Seed Metabolomic Landscape Reflecting Key Differential Metabolic Profiles Among Different Wheat Cultivars
Abstract
1. Introduction
2. Materials and Methods
2.1. Metabolite Extraction and Sample Preparation
2.2. Liquid Chromatography-Quadrupole Time-of-Flight Tandem Mass Spectrometry
2.3. Data Mining: Data Pre-Processing and Chemometrics Analyses
2.4. Metabolite Annotation and Biological Interpretation
3. Results
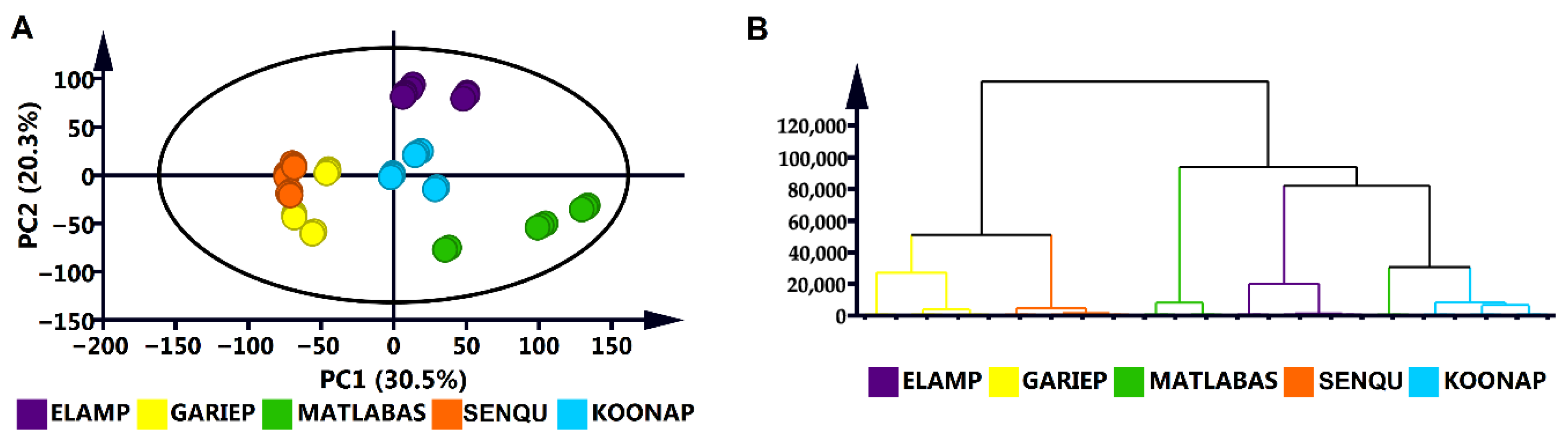
3.1. Chemometric Analyses and Metabolic Profiling of Wheat Cultivar Seed Metabolomes
3.2. Differentiation of the Different Wheat Cultivar Seeds
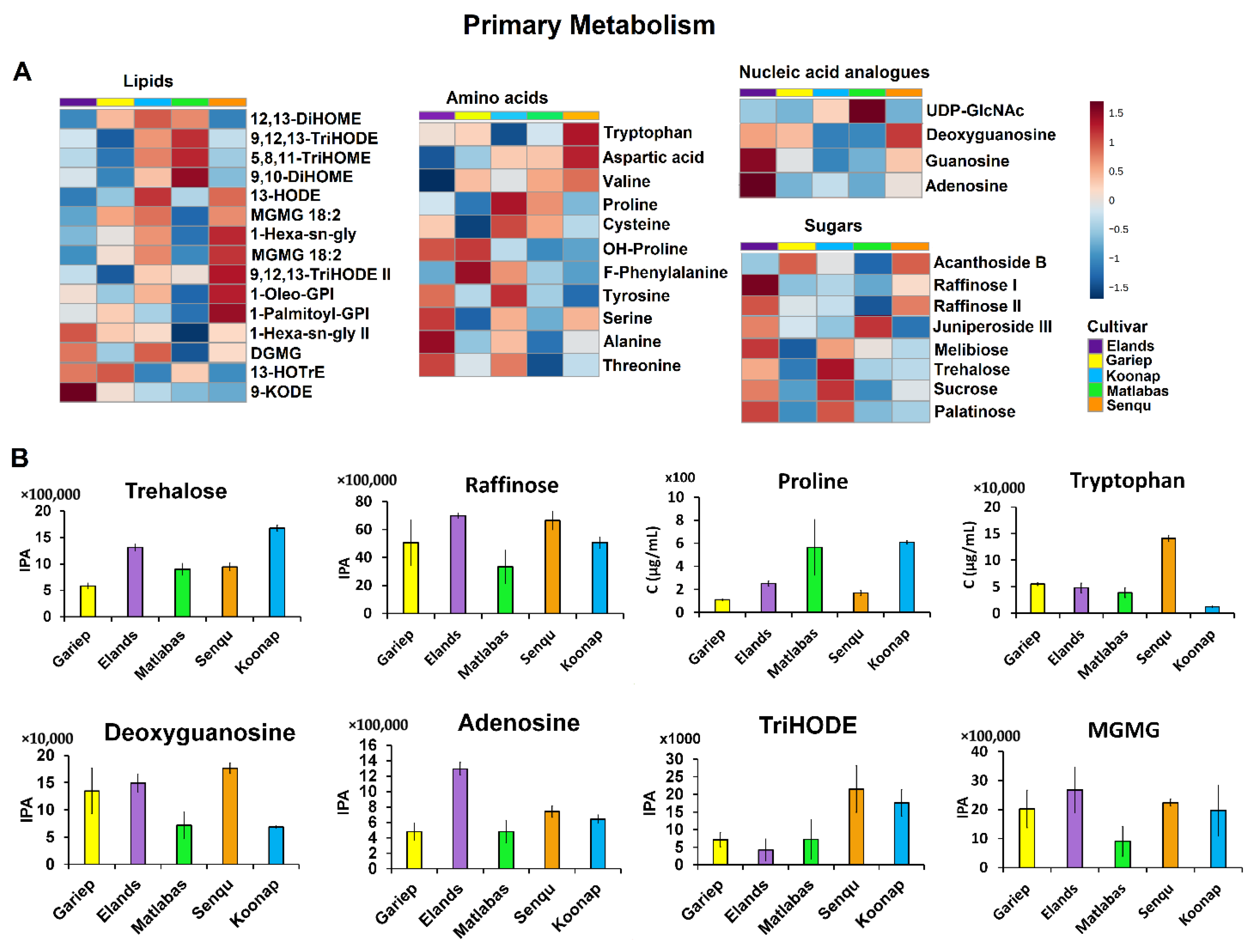
3.2.1. Primary Metabolism in Dry Wheat Seeds
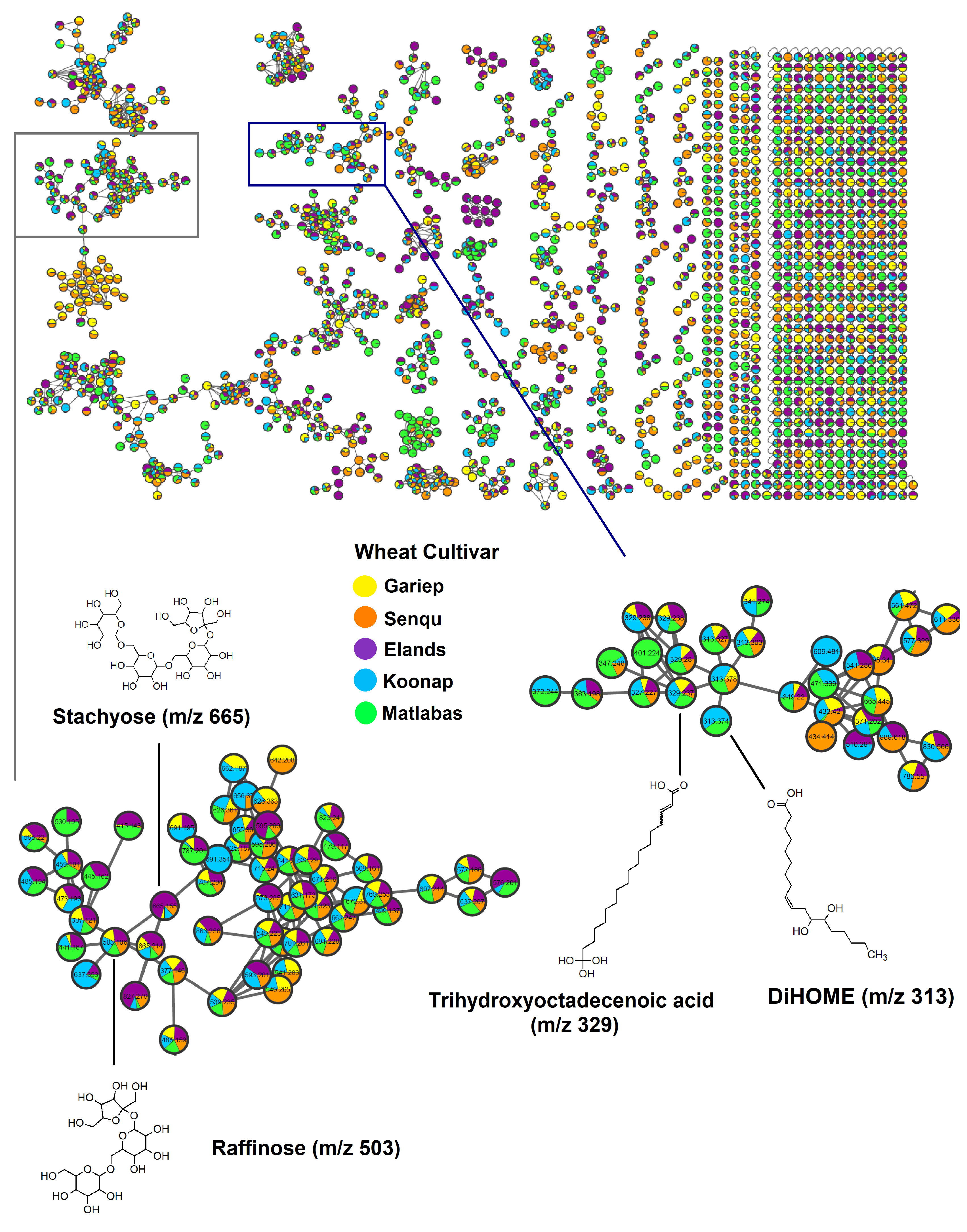
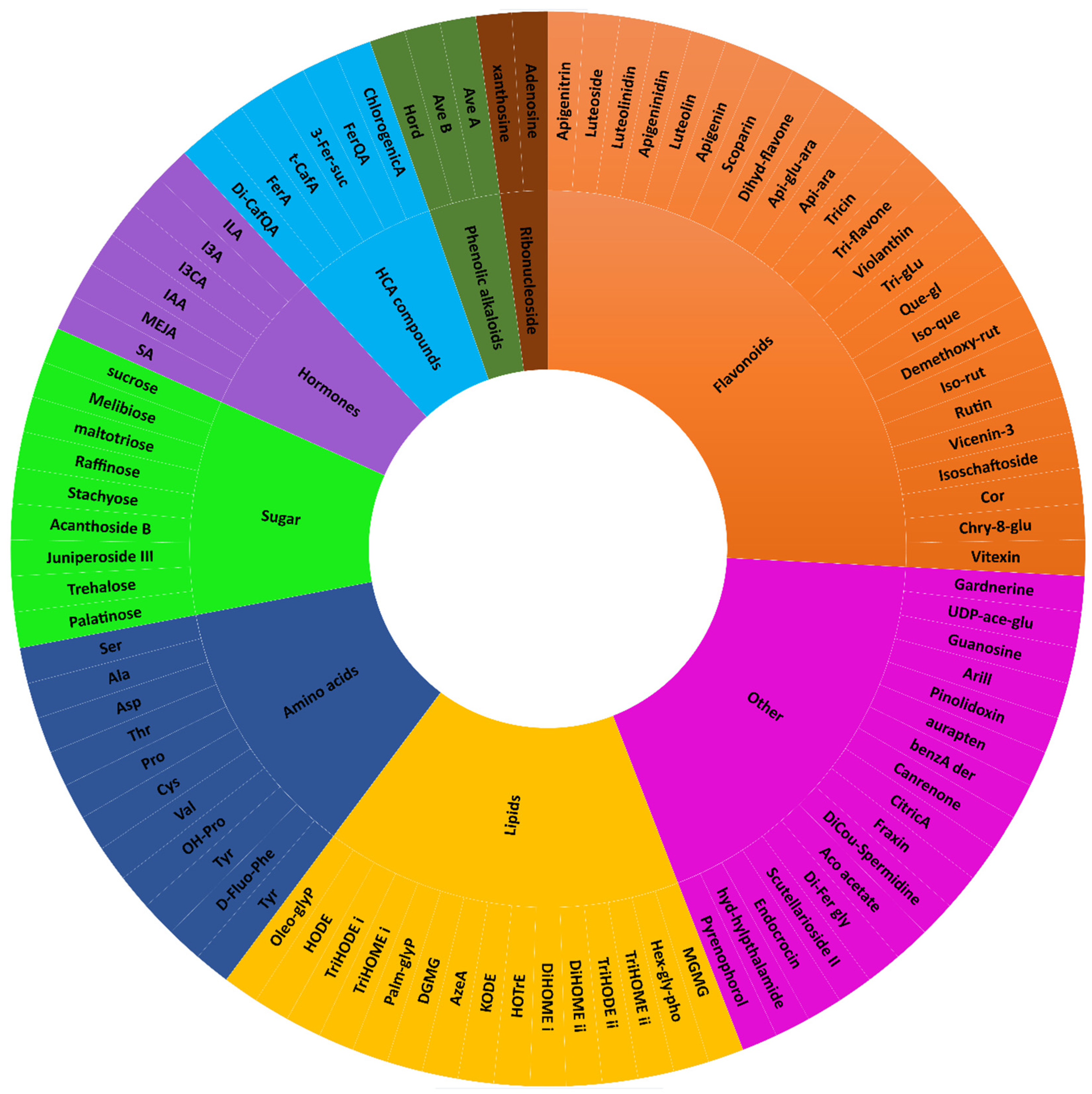
3.2.2. Specialised Metabolism in Dry Wheat Seeds
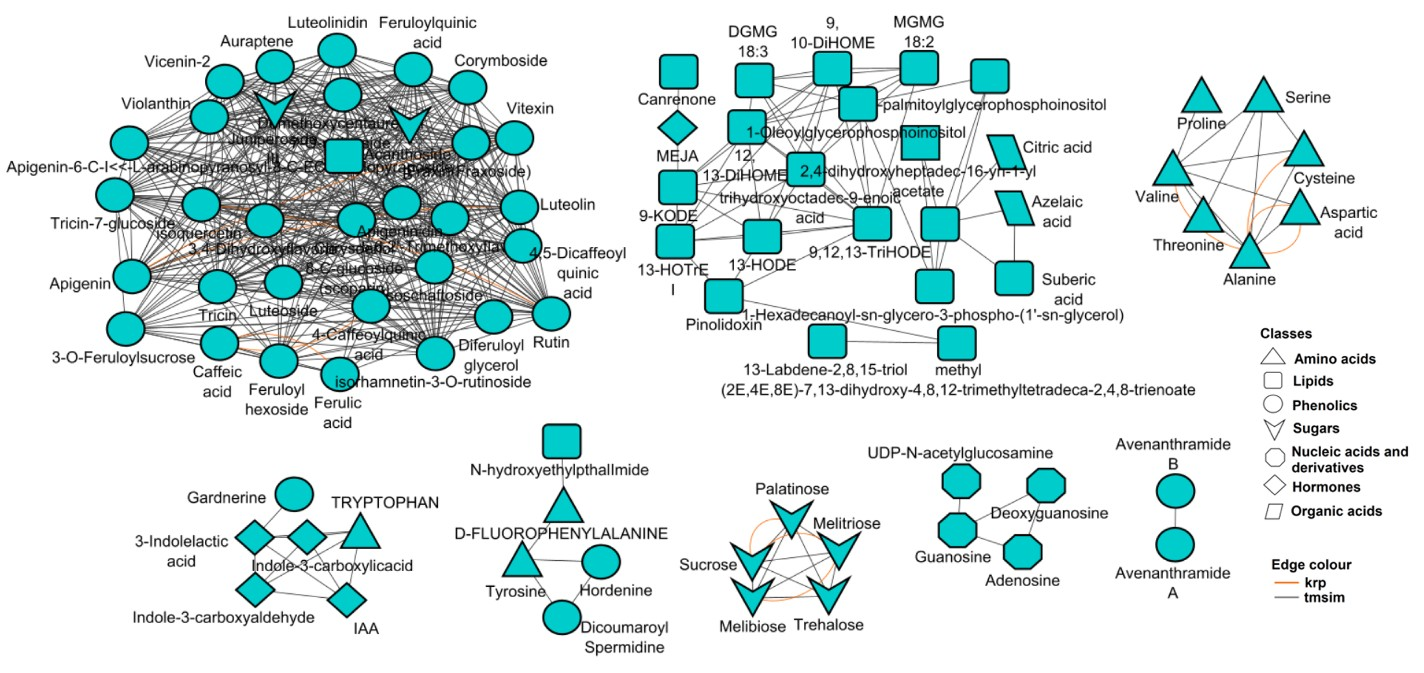
3.3. Metabolic Pathway and Network Analyses of the Annotated Wheat Cultivar Seed Metabolomes
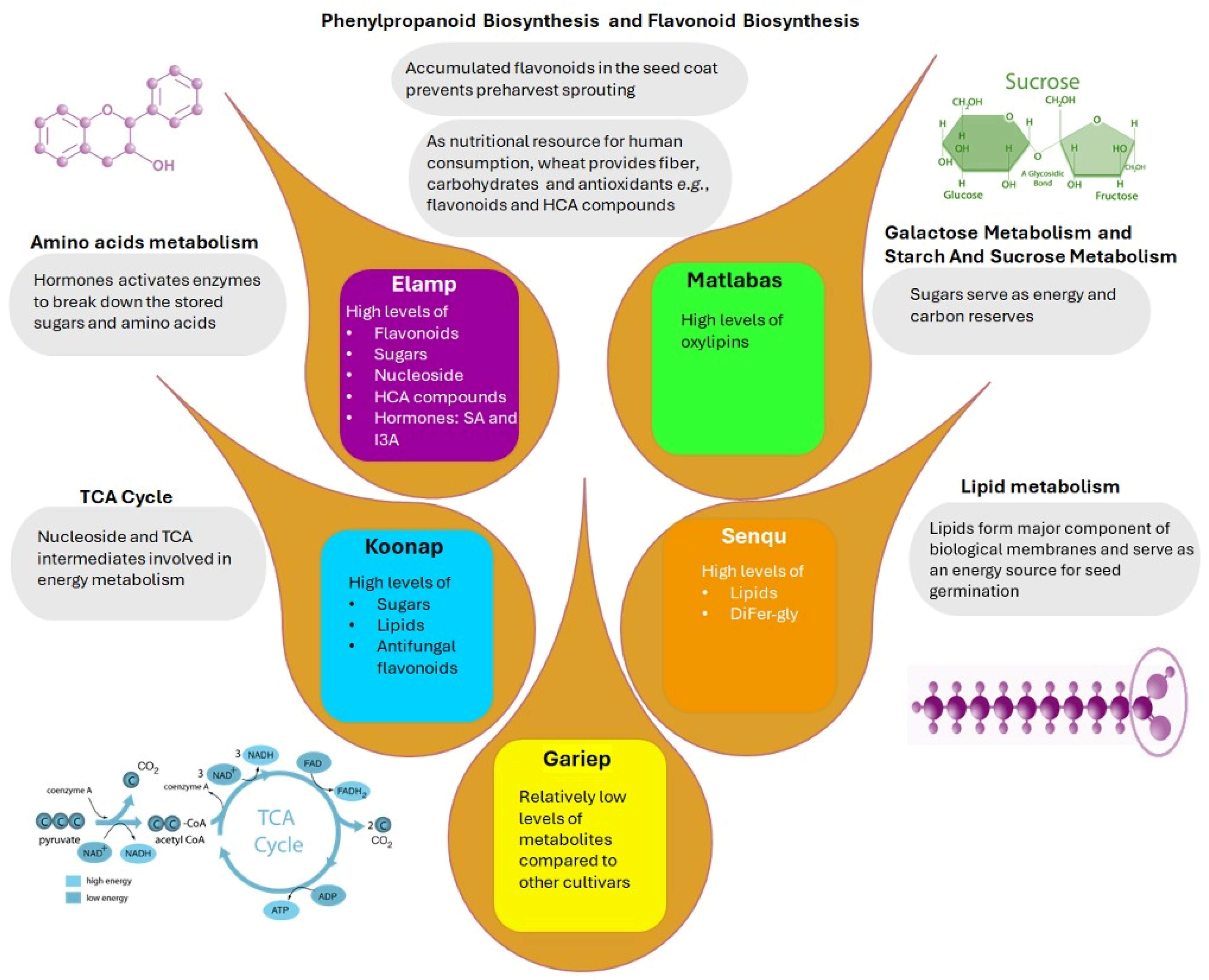
4. Discussion
4.1. The Primary Metabolism Charts in Dry Wheat Seeds
4.2. The Specialised Metabolism Charts in Dry Wheat Seeds
4.3. Biological Insights into Wheat Seed Metabolic Profiles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jankielsohn, A.; Miles, C. How Do Older Wheat Cultivars Compare to Modern Wheat Cultivars Currently on the Market in South Africa? J. Hortic. Sci. Res. 2017, 1, 42–47. [Google Scholar] [CrossRef]
- Vu, L.D.; Verstraeten, I.; Stes, E.; Van Bel, M.; Coppens, F.; Gevaert, K.; De Smet, I. Proteome Profiling of Wheat Shoots from Different Cultivars. Front. Plant Sci. 2017, 8, 332. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The Contribution of Wheat to Human Diet and Health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Atack, J.; Coclanis, P.; Grantham, G. Creating Abundance: Biological Innovation and American Agricultural Development-An Appreciation and Research Agenda. Explor. Econ. Hist. 2009, 46, 160–167. [Google Scholar] [CrossRef]
- Lachman, J.; Hejtmánková, K.; Kotíková, Z. Tocols and Carotenoids of Einkorn, Emmer and Spring Wheat Varieties: Selection for Breeding and Production. J. Cereal Sci. 2013, 57, 207–214. [Google Scholar] [CrossRef]
