Metabolomic Analysis of Feces vs. Cecum Content in Animals: A Comparative Study Investigated by 1H-NMR
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Metabolomic Analysis
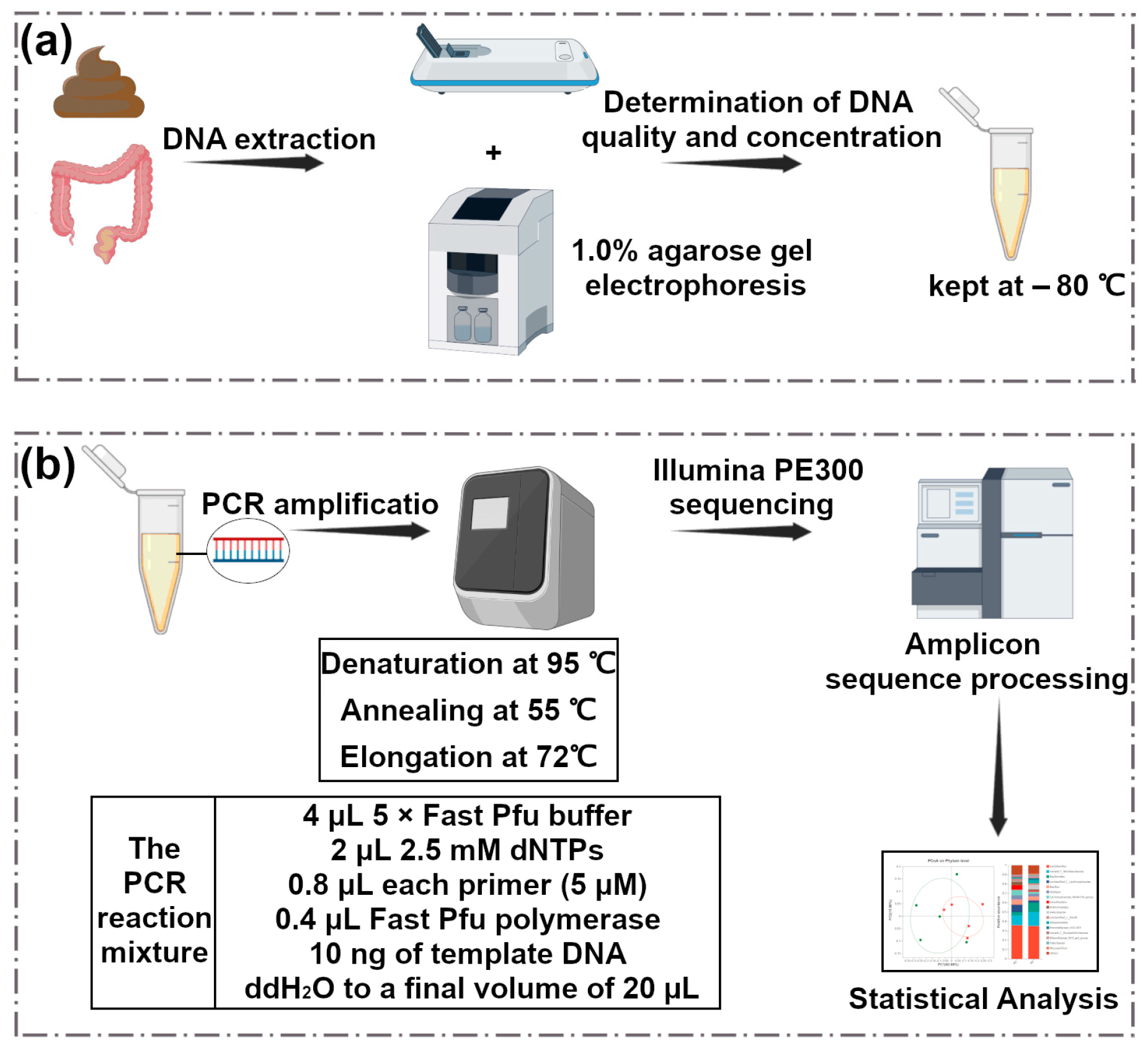
2.3. Microbiome Analysis
2.4. Statistical Analysis
3. Results
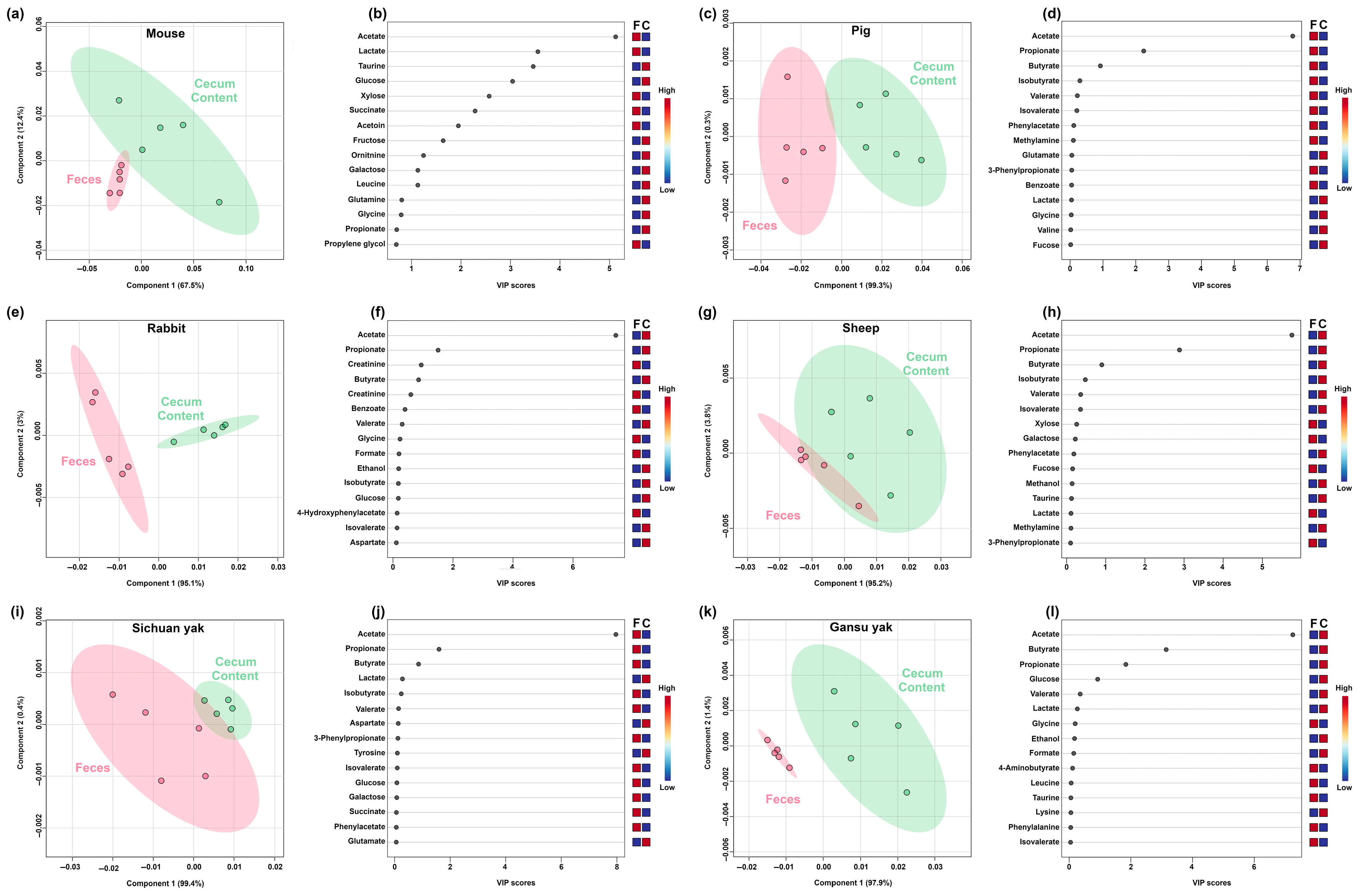
3.1. Metabolomic Characterizations of Feces and Cecum Content
3.2. Comparative Metabolomic Analysis of Feces and Cecum Content in Mammals and Non-Mammals
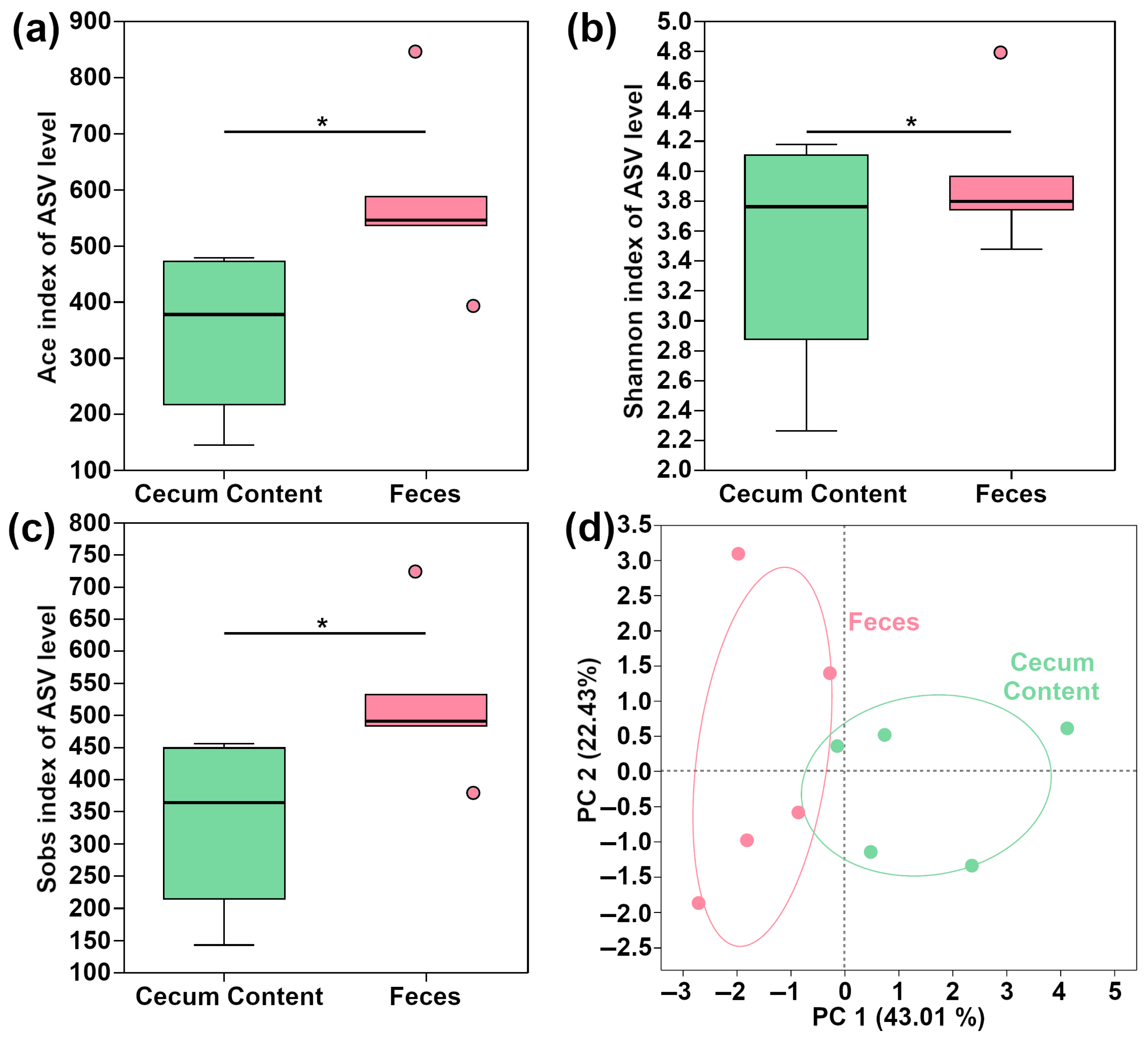
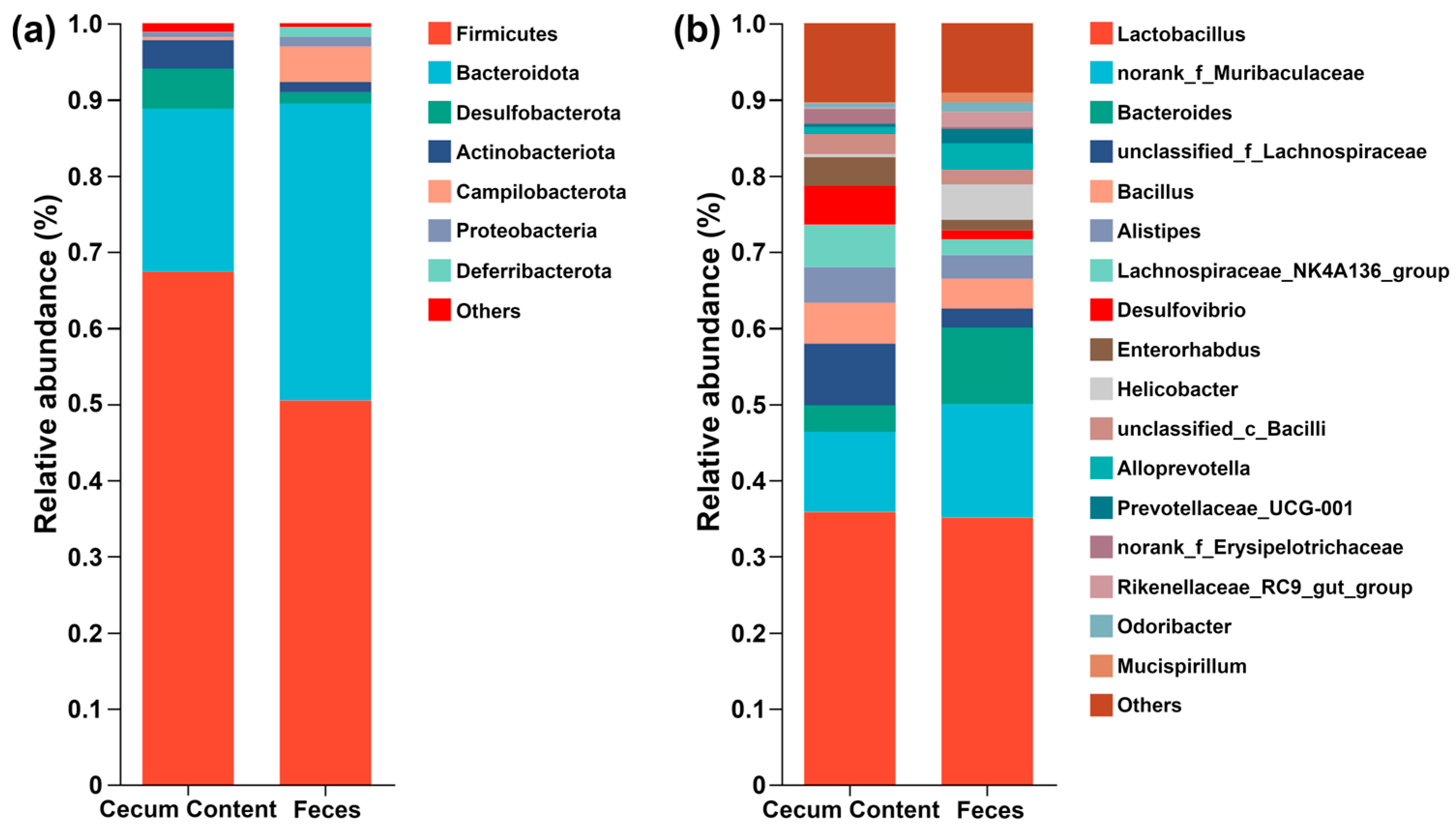
3.3. Microbiome Characterizations of Feces and Cecum Content in Mouse
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, T.; Tan, Y.; Tsui, M.S.; Yi, H.; Fu, X.Q.; Li, T.; Chan, C.L.; Guo, H.; Li, Y.X.; Zhu, P.L.; et al. Metabolomics Reveals the Mechanisms for the Cardiotoxicity of Pinelliae Rhizoma and the Toxicity-Reducing Effect of Processing. Sci. Rep. 2016, 6, 34692. [Google Scholar] [CrossRef]
- Shi, H.; Hu, L.; Chen, S.; Bao, W.; Yang, S.; Zhao, X.; Sun, C. Metabolomics Analysis of Urine from Rats Administered with Long-Term, Low-Dose Acrylamide by Ultra-Performance Liquid Chromatography-Mass Spectrometry. Xenobiotica 2017, 47, 439–449. [Google Scholar] [CrossRef]
- Jin, X.; Yang, H.; Coldea, T.E.; Xu, Y.; Zhao, H. Metabonomic Analysis Reveals Enhanced Growth and Ethanol Production of Brewer’s Yeast by Wheat Gluten Hydrolysates and Potassium Supplementation. LWT 2021, 145, 111387. [Google Scholar] [CrossRef]
