth, N.; Mattanovich, D. Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris. Biotechnol. Bioeng. 2004, 85, 367–375. [Google Scholar] [CrossRef]
- Gasser, B.; Maurer, M.; Rautio, J.; Sauer, M.; Bhattacharyya, A.; Saloheimo, M.; Penttilä, M.; Mattanovich, D. Monitoring of transcriptional regulation in Pichia pastoris under protein production conditions. BMC Genomics 2007, 8, 179. [Google Scholar] [CrossRef]
- Hesketh, A.R.; Castrillo, J.I.; Sawyer, T.; Archer, D.B.; Oliver, S.G. Investigating the physiological response of Pichia (Komagataella) pastoris GS115 to the heterologous expression of misfolded proteins using chemostat cultures. Appl. Microbiol. Biotechnol. 2013, 97, 9747–9762. [Google Scholar] [CrossRef]
- Resina, D.; Bollok, M.; Khatri, N.K.; Valero, F.; Neubauer, P.; Ferrer, P. Transcriptional response of P. pastoris in fed-batch cultivations to Rhizopus oryzae lipase production reveals UPR induction. Microb. Cell Fact. 2007, 6, 21. [Google Scholar] [CrossRef]
- Cos, O.; Ramon, R.; Montesinos, J.L.; Valero, F. Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: A review. Microb. Cell Fact 2006, 5, 17. [Google Scholar] [CrossRef]
- Inan, M.; Meagher, M.M. Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J. Biosci. Bioeng. 2001, 92, 585–589. [Google Scholar] [CrossRef]
- Jungo, C.; Schenk, J.; Pasquier, M.; Marison, I.W.; von Stockar, U. A quantitative analysis of the benefits of mixed feeds of sorbitol and methanol for the production of recombinant avidin with Pichia pastoris. J. Biotechnol. 2007, 131, 57–66. [Google Scholar] [CrossRef]
- Ramon, R.; Ferrer, P.; Valero, F. Sorbitol co-feeding reduces metabolic burden caused by the overexpression of a Rhizopus oryzae lipase in Pichia pastoris. J. Biotechnol. 2007, 130, 39–46. [Google Scholar] [CrossRef]
- Arnau, C.; Ramon, R.; Casas, C.; Valero, F. Optimization of the heterologous production of a Rhizopus oryzae lipase in Pichia pastoris system using mixed substrates on controlled fed-batch bioprocess. Enzyme Microb. Technol. 2010, 46, 494–500. [Google Scholar] [CrossRef]
- Dragosits, M.; Stadlmann, J.; Albiol, J.; Baumann, K.; Maurer, M.; Gasser, B.; Sauer, M.; Altmann, F.; Ferrer, P.; Mattanovich, D. The effect of temperature on the proteome of recombinant Pichia pastoris. J. Proteome Res. 2009, 8, 1380–1392. [Google Scholar] [CrossRef]
- Heyland, J.; Fu, J.; Blank, L.M.; Schmid, A. Quantitative physiology of Pichia pastoris during glucose-limited high-cell density fed-batch cultivation for recombinant protein production. Biotechnol. Bioeng. 2010, 107, 357–368. [Google Scholar] [CrossRef]
- Driouch, H.; Melzer, G.; Wittmann, C. Integration of in vivo and in silico metabolic fluxes for improvement of recombinant protein production. Metab. Eng. 2012, 14, 47–58. [Google Scholar] [CrossRef]
- Jordà, J.; Jouhten, P.; Cámara, E.; Maaheimo, H.; Albiol, J.; Ferrer, P. Metabolic flux profiling of recombinant protein secreting Pichia pastoris growing on glucose: Methanol mixtures. Microb. Cell Fact 2012, 11, 57. [Google Scholar] [CrossRef]
- Bolten, C.J.; Kiefer, P.; Letisse, F.; Portais, J.C.; Wittmann, C. Sampling for metabolome analysis of microorganisms. Anal. Chem. 2007, 79, 3843–3849. [Google Scholar] [CrossRef]
- Van Gulik, W.M. Fast sampling for quantitative microbial metabolomics. Curr. Opin. Biotechnol. 2010, 21, 27–34. [Google Scholar] [CrossRef]
- Cascante, M.; Marin, S. Metabolomics and fluxomics approaches. Essays Biochem 2008, 45, 67–81. [Google Scholar] [CrossRef]
- Wiechert, W.; Wurzel, M. Metabolic isotopomer labeling systems. Part I: Global dynamic behavior. Math. Biosci. 2001, 169, 173–205. [Google Scholar] [CrossRef]
- Sauer, U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2006, 2, 62. [Google Scholar] [CrossRef]
- Wiechert, W.; Nöh, K. Isotopically non-stationary metabolic flux analysis: Complex yet highly informative. Curr. Opin. Biotechnol. 2013, 24, 979–986. [Google Scholar] [CrossRef]
