An NMR Metabolomics Analysis Pipeline for Human Neutrophil Samples with Limited Source Material
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Neutrophil Isolation
2.3. Preparation of Samples for NMR Metabolomics
2.4. Metabolite Extraction
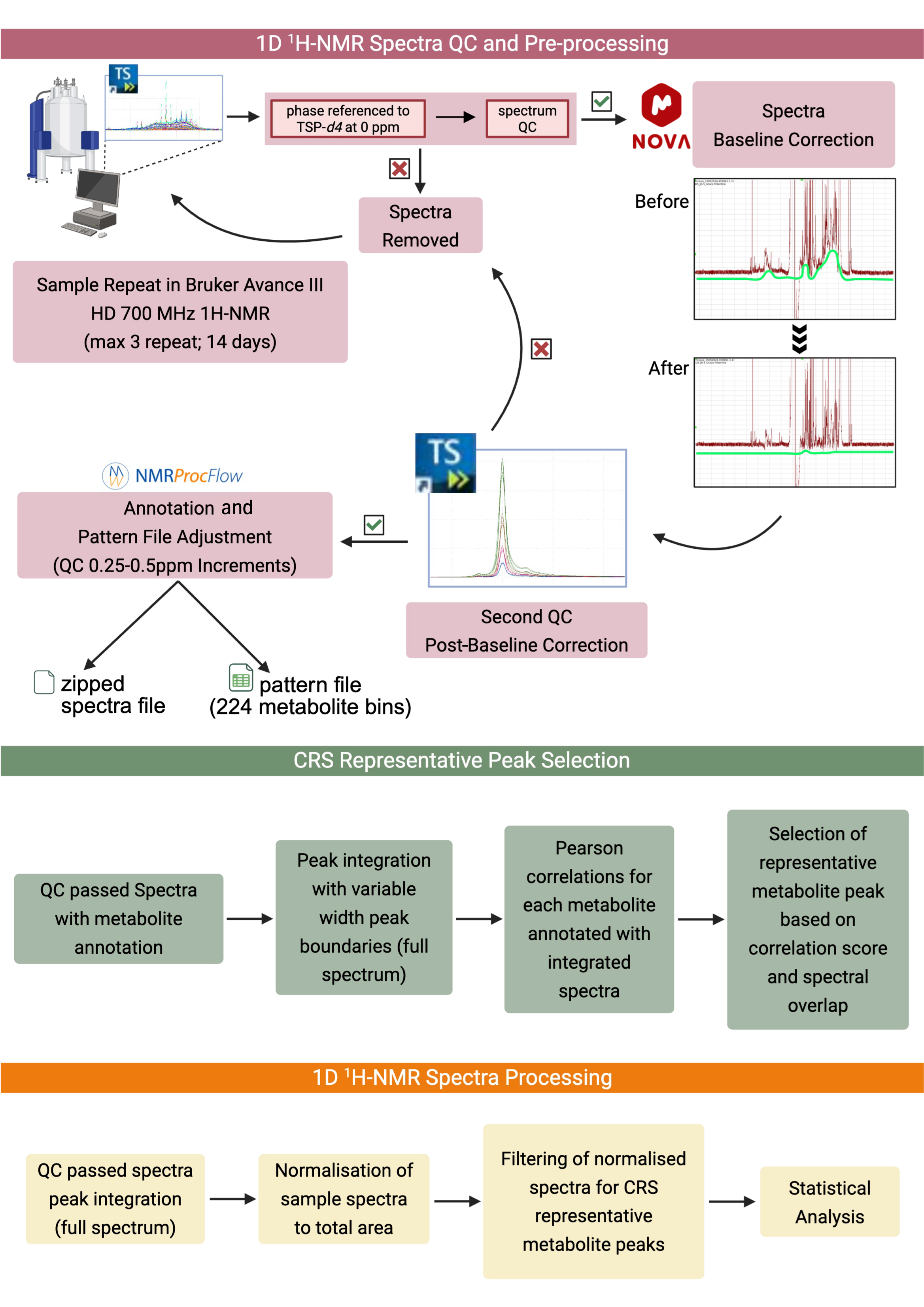
2.5. NMR Spectral Acquisition and Quality Assurance of Acquired Spectra
2.6. Annotation for Neutrophil Intracellular Metabolomic Profiling Using CRS-Selected Bins
2.7. Data Availability
2.8. Statistical Analysis
3. Results
3.1. Spectral Binning and Metabolite Annotation
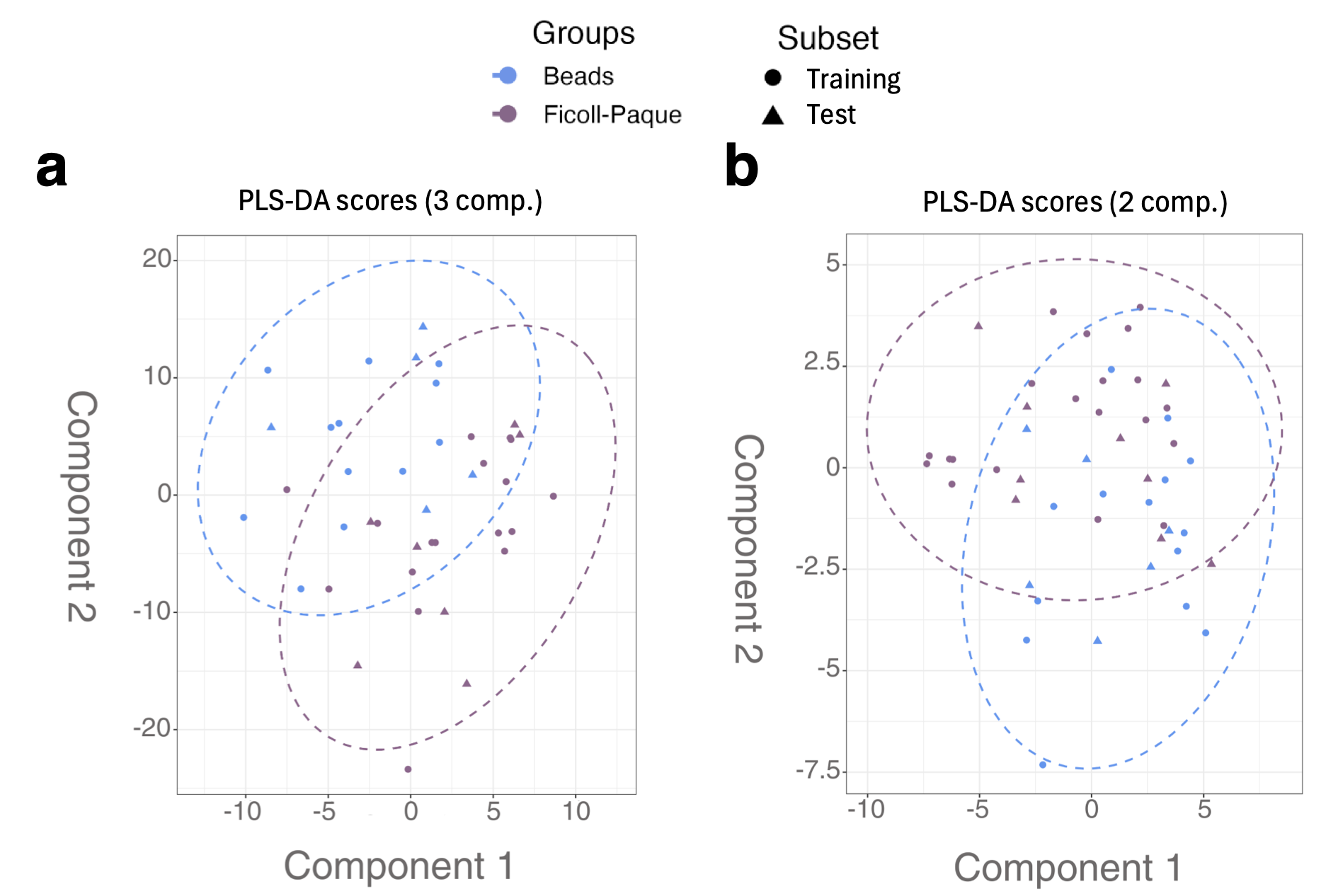
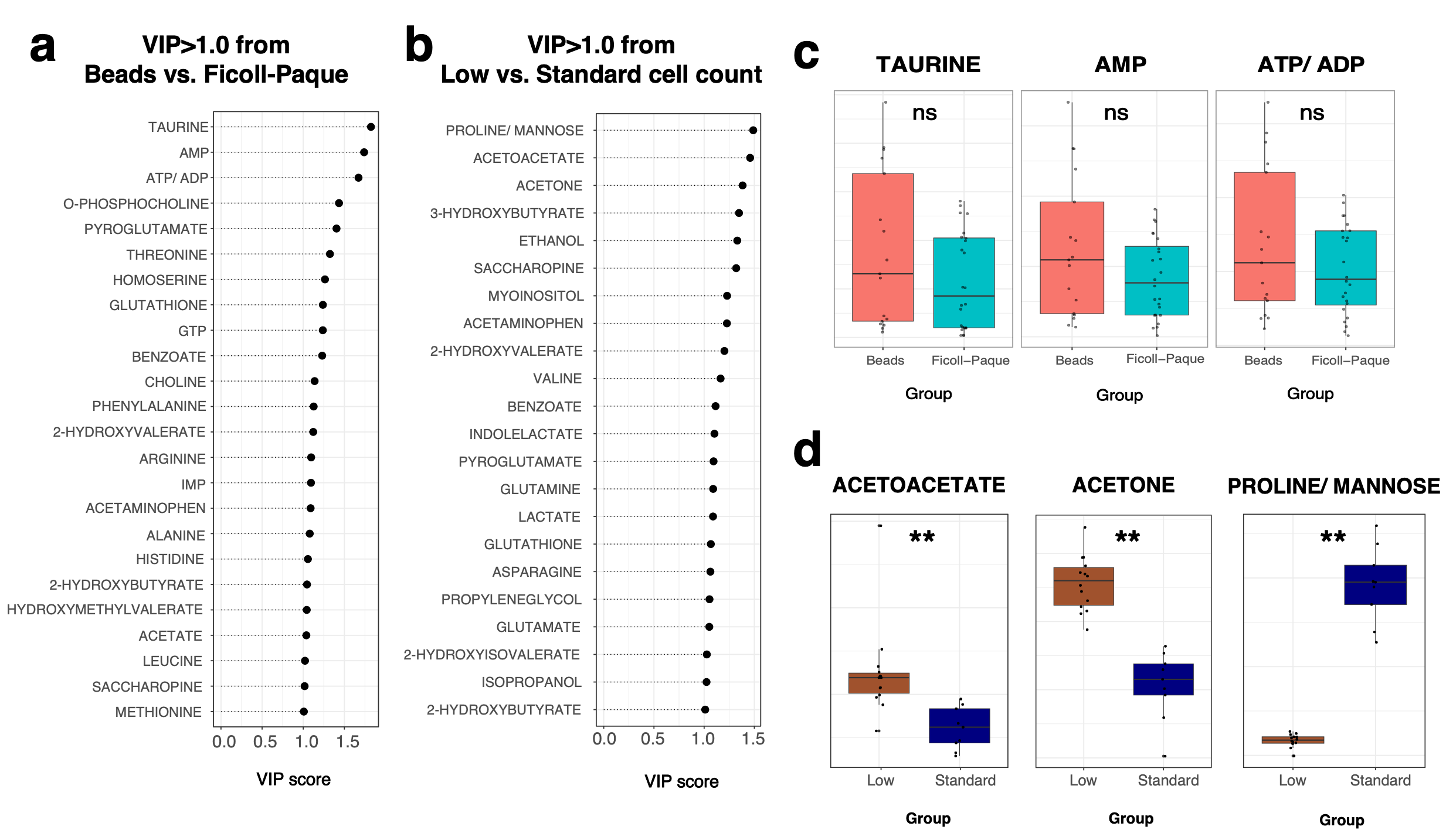
3.2. Comparing Different Isolation Techniques for Samples
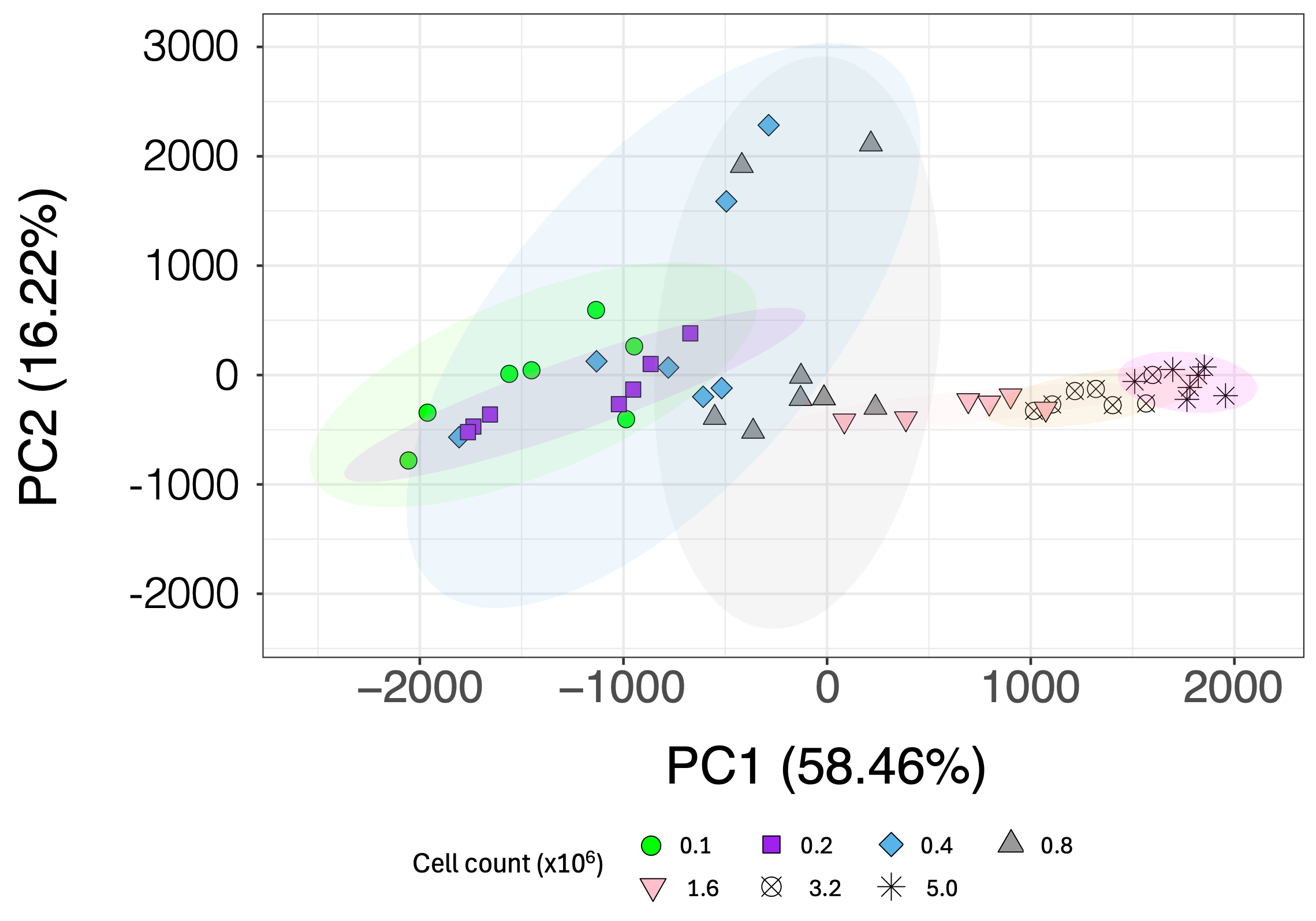
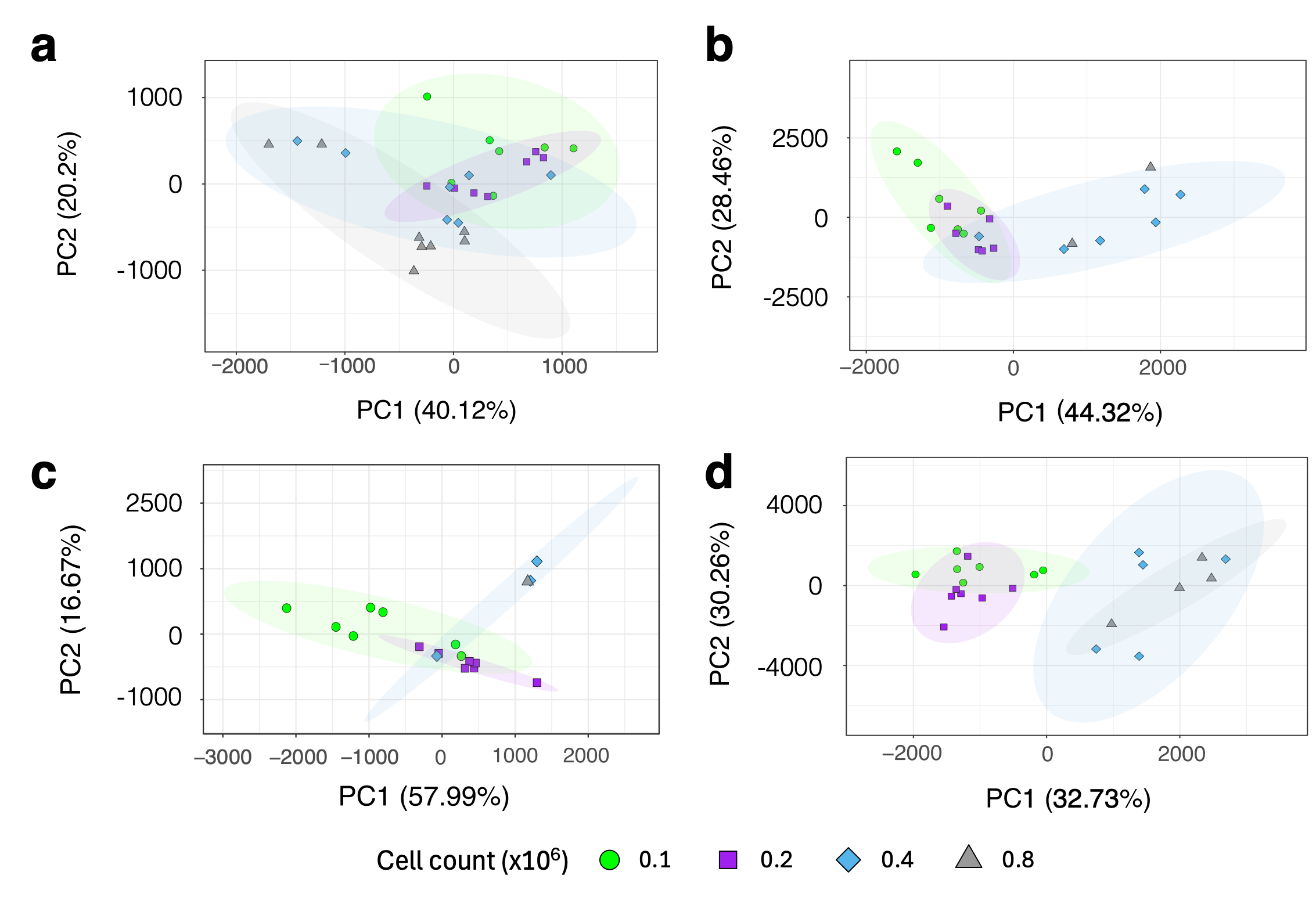
3.3. Effect of Cell Count on Metabolite Profile
3.4. Comparing the Effect of Increasing the Number of Scans on Low-Cell-Count Neutrophil NMR Metabolic Profiles
3.5. Comparison of Neutrophil Isolation Techniques Using Supervised Multivariate Analysis by PLS-DA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADP | adenosine diphosphate |
| AMP | adenosine monophosphate |
| ATP | adenosine triphosphate |
| COMETS | Consortium of Metabolomics Studies |
| CPMG | Carr–Purcell–Meiboom–Gill |
| CRS | correlation reliability score |
| FDR | false discovery rate |
| LOOCV | leave-one-out cross-validation |
| NAD | nicotinamide adenine dinucleotide |
| NADP | nicotinamide adenine dinucleotide phosphate |
| NMR | nuclear magnetic resonance |
| NOESY | nuclear Overhauser effect spectroscopy |
| NS | number of scans |
| PCA | principal component analysis |
| PLS-DA | partial least squares discriminant analysis |
| S/N | signal-to-noise ratio |