- Massawe, F.; Mayes, S.; Cheng, A. Crop Diversity: An Unexploited Treasure Trove for Food Security. Trends Plant Sci. 2016, 21, 365–368. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding Crops to Feed 10 Billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef]
- Ranilla, L. The Application of Metabolomics for the Study of Cereal Corn (Zea mays L.). Metabolites 2020, 10, 300. [Google Scholar] [CrossRef]
- Bishaw, Z.; Struik, P.C.; van Gastel, A.J.G. Farmers’ Seed Sources and Seed Quality: 1. Physical and Physiological Quality. J. Crop Improv. 2012, 26, 655–692. [Google Scholar] [CrossRef]
- Corbineau, F. Markers of Seed Quality: From Present to Future. Seed Sci. Res. 2012, 22, S61–S68. [Google Scholar] [CrossRef]
- Singh, N.; Rai, V.; Kumar, N. Multi Omics Strategies and Prospects to Enhance Seed Quality and Nutritional Traits in Pigeonpea. Nucleus 2020, 63, 249–256. [Google Scholar] [CrossRef]
- Pfeiffer, W.H.; Mcclafferty, B. HarvestPlus: Breeding Crops for Better Nutrition. Crop Sci. 2007, 47, S88–S105. [Google Scholar] [CrossRef]
- Chen, J.; Xue, M.; Liu, H.; Fernie, A.; Chen, W. Exploring the Genic Resources Underlying Metabolites through MGWAS and MQTL in Wheat: From Large-Scale Gene Identification and Pathway Elucidation to Crop Improvement. Plant Commun. 2021. [Google Scholar] [CrossRef]
- Francki, M.G.; Hayton, S.; Gummer, J.P.A.; Rawlinson, C.; Trengove, R.D. Metabolomic Profiling and Genomic Analysis of Wheat Aneuploid Lines to Identify Genes Controlling Biochemical Pathways in Mature Grain. Plant Biotechnol. J. 2016. [Google Scholar] [CrossRef]
- Schauer, N.; Fernie, A.R. Plant Metabolomics: Towards Biological Function and Mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Lyv, Y.; Zheng, W.; Yang, C.; Li, Y.; Wang, X.; Chen, R.; Wang, C.; Luo, J.; Qu, L. Comparative Metabolomics Reveals Two Metabolic Modules Affecting Seed Germination in Rice (Oryza sativa). Metabolites 2021, 11, 880. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Huang, W.; Gao, J.; Fu, H.; Liu, J. Comparative Metabolomic Analysis of Seed Metabolites Associated with Seed Storability in Rice (Oryza sativa L.) during Natural Aging. Plant Physiol. Biochem. 2018, 127, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Kim, D.W.; Khadka, P.; Rakwal, R.; Rohila, J.S. Unraveling Key Metabolomic Alterations in Wheat Embryos Derived from Freshly Harvested and Water-Imbibed Seeds of Two Wheat Cultivars with Contrasting Dormancy Status. Front. Plant Sci. 2017, 8, 1203. [Google Scholar] [CrossRef]