- Nelson, A.D.; Camilleri, M.; Chirapongsathorn, S.; Vijayvargiya, P.; Valentin, N.; Shin, A.; Erwin, P.J.; Wang, Z.; Hassan Murad, M. Comparison of Efficacy of Pharmacological Treatments for Chronic Idiopathic Constipation: A Systematic Review and Network Meta-Analysis. Gut 2017, 66, 1611–1622. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, Q.; Zhao, X.; Yang, Z.; Yang, F.; Yang, Y.; Tang, J.; Laghi, L. Metabolomic Analysis of Multiple Biological Specimens (Feces, Serum, and Urine) by 1H-NMR Spectroscopy from Dairy Cows with Clinical Mastitis. Animals 2023, 13, 741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, J.; Jiang, X.; Tang, J.; Zhu, C.; Chen, H.; Laghi, L. Metabolomic Characteristics of Cecum Contents in High-Fat-Diet-Induced Obese Mice Intervened with Different Fibers. Foods 2023, 12, 1403. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Chen, H.; Hughes, R.J.; Moore, R.J. Comparison of Fecal and Cecal Microbiotas Reveals Qualitative Similarities but Quantitative Differences. BMC Microbiol. 2015, 15, 51. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, X.; Cui, B. Modulation of the Fecal Microbiome and Metabolome by Resistant Dextrin Ameliorates Hepatic Steatosis and Mitochondrial Abnormalities in Mice. Food Funct. 2021, 12, 4504–4518. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhang, B.; Li, H.; Huang, G.; Lv, H.; Jiang, K. Differentiation of Obese and Healthy Mice by Analyzing the Carboxylic Acids in the TCA Cycle in Their Feces: Determination of Chelating Carboxylic Acids in Feces. Talanta Open 2023, 7, 100230. [Google Scholar] [CrossRef]