| VIP | variable importance in projection |
References
- Emwas, A.M.; Salek, R.M.; Griffin, J.L.; Merzaban, J. NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics 2013, 9, 1048–1072. [Google Scholar] [CrossRef]
- Amaral, P.; Carbonell-Sala, S.; De La Vega, F.M.; Faial, T.; Frankish, A.; Gingeras, T.; Guigo, R.; Harrow, J.L.; Hatzigeorgiou, A.G.; Johnson, R.; et al. The status of the human gene catalogue. Nature 2023, 622, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Hassan, M.A.; Al-Sakkaf, K.; Shait Mohammed, M.R.; Dallol, A.; Al-Maghrabi, J.; Aldahlawi, A.; Ashoor, S.; Maamra, M.; Ragoussis, J.; Wu, W.; et al. Integration of Transcriptome and Metabolome Provides Unique Insights to Pathways Associated With Obese Breast Cancer Patients. Front. Oncol. 2020, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Kouznetsova, V.L.; Kim, E.; Romm, E.L.; Zhu, A.; Tsigelny, I.F. Recognition of early and late stages of bladder cancer using metabolites and machine learning. Metabolomics 2019, 15, 94. [Google Scholar] [CrossRef]
- Kim, W.T.; Yun, S.J.; Yan, C.; Jeong, P.; Kim, Y.H.; Lee, I.S.; Kang, H.W.; Park, S.; Moon, S.K.; Choi, Y.H.; et al. Metabolic Pathway Signatures Associated with Urinary Metabolite Biomarkers Differentiate Bladder Cancer Patients from Healthy Controls. Yonsei Med. J. 2016, 57, 865–871. [Google Scholar] [CrossRef]
- Jin, X.; Yun, S.J.; Jeong, P.; Kim, I.Y.; Kim, W.J.; Park, S. Diagnosis of bladder cancer and prediction of survival by urinary metabolomics. Oncotarget 2014, 5, 1635–1645. [Google Scholar] [CrossRef]
- Wright, H.L.; Moots, R.J.; Bucknall, R.C.; Edwards, S.W. Neutrophil function in inflammation and inflammatory diseases. Rheumatology 2010, 49, 1618–1631. [Google Scholar] [CrossRef]