- Jordà, J.; Suarez, C.; Carnicer, M.; ten Pierick, A.; Heijnen, J.J.; van Gulik, W.; Ferrer, P.; Albiol, J.; Wahl, A. Glucose-methanol co-utilization in Pichia pastoris studied by metabolomics and instationary 13C flux analysis. BMC Syst. Biol. 2013, 7, 17. [Google Scholar] [CrossRef]
- Van der Heijden, R.T.; Heijnen, J.J.; Hellinga, C.; Romein, B.; Luyben, K.C. Linear constraint relations in biochemical reaction systems: I. Classification of the calculability and the balanceability of conversion rates. Biotechnol. Bioeng. 1994, 43, 3–10. [Google Scholar]
- Verheijen, P.J.T. Data Reconciliation and Error Detection. In The Metabolic Pathway Engineering Handbook; Smoke, C., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 8.1–8.13. [Google Scholar]
- Edwards, K.; Urban, J.; Schreiber, G. Relationship between protein synthesis and secretion in liver cells and the state of the adenine nucleotide system. Aust. J. Biol. Sci. 1979, 32, 299–307. [Google Scholar]
- Plantz, B.A.; Sinha, J.; Villarete, L.; Nickerson, K.W.; Schlegel, V.L. Pichia pastoris fermentation optimization: Energy state and testing a growth-associated model. Appl. Microbiol. Biotechnol. 2006, 72, 297–305. [Google Scholar] [CrossRef]
- Carnicer, M. Systematic metabolic analysis of recombinant Pichia pastoris under different oxygen conditions A Metabolome and Fluxome Based Study. Ph.D. Thesis, Autonomous University of Barcelona, Bellaterra (Cerdanyola del Vallès), Catalonia, Spain, 2012. [Google Scholar]
- Katakura, Y.; Zhang, W.; Guoqiang, O.; Takeshi, K.; Kishimoto, M.; Goto, Y.; Suga, K.-I. Effect of methanol concentration on the production of β2-glycoprotein I domain V by a recombinant Pichia pastoris: A simple system for the control of methanol concentration using a semiconductor gas sensor. J. Ferment. Bioeng. 1998, 86, 482–487. [Google Scholar] [CrossRef]
- Parrou, J.L.; Teste, M.A.; Francois, J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: Genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 1997, 143, 1891–1900. [Google Scholar] [CrossRef]
- Francois, J.; Parrou, J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 125–145. [Google Scholar] [CrossRef]
- Carnicer, M.; ten Pierick, A.; van Dam, J.; Heijnen, J.J.; Albiol, J.; van Gulik, W.; Ferrer, P. Quantitative metabolomics analysis of amino acid metabolism in recombinant Pichia pastoris under different oxygen availability conditions. Microb. Cell Fact 2012, 11, 83. [Google Scholar] [CrossRef]
- Pfeffer, M.; Maurer, M.; Kollensperger, G.; Hann, S.; Graf, A.B.; Mattanovich, D. Modeling and measuring intracellular fluxes of secreted recombinant protein in Pichia pastoris with a novel 34S labeling procedure. Microb. Cell Fact 2011, 10, 47. [Google Scholar] [CrossRef]
- Aboka, F.O.; Heijnen, J.J.; van Winden, W.A. Dynamic 13C-tracer study of storage carbohydrate pools in aerobic glucose-limited Saccharomyces cerevisiae confirms a rapid steady-state turnover and fast mobilization during a modest stepup in the glucose uptake rate. FEMS Yeast Res. 2009, 9, 191–201. [Google Scholar] [CrossRef]
- Zhao, Z.; ten Pierick, A.; de Jonge, L.; Heijnen, J.J.; Wahl, A. Substrate cycles in Penicillium chrysogenum quantified by isotopic non-stationary flux analysis. Microb. Cell Fact 2012, 11, 140. [Google Scholar] [CrossRef]
- Blank, L.M.; Lehmbeck, F.; Sauer, U. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 2005, 5, 545–558. [Google Scholar] [CrossRef]
- Gonzalez-Siso, M.I.; Garcia-Leiro, A.; Tarrio, N.; Cerdan, M.E. Sugar metabolism, redox balance and oxidative stress response in the respiratory yeast Kluyveromyces lactis. Microb. Cell Fact 2009, 8, 46. [Google Scholar] [CrossRef]
- Gasser, B.; Saloheimo, M.; Rinas, U.; Dragosits, M.; Rodriguez-Carmona, E.; Baumann, K.; Giuliani, M.; Parrilli, E.; Branduardi, P.; Lang, C.; et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: A host comparative overview. Microb. Cell Fact 2008, 7, 11. [Google Scholar] [CrossRef]
- Schröder, M.; Clark, R.; Kaufman, R.J. IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol. Microbiol. 2003, 49, 591–606. [Google Scholar]