- Chen, Y.; Dinges, M.M.; Green, A.; Cramer, S.E.; Larive, C.K.; Lytle, C. Absorptive Transport of Amino Acids by the Rat Colon. Am. J. Physiol. Gastrointest Liver Physiol. 2020, 318, G189–G202. [Google Scholar] [CrossRef]
- Heinzmann, S.S.; Schmitt-Kopplin, P. Deep Metabotyping of the Murine Gastrointestinal Tract for the Visualization of Digestion and Microbial Metabolism. J. Proteome Res. 2015, 14, 2267–2277. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, L.; Wang, Y.; Tang, H. Age-Related Topographical Metabolic Signatures for the Rat Gastrointestinal Contents. J. Proteome Res. 2012, 11, 1397–1411. [Google Scholar] [CrossRef]
- Dougal, K.; Harris, P.A.; Edwards, A.; Pachebat, J.A.; Blackmore, T.M.; Worgan, H.J.; Newbold, C.J. A Comparison of the Microbiome and the Metabolome of Different Regions of the Equine Hindgut. FEMS Microbiol. Ecol. 2012, 82, 642–652. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Hou, G.; Wang, J.; Zhou, H.; Wang, D. Effects of Vine Tea Extract on Meat Quality, Gut Microbiota and Metabolome of Wenchang Broiler. Animals 2022, 12, 1661. [Google Scholar] [CrossRef]
- Van Hul, M.; Karnik, K.; Canene-Adams, K.; De Souza, M.; Van den Abbeele, P.; Marzorati, M.; Delzenne, N.M.; Everard, A.; Cani, P.D. Comparison of the Effects of Soluble Corn Fiber and Fructooligosaccharides on Metabolism, Inflammation, and Gut Microbiome of High-Fat Diet-Fed Mice. Am. J. Physiol.-Endocrinol. Metab. 2020, 319, E779–E791. [Google Scholar] [CrossRef]