- Fresneda Alarcon, M.; McLaren, Z.; Wright, H.L. Neutrophils in the Pathogenesis of Rheumatoid Arthritis and Systemic Lupus Erythematosus: Same Foe Different M.O. Front. Immunol. 2021, 12, 649693. [Google Scholar] [CrossRef]
- Yu, B.; Zanetti, K.A.; Temprosa, M.; Albanes, D.; Appel, N.; Barrera, C.B.; Ben-Shlomo, Y.; Boerwinkle, E.; Casas, J.P.; Clish, C.; et al. The Consortium of Metabolomics Studies (COMETS): Metabolomics in 47 Prospective Cohort Studies. Am. J. Epidemiol. 2019, 188, 991–1012. [Google Scholar] [CrossRef] [PubMed]
- Richer, B.C.; Salei, N.; Laskay, T.; Seeger, K. Changes in Neutrophil Metabolism upon Activation and Aging. Inflammation 2018, 41, 710–721. [Google Scholar] [CrossRef]
- Chokesuwattanaskul, S.; Phelan, M.M.; Edwards, S.W.; Wright, H.L. A robust intracellular metabolite extraction protocol for human neutrophil metabolic profiling. PLoS ONE 2018, 13, e0209270. [Google Scholar] [CrossRef]
- Chokesuwattanaskul, S.; Fresneda Alarcon, M.; Mangalakumaran, S.; Grosman, R.; Cross, A.L.; Chapman, E.A.; Mason, D.; Moots, R.J.; Phelan, M.M.; Wright, H.L. Metabolic Profiling of Rheumatoid Arthritis Neutrophils Reveals Altered Energy Metabolism That Is Not Affected by JAK Inhibition. Metabolites 2022, 12, 650. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Findeisen, M.; Brand, T.; Berger, S. A 1H-NMR thermometer suitable for cryoprobes. Magn. Reson. Chem. 2007, 45, 175–178. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Ebbels, T.M.D.; Karaman, I.; Graca, G. Processing and Analysis of Untargeted Multicohort NMR Data. Methods Mol. Biol. 2019, 2037, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Vu, T.N.; Valkenborg, D.; Smets, K.; Verwaest, K.A.; Dommisse, R.; Lemiere, F.; Verschoren, A.; Goethals, B.; Laukens, K. An integrated workflow for robust alignment and simplified quantitative analysis of NMR spectrometry data. BMC Bioinform. 