- Matsuda, F.; Okazaki, Y.; Oikawa, A.; Kusano, M.; Nakabayashi, R.; Kikuchi, J.; Yonemaru, J.; Ebana, K.; Yano, M.; Saito, K. Dissection of Genotype—Phenotype Associations in Rice Grains Using Metabolome Quantitative Trait Loci Analysis. Plant J. 2012, 70, 624–636. [Google Scholar] [CrossRef]
- Toubiana, D.; Semel, Y.; Tohge, T.; Beleggia, R.; Cattivelli, L.; Rosental, L.; Nikoloski, Z.; Zamir, D.; Fernie, A.R.; Fait, A. Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations. PLoS Genet. 2012, 8, e1002612. [Google Scholar] [CrossRef]
- Rao, J.; Cheng, F.; Hu, C.; Quan, S.; Lin, H.; Wang, J.; Chen, G.; Zhao, X.; Alexander, D.; Guo, L.; et al. Metabolic Map of Mature Maize Kernels. Metabolomics 2014, 10, 775–787. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Piater, L.A.; Steenkamp, P.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Comparative Metabolite Profiling of Wheat Cultivars (Triticum Aestivum) Reveals Signatory Markers for Resistance and Susceptibility to Stripe Rust and Aluminium (Al3+) Toxicity. Metabolites 2022, 12, 98. [Google Scholar] [CrossRef]
- Khanijou, J.K.; Kulyk, H.; Bergès, C.; Khoo, L.W.; Ng, P.; Yeo, H.C.; Helmy, M.; Bellvert, F.; Chew, W.; Selvarajoo, K. Metabolomics and Modelling Approaches for Systems Metabolic Engineering. Metab. Eng. Commun. 2022. [Google Scholar] [CrossRef]
- Ramabulana, A.; Petras, D.; Madala, N.E.; Tugizimana, F. Metabolomics and Molecular Networking to Characterize the Chemical Space of Four Momordica Plant Species. Metabolites 2021, 11, 763. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; Maille, G.O.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, Scaling, and Transformations: Improving the Biological Information Content of Metabolomics Data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; Anderson, D.; Morais, D.L.; Chang, L.; Barrette, M.; Gauthier, C.; Etienne, P.-É; Li, S.; et al. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, 388–396. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A Conversation on Data Mining Strategies in LC-MS Untargeted Metabolomics: Pre-Processing and Pre-Treatment Steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Othibeng, K.; Nephali, L.; Ramabulana, A.T.; Steenkamp, P.; Petras, D.; Kang, K.B.; Opperman, H.; Huyser, J.; Tugizimana, F. A Metabolic Choreography of Maize Plants Treated with a Humic Substance-Based Biostimulant under Normal and Starved Conditions. Metabolites 2021, 11, 403. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Tugizimana, F.; Djami-Tchatchou, A.T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic Analysis of Defense-Related Reprogramming in Sorghum Bicolor in Response to Colletotrichum Sublineolum Infection Reveals a Functional Metabolic Web of Phenylpropanoid and Flavonoid Pathways. Front. Plant Sci. 2019, 9, 1840. [Google Scholar] [CrossRef]
- Barupal, D.K.; Haldiy, P.K.; Wohlgemuth, G.; Kind, T.; Kothari, S.L.; Pinkerton, K.E.; Fiehn, O. MetaMapp: Mapping and Visualizing Metabolomic Data by Integrating Information from Biochemical Pathways and Chemical and Mass Spectral Similarity. BMC Bioinform. 2012, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Rao, J.; Shi, J.; Hu, C.; Cheng, F.; Wilson, Z.A.; Zhang, D.; Quan, S. Seed Metabolomic Study Reveals Significant Metabolite Variations and Correlations among Different Soybean Cultivars. J. Integr. Plant Biol. 2014, 56, 826–836. [Google Scholar] [CrossRef]
- Toubiana, D.; Puzis, R.; Wen, L.; Sikron, N.; Kurmanbayeva, A.; Soltabayeva, A.; Wilhelmi, M.D.M.R.; Sade, N.; Fait, A.; Sagi, M.; et al. Combined network analysis and machine learning allows the prediction of metabolic pathways from tomato metabolomics data. Commun. Biol. 2019, 2, 214. [Google Scholar] [CrossRef]
- Amara, A.; Frainary, C.; Jourdan, F.; Naake, T.; Neumann, S.; Novoa-del-Toro, E.M.; Salek, R.M.; Salzer, L.; Scharfenberg, S.; Witting, M. Networks and Graphs Discovery in Metabolomics Data Analysis and Interpretation. Front. Mol. Biosci. 2022. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Zhu, H.; Liu, Y.; Pan, S.; Chen, X.; Wu, T. Identifying Distinct Markers in Two Sorghum Varieties for Baijiu Fermentation Using Untargeted Metabolomics and Molecular Network Approaches. Food Chem. 2024. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- van der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic Modeling for Untargeted Substructure Exploration in Metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.; Wei Ong, C.; Wandy, J.; Ernst, M.; Ridder, L.; van der Hooft, J.J.J. Deciphering Complex Metabolite Mixtures by Unsupervised and Supervised Substructure Discovery and Semi-Automated Annotation from MS/MS Spectra. Faraday Discuss. 2019, 218, 284–302. [Google Scholar] [CrossRef]