- Zhu, C.; Jin, L.; Luo, B.; Zhou, Q.; Dong, L.; Li, X.; Zhang, H.; Huang, Y.; Li, C.; Zou, L.; et al. Dominant Components of the Giant Panda Seminal Plasma Metabolome, Characterized By1 H-NMR Spectroscopy. Animals 2022, 12, 1536. [Google Scholar] [CrossRef]
- Zhu, C.; Petracci, M.; Li, C.; Fiore, E.; Laghi, L. An Untargeted Metabolomics Investigation of Jiulong Yak (Bos Grunniens) Meat by 1H-NMR. Foods 2020, 9, 481. [Google Scholar] [CrossRef]
- Kneen, M.A.; Annegarn, H.J. Algorithm for Fitting XRF, SEM and PIXE X-Ray Spectra Backgrounds. Nucl. Instrum. Methods Phys. Res. B. 1996, 109–110, 209–213. [Google Scholar] [CrossRef]
- Liland, K.H.; Almøy, T.; Mevik, B.H. Optimal Choice of Baseline Correction for Multivariate Calibration of Spectra. Appl. Spectrosc. 2010, 64, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application In1H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Laghi, L.; Zhu, C.; Campagna, G.; Rossi, G.; Bazzano, M.; Laus, F. Probiotic Supplementation in Trained Trotter Horses: Effect on Blood Clinical Pathology Data and Urine Metabolomic Assessed in Field. J. Appl. Physiol. 2018, 125, 654–660. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Wang, A.; Wang, Q.; Lee, D.J. Denitrifying Sulfide Removal Process on High-Salinity Wastewaters in the Presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011; Volume 1, ISBN 3900051070. [Google Scholar]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Soc. Ser. B. (Methodol.) 1964, 26, 211–243. [Google Scholar] [CrossRef]