2011, 12, 405. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.; Andersson, E.R.; Bayless, A.L.; Brua, R.B.; Chang, M.C.; Cheng, L.L.; Clendinen, C.S.; Cochran, D.; Copié, V.; Cort, J.R.; et al. Best practices in NMR metabolomics: Current state. TrAC Trends Anal. Chem. 2024, 171, 117478. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Thomas, N.; Gooley, P.R.; Armstrong, C.W. Systematic Review of NMR-Based Metabolomics Practices in Human Disease Research. Metabolites 2022, 12, 963. [Google Scholar] [CrossRef]
- Bravo, J.A.; Vila, J.; Flores, Y. NMR Mestrenova, short manual for beginners. Boliv. J. Chem. 2018, 34, 123–131. [Google Scholar]
- Forezi, L.S.M.; Branco, F.S.C. Editing NMR spectra eith MestReNova software: A practical guide. Revisita Virtual De Quim. 2017, 9, 2650–2672. [Google Scholar] [CrossRef]
- Maciejewski, M.W.; Schuyler, A.D.; Gryk, M.R.; Moraru, I.I.; Romero, P.R.; Ulrich, E.L.; Eghbalnia, H.R.; Livny, M.; Delaglio, F.; Hoch, J.C. NMRbox: A Resource for Biomolecular NMR Computation. Biophys. J. 2017, 112, 1529–1534. [Google Scholar] [CrossRef]
- Garcia-Perez, I.; Posma, J.M.; Serrano-Contreras, J.I.; Boulange, C.L.; Chan, Q.; Frost, G.; Stamler, J.; Elliott, P.; Lindon, J.C.; Holmes, E.; et al. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat. Protoc. 2020, 15, 2538–2567. [Google Scholar] [CrossRef]
- Izquierdo-Garcia, J.L.; Villa, P.; Kyriazis, A.; del Puerto-Nevado, L.; Perez-Rial, S.; Rodriguez, I.; Hernandez, N.; Ruiz-Cabello, J. Descriptive review of current NMR-based metabolomic data analysis packages. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 263–270. [Google Scholar] [CrossRef]