- Beniddir, M.A.; Kang, K.B.; Genta-Jouve, G.; Huber, F.; Rogers, S.; van der Hooft, J.J.J.J. Advances in Decomposing Complex Metabolite Mixtures Using Substructure- and Network-Based Computational Metabolomics Approaches. Nat. Prod. Rep. 2021, 38, 1967–1993. [Google Scholar] [CrossRef] [PubMed]
- Shirley, B.W. Flavonoids in Seeds and Grains: Physiological Function, Agronomic Importance and the Genetics of Biosynthesis. Seed Sci. Res. 1998, 8, 415–422. [Google Scholar] [CrossRef]
- Kim, H.U. Lipid Metabolism in Plants. Plants 2020, 9, 871. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.T.; Kapoor, R.; Datta, A.; Okumoto, S. Tissue Specific Expression of UMAMIT Amino Acid Transporters in Wheat. Sci. Rep. 2022, 12, 348. [Google Scholar] [CrossRef]
- Masoni, A.; Ercoli, L.; Mariotti, M.; Arduini, I. Post-Anthesis Accumulation and Remobilization of Dry Matter, Nitrogen and Phosphorus in Durum Wheat as Affected by Soil Type. Eur. J. Agron. 2007, 26, 179–186. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Liu, N.; Su, D.; Xue, Q.; Stewart, B.A.; Wang, Z. Effect of Source—Sink Manipulation on Accumulation of Micronutrients and Protein in Wheat Grains. J. Plant Nutr. Soil Sci. 2012, 175, 622–629. [Google Scholar] [CrossRef]
- Sun, S.; Fang, J.; Lin, M.; Hu, C.; Qi, X.; Chen, J.; Zhong, Y.; Muhammad, A.; Li, Z.; Li, Y. Comparative Metabolomic and Transcriptomic Studies Reveal Key Metabolism Pathways Contributing to Freezing Tolerance Under Cold Stress in Kiwifruit. Front. Plant Sci. 2021, 12, 628969. [Google Scholar] [CrossRef]
- Frimpong, F.; Anokye, M.; Windt, C.W.; Naz, A.A.; Frei, M.; van Dusschoten, D.; Fiorani, F. Proline-Mediated Drought Tolerance in the Barley (Hordeum Vulgare l.) Isogenic Line Is Associated with Lateral Root Growth at the Early Seedling Stage. Plants 2021, 10, 2177. [Google Scholar] [CrossRef]
- Dien, D.C.; Mochizuki, T.; Yamakawa, T. Effect of Various Drought Stresses and Subsequent Recovery on Proline, Total Soluble Sugar and Starch Metabolisms in Rice (Oryza Sativa L.) Varieties. Plant Prod. Sci. 2019. [Google Scholar] [CrossRef]