- Hubert, M.; Rousseeuw, P.J.; Vanden Branden, K. ROBPCA: A New Approach to Robust Principal Component Analysis. Technometrics 2005, 47, 64–79. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A Composable Bioinformatics Cloud Platform with Real-Time Feedback That Can Generate High-Quality Graphs for Publication. iMeta 2023, 2, e85. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Jaeger, M.; Beumer, J.; Bakker, O.B.; Aguirre-Gamboa, R.; Oosting, M.; Smeekens, S.P.; Moorlag, S.; Mourits, V.P.; Koeken, V.A.C.M.; et al. Integration of Metabolomics, Genomics, and Immune Phenotypes Reveals the Causal Roles of Metabolites in Disease. Genome Biol. 2021, 22, 198. [Google Scholar] [CrossRef]
- Li, Z.; He, H.; Ni, M.; Wang, Z.; Guo, C.; Niu, Y.; Xing, S.; Song, M.; Wang, Y.; Jiang, Y.; et al. Microbiome-Metabolome Analysis of the Immune Microenvironment of the Cecal Contents, Soft Feces, and Hard Feces of Hyplus Rabbits. Oxid. Med. Cell Longev. 2022, 2022, 5725442. [Google Scholar] [CrossRef]
- Zhou, J.; Ding, L.; Chen, W.; Wang, Y. Green Tea Catechin Epigallocatechin Gallate Alleviates High-Fat Diet-Induced Obesity in Mice by Regulating the Gut–Brain Axis. Food Front. 2023, 4, 1413–1425. [Google Scholar] [CrossRef]