- Sawall, M.; von Harbou, E.; Moog, A.; Behrens, R.; Schroder, H.; Simoneau, J.; Steimers, E.; Neymeyr, K. Multi-objective optimization for an automated and simultaneous phase and baseline correction of NMR spectral data. J. Magn. Reson. 2018, 289, 132–141. [Google Scholar] [CrossRef]
- Wang, K.C.; Wang, S.Y.; Kuo, C.H.; Tseng, Y.J. Distribution-based classification method for baseline correction of metabolomic 1D proton nuclear magnetic resonance spectra. Anal. Chem. 2013, 85, 1231–1239. [Google Scholar] [CrossRef]
- Lorentz. Bernstein Polynomials; American Mathematical Society: Providence, RI, USA, 2012. [Google Scholar]
- Ralston, S.L.; Pappalardo, L.; Pelczer, I. Breed and age effects on metabolic profiles of young horses using NMR-based Metabonomic analyses of serum. J. Equine Vet. Sci. 2011, 31, 304–305. [Google Scholar] [CrossRef]
- Corona-Vázquez, M.; Hernández-Bolio, G.I.; Muñoz-Cázares, N.; Peña-González, M.C.; Derbré, S.; Richomme, P.; Peña-Rodríguez, L.M. 1H-NMR-Based Chemometrics and 13C-NMR Dereplication Analysis Applied to the Bioprospecting of the Yucatecan Flora: Identification of 3-O-Acetyl-Ceanotic Acid as an Inhibitor of Bacterial Virulence Factors from Colubrina yucatanensis. Rev. Bras. Farm. 2024, 35, 134–145. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Hoch, J.C.; Baskaran, K.; Burr, H.; Chin, J.; Eghbalnia, H.R.; Fujiwara, T.; Gryk, M.R.; Iwata, T.; Kojima, C.; Kurisu, G.; et al. Biological Magnetic Resonance Data Bank. Nucleic Acids Res. 2023, 51, D368–D376. [Google Scholar] [CrossRef]
- Grosman, R. NMR Metabolomic Profiling of Mosquito Species to Understand Insecticide Resistance; University of Liverpool: Liverpool, UK, 2019. [Google Scholar]