- Peterbauer, T.; Richter, A. Biochemistry and Physiology of Raffinose Family Oligosaccharides and Galactosyl Cyclitols in Seeds. Seed Sci. Res. 2001, 11, 185–197. [Google Scholar] [CrossRef]
- Salvi, P.; Varshney, V.; Majee, M. Raffinose Family Oligosaccharides (RFOs): Role in Seed Vigor and Longevity. Biosci. Rep. 2022, 42, BSR20220198. [Google Scholar] [CrossRef]
- González-Thuillier, I.; Salt, L.; Chope, G.; Penson, S.; Skeggs, P.; Tosi, P.; Powers, S.J.; Ward, J.L.; Wilde, P.; Shewry, P.R.; et al. Distribution of Lipids in the Grain of Wheat (Cv. Hereward) Determined by Lipidomic Analysis of Milling and Pearling Fractions. J. Agric. Food Chem. 2015, 63, 10705–10716. [Google Scholar] [CrossRef]
- Delgado-garcía, E.; Piedras, P.; García-magdaleno, I.M.; Pineda, M.; Gálvez-valdivieso, G.; Gálvez-valdivieso, G. Nucleoside Metabolism Is Induced in Common Bean During Early Seedling Development. Front. Plant Sci. 2021, 12, 651015. [Google Scholar] [CrossRef]
- Stasolla, C.; Katahira, R.; Thorpe, T.A.; Ashihara, H. Purine and Pyrimidine Nucleotide Metabolism in Higher Plants. J. Plant Physiol. 2003, 160, 1271–1295. [Google Scholar] [CrossRef] [PubMed]
- Eastmond, P.J.; Graham, I.A.; Graham, I.A. Re-Examining the Role of the Glyoxylate Cycle in Oilseeds. Trends Plant Sci. 2001, 6, 72–77. [Google Scholar] [CrossRef]
- Dahncke, K.; Witte, C. Plant Purine Nucleoside Catabolism Employs a Guanosine Deaminase Required for the Generation of Xanthosine in Arabidopsis. Plant Cell 2013, 25, 4101–4109. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Zhang, S.; Zhao, H.; Chen, J.; Zuo, Z.; Sun, X.; Cheng, Y.; Xu, Z.; Liu, G. Analysis of the Energy Source at the Early Stage of Poplar Seed Germination: Verification of Perl’ s Pathway. 3 Biotech 2020, 10, 418. [Google Scholar] [CrossRef]
- Vishwanath, S.J.; Kosma, D.K.; Pulsifer, I.P.; Scandola, S.; Pascal, S.; Joubès, J.; Dittrich-domergue, F.; Lessire, R.; Rowland, O.; Domergue, F. Suberin-Associated Fatty Alcohols in Arabidopsis: Distributions in Roots and Contributions to Seed Coat. Plant Physiol. 2013, 163, 1118–1132. [Google Scholar] [CrossRef]
- Butts-Wilmsmeyer, C.J.; Mumm, R.H.; Bohn, M.O. Quantitative Genetic Analysis of Hydroxycinnamic Acids in Maize (Zea mays L.) for Plant Improvement and Production of Health-Promoting Compounds. J. Agric. Food Chem. 2020, 68, 9585–9593. [Google Scholar] [CrossRef]