- Bakker, B.M.; Overkamp, K.M.; van Maris, A.J.; Kotter, P.; Luttik, M.A.; van Dijken, J.P.; Pronk, J.T. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 2001, 25, 15–37. [Google Scholar] [CrossRef]
- Carnicer, M.; Baumann, K.; Töplitz, I.; Sanchez-Ferrando, F.; Mattanovich, D.; Ferrer, P.; Albiol, J. Macromolecular and elemental composition analysis and extracellular metabolite balances of Pichia pastoris growing at different oxygen levels. Microb. Cell Fact 2009, 8, 65. [Google Scholar] [CrossRef]
- Minning, S.; Serrano, A.; Ferrer, P.; Solà, C.; Schmid, R.D.; Valero, F. Optimization of the high-level production of Rhizopus oryzae lipase in Pichia pastoris. J. Biotechnol. 2001, 86, 59–70. [Google Scholar] [CrossRef]
- Lange, H.C.; Eman, M.; van Zuijlen, G.; Visser, D.; van Dam, J.C.; Frank, J.; de Mattos, M.J.; Heijnen, J.J. Improved rapid sampling for in vivo kinetics of intracellular metabolites in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2001, 75, 406–415. [Google Scholar] [CrossRef]
- Carnicer, M.; Canelas, A.B.; ten Pierick, A.; Zeng, Z.; van Dam, J.; Albiol, J.; Ferrer, P.; Heijnen, J.J.; van Gulik, W. Development of quantitative metabolomics for Pichia pastoris. Metabolomics 2012, 8, 284–298. [Google Scholar] [CrossRef]
- Canelas, A.B.; ten Pierick, A.; Ras, C.; Seifar, R.M.; van Dam, J.C.; Van Gulik, W.M.; Heijnen, J.J. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal. Chem. 2009, 81, 7379–7389. [Google Scholar] [CrossRef]
- Wahl, A.; Seifar, R.M.; ten Pierick, A.; Ras, C.; van Dam, J.C.; Heijnen, J.J.; van Gulik, W.M. Quantitative Metabolomics using ID MS. Methods Mol. Biol. 2014. to be published. [Google Scholar]
- Wahl, S.A.; Dauner, M.; Wiechert, W. New tools for mass isotopomer data evaluation in 13C flux analysis: Mass isotope correction, data consistency checking, and precursor relationships. Biotechnol. Bioeng. 2004, 85, 259–268. [Google Scholar] [CrossRef]
- Wang, N.S.; Stephanopoulos, G. Application of macroscopic balances to the identification of gross measurement errors. Biotechnol. Bioeng. 1983, 25, 2177–2208. [Google Scholar] [CrossRef]
- Kümmel, A.; Panke, S.; Heinemann, M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2006, 2, 2006. [Google Scholar]
- Zamboni, N.; Kümmel, A.; Heinemann, M. anNET: A tool for network-embedded thermodynamic analysis of quantitative metabolome data. BMC Bioinformatics 2008, 9, 199. [Google Scholar] [CrossRef]
- Cámara, E.; Jordà, J.; Jouhten, P.; Maaheimo, H.; Albiol, J.; Ferrer, P. Impact of recombinant protein production on the energy metabolism of Pichia pastoris growing on glucose: Methanol mixtures. In Proceedings of the 2nd Conference on Microbial Stress: From Molecules to Systems, Belgirate, Italy, 10–13 May 2012.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jordà, J.; Rojas, H.C.; Carnicer, M.; Wahl, A.; Ferrer, P.; Albiol, J. Quantitative Metabolomics and Instationary 13C-Metabolic Flux Analysis Reveals Impact of Recombinant Protein Production on Trehalose and Energy Metabolism in Pichia pastoris. Metabolites 2014, 4, 281-299. https://doi.org/10.3390/metabo4020281
Jordà J, Rojas HC, Carnicer M, Wahl A, Ferrer P, Albiol J. Quantitative Metabolomics and Instationary 13C-Metabolic Flux Analysis Reveals Impact of Recombinant Protein Production on Trehalose and Energy Metabolism in Pichia pastoris. Metabolites. 2014; 4(2):281-299. https://doi.org/10.3390/metabo4020281
Chicago/Turabian StyleJordà, Joel, Hugo Cueto Rojas, Marc Carnicer, Aljoscha Wahl, Pau Ferrer, and Joan Albiol. 2014. "Quantitative Metabolomics and Instationary 13C-Metabolic Flux Analysis Reveals Impact of Recombinant Protein Production on Trehalose and Energy Metabolism in Pichia pastoris" Metabolites 4, no. 2: 281-299. https://doi.org/10.3390/metabo4020281
APA StyleJordà, J., Rojas, H. C., Carnicer, M., Wahl, A., Ferrer, P., & Albiol, J. (2014). Quantitative Metabolomics and Instationary 13C-Metabolic Flux Analysis Reveals Impact of Recombinant Protein Production on Trehalose and Energy Metabolism in Pichia pastoris. Metabolites, 4(2), 281-299. https://doi.org/10.3390/metabo4020281