- Yurekten, O.; Payne, T.; Tejera, N.; Amaladoss, F.X.; Martin, C.; Williams, M.; O’Donovan, C. MetaboLights: Open data repository for metabolomics. Nucleic Acids Res. 2024, 52, D640–D646. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Goodacre, R. On Splitting Training and Validation Set: A Comparative Study of Cross-Validation, Bootstrap and Systematic Sampling for Estimating the Generalization Performance of Supervised Learning. J. Anal. Test. 2018, 2, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Perez, R.; Fernandez, L.; Marco, S. Overoptimism in cross-validation when using partial least squares-discriminant analysis for omics data: A systematic study. Anal. Bioanal. Chem. 2018, 410, 5981–5992. [Google Scholar] [CrossRef]
- Gromski, P.S.; Correa, E.; Vaughan, A.A.; Wedge, D.C.; Turner, M.L.; Goodacre, R. A comparison of different chemometrics approaches for the robust classification of electronic nose data. Anal. Bioanal. Chem. 2014, 406, 7581–7590. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis--a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8 (Suppl. 1), 3–16. [Google Scholar] [CrossRef]
- Thomas, H.B.; Moots, R.J.; Edwards, S.W.; Wright, H.L. Whose Gene Is It Anyway? The Effect of Preparation Purity on Neutrophil Transcriptome Studies. PLoS ONE 2015, 10, e0138982. [Google Scholar] [CrossRef]
- Pelletier, M.; Maggi, L.; Micheletti, A.; Lazzeri, E.; Tamassia, N.; Costantini, C.; Cosmi, L.; Lunardi, C.; Annunziato, F.; Romagnani, S.; et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood 2010, 115, 335–343. [Google Scholar] [CrossRef]