- Khlestkina, E. The Adaptive Role of Flavonoids: Emphasis on Cereals. Cereal Res. Commun. 2013, 41, 185–198. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, C.; Shu, X.; Hu, H.; Cao, M.; Ma, Y.; Zhao, W.; Wu, Z.; Li, M.; Cai, D.; et al. Controlling Preharvest Sprouting of Wheat through Nanonetworks. ACS Sustain. Chem. Eng. 2018, 6, 11050–11057. [Google Scholar] [CrossRef]
- El-mergawi, R.A.; El-Wahed, M.S.A.A. Effect of Exogenous Salicylic Acid or Indole Acetic Acid on Their Endogenous Levels, Germination, and Growth in Maize. Bull. Natl. Res. Cent. 2020, 44, 167. [Google Scholar] [CrossRef]
- Vicente, M.R.; Plasencia, J. Salicylic Acid beyond Defence: Its Role in Plant Growth and Development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.B.; Hemlata, C.; Wani, A.H.; Singh, S.; Upadhyay, N. Salicylic Acid to Decrease Plant Stress. Environ. Chem. Lett. 2016, 15, 101–123. [Google Scholar] [CrossRef]
- Karimi, M.R.; Sabokdast, M.; Korang, B.H.; Abbasi, A.R.; Bihamta, M.R. Seed Priming with Salicylic Acid Enhances Salt Stress Tolerance by Boosting Antioxidant Defense in Phaseolus Vulgaris Genotypes. BMC Plant Biol. 2025. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Ali, B.; Rani, I.; Hayat, S.; Ahmad, A. Effect of 4-Cl-Indole-3-Acetic Acid on the Seed Germination of Cicer Arietinum Exposed to Cadmium. Acta Bot. Croat. 2007, 66, 57–65. [Google Scholar]
- Schenck, C.A.; Maeda, H.A. Tyrosine Biosynthesis, Metabolism, and Catabolism in Plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef]
- Al-mohammad, M.H.S.; Al-taey, D.K. Effect of Tyrosine and Sulfur on Growth, Yield and Antioxidant Compounds in Arugula Leaves and Seeds. Res. Crops 2019, 20, 116–120. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Saxena, P. Auxin Driven Indoleamine Biosynthesis and the Role of Tryptophan as an Inductive Signal in Hypericum perforatum (L.). PLoS ONE 2019, 14, e0223878. [Google Scholar] [CrossRef]