- Mitchell, T.S.; Moots, R.J.; Wright, H.L. Janus kinase inhibitors prevent migration of rheumatoid arthritis neutrophils towards interleukin-8, but do not inhibit priming of the respiratory burst or reactive oxygen species production. Clin. Exp. Immunol. 2017, 189, 250–258. [Google Scholar] [CrossRef]
- Wright, H.L.; Thomas, H.B.; Moots, R.J.; Edwards, S.W. RNA-Seq Reveals Activation of Both Common and Cytokine-Specific Pathways following Neutrophil Priming. PLoS ONE 2013, 8, e58598. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Dona, A.C.; Kyriakides, M.; Scott, F.; Shephard, E.A.; Varshavi, D.; Veselkov, K.; Everett, J.R. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput. Struct. Biotechnol. J. 2016, 14, 135–153. [Google Scholar] [CrossRef]
- Debik, J.; Sangermani, M.; Wang, F.; Madssen, T.S.; Giskeødegård, G.F. Multivariate analysis of NMR-based metabolomic data. NMR Biomed. 2022, 35, e4638. [Google Scholar] [CrossRef]
- Bharti, S.K.; Roy, R. Quantitative 1H NMR spectroscopy. TrAC Trends Anal. Chem. 2012, 35, 5–26. [Google Scholar] [CrossRef]
- Skinner, S.P.; Fogh, RH.; Boucher, W.; Ragan, T.J.; Mureddu, L.G.; Vuister, G.W. CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J Biomol NMR 2016, 66, 111–124. [Google Scholar] [CrossRef]
- Hayward, M.W.; Mureddu, L.G.; Thompson, G.; Phelan, M.; Brooksbank, E.J.; Vuister, G.W. The CcpNmr Analysis Simulated Metabolomics Database (CASMDB): An Open-Source Collection of Metabolite Annotation Data for 1D 1H NMR-Based Metabolomics. bioRxiv 2024. [Google Scholar] [CrossRef]


| Dataset | Bin | Components | Accuracy | Sensitivity | Specificity | Precision | F1 Score |
|---|---|---|---|---|---|---|---|
| Magnetic beads vs. Ficoll-Paque | Original | 3 | 0.87 ± 0.11 | 0.89 ± 0.12 | 0.82 ± 0.15 | 0.88 ± 0.10 | 0.88 ± 0.09 |
| Post-CRS | 2 | 0.74 ± 0.12 | 0.83 ± 0.15 | 0.64 ± 0.19 | 0.74 ± 0.14 | 0.78 ± 0.12 | |
| NS 256 100 + 200 (low) vs. 400 + 800 (standard) cell count | Original | 1 | 0.82 ± 0.12 | 0.86 ± 0.16 | 0.77 ± 0.21 | 0.81 ± 0.19 | 0.82 ± 0.14 |
| Post-CRS | 1 | 0.85 ± 0.09 | 0.93 ± 0.13 | 0.79 ± 0.19 | 0.83 ± 0.17 | 0.86 ± 0.09 | |
| NS 2048 100 + 200 (low) vs. 400 + 800 (standard) cell count | Original | 2 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| Post-CRS | 2 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filbertine, G.; Abdullah, G.A.; Gill, L.; Grosman, R.; Phelan, M.M.; Chiewchengchol, D.; Hirankarn, N.; Wright, H.L. An NMR Metabolomics Analysis Pipeline for Human Neutrophil Samples with Limited Source Material. Metabolites 2025, 15, 612. https://doi.org/10.3390/metabo15090612
Filbertine G, Abdullah GA, Gill L, Grosman R, Phelan MM, Chiewchengchol D, Hirankarn N, Wright HL. An NMR Metabolomics Analysis Pipeline for Human Neutrophil Samples with Limited Source Material. Metabolites. 2025; 15(9):612. https://doi.org/10.3390/metabo15090612
Chicago/Turabian StyleFilbertine, Grace, Genna A. Abdullah, Lucy Gill, Rudi Grosman, Marie M. Phelan, Direkrit Chiewchengchol, Nattiya Hirankarn, and Helen L. Wright. 2025. "An NMR Metabolomics Analysis Pipeline for Human Neutrophil Samples with Limited Source Material" Metabolites 15, no. 9: 612. https://doi.org/10.3390/metabo15090612
APA StyleFilbertine, G., Abdullah, G. A., Gill, L., Grosman, R., Phelan, M. M., Chiewchengchol, D., Hirankarn, N., & Wright, H. L. (2025). An NMR Metabolomics Analysis Pipeline for Human Neutrophil Samples with Limited Source Material. Metabolites, 15(9), 612. https://doi.org/10.3390/metabo15090612