- Pirasteh-Anosheh, H.; Emam, Y.; Pessarakli, M. Changes in Endogenous Hormonal Status in Corn (Zea mays) Hybrids Under Drought Stress. J. Plant Nutr. 2013, 36, 1695–1707. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.J.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A Three-Ring Circus: Metabolism of the Three Proteogenic Aromatic Amino Acids and Their Role in the Health of Plants and Animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Frainay, C.; Jourdan, F. Computational Methods to Identify Metabolic Sub-Networks Based on Metabolomic Profiles. Brief. Bioinform. 2017, 18, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Kim, T.Y.; Lee, S.Y. Framework for Network Modularization and Bayesian Network Analysis to Investigate the Perturbed Metabolic Network. BMC Syst. Biol. 2011, 5, S14–S26. [Google Scholar] [CrossRef] [PubMed]
- Wanichthanarak, K.; Fahrmann, J.F.; Grapov, D. Genomic, Proteomic, and Metabolomic Data Integration Strategies. Biomark. Insights 2015, 10, BMI.S29511. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, S.P.; Pan, X.; Hong, Y.; Roth, M.; Welti, R.; Wang, X. Enhancing Seed Quality and Viability by Suppressing Phospholipase D in Arabidopsis. Plant J. 2007, 50, 950–957. [Google Scholar] [CrossRef]


| Wheat Cultivar | Wheat Type | Resistance/Susceptibility |
|---|---|---|
| Gariep | Intermediate-type | Susceptible to both |
| Elands | Intermediate-type | Susceptible to both |
| Matlabas | Winter-type | Susceptible to both |
| Koonap | Intermediate-type | Resistant to both |
| Senqu | Intermediate-type | Susceptible to aluminium toxicity and resistant to stripe rust |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othibeng, K.; Nephali, L.; Tugizimana, F. Seed Metabolomic Landscape Reflecting Key Differential Metabolic Profiles Among Different Wheat Cultivars. Metabolites 2025, 15, 603. https://doi.org/10.3390/metabo15090603
Othibeng K, Nephali L, Tugizimana F. Seed Metabolomic Landscape Reflecting Key Differential Metabolic Profiles Among Different Wheat Cultivars. Metabolites. 2025; 15(9):603. https://doi.org/10.3390/metabo15090603
Chicago/Turabian StyleOthibeng, Kgalaletso, Lerato Nephali, and Fidele Tugizimana. 2025. "Seed Metabolomic Landscape Reflecting Key Differential Metabolic Profiles Among Different Wheat Cultivars" Metabolites 15, no. 9: 603. https://doi.org/10.3390/metabo15090603
APA StyleOthibeng, K., Nephali, L., & Tugizimana, F. (2025). Seed Metabolomic Landscape Reflecting Key Differential Metabolic Profiles Among Different Wheat Cultivars. Metabolites, 15(9), 603. https://doi.org/10.3390/metabo15090603