- Liu, J.; Hao, W.; He, Z.; Kwek, E.; Zhao, Y.; Zhu, H.; Liang, N.; Ma, K.Y.; Lei, L.; He, W.S.; et al. Beneficial Effects of Tea Water Extracts on the Body Weight and Gut Microbiota in C57BL/6J Mice Fed with a High-Fat Diet. Food Funct. 2019, 10, 2847–2860. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.; Romano, S.; Ansorge, R.; Aboelnour, A.; Le Gall, G.; Savva, G.M.; Pontifex, M.G.; Telatin, A.; Baker, D.; Jones, E.; et al. Fecal Microbiota Transfer between Young and Aged Mice Reverses Hallmarks of the Aging Gut, Eye, and Brain. Microbiome 2022, 10, 68. [Google Scholar] [CrossRef]
- Meng, Z.; Huang, S.; Sun, W.; Yan, S.; Chen, X.; Diao, J.; Zhou, Z.; Zhu, W. A Typical Fungicide and Its Main Metabolite Promote Liver Damage in Mice through Impacting Gut Microbiota and Intestinal Barrier Function. J. Agric. Food Chem. 2021, 69, 13436–13447. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.; Cauquil, L.; Bertide, A.; Ahn, I.; Barilly, C.; Gil, L.; Canlet, C.; Zemb, O.; Pascal, G.; Samson, A.; et al. Gut Microbiota-Derived Metabolite Signature in Suckling and Weaned Piglets. J. Proteome Res. 2021, 20, 982–994. [Google Scholar] [CrossRef]
- Beaumont, M.; Paës, C.; Mussard, E.; Knudsen, C.; Cauquil, L.; Aymard, P.; Barilly, C.; Gabinaud, B.; Zemb, O.; Fourre, S.; et al. Gut Microbiota Derived Metabolites Contribute to Intestinal Barrier Maturation at the Suckling-to-Weaning Transition. Gut Microbes 2020, 11, 1268–1286. [Google Scholar] [CrossRef]
- Martias, C.; Gatien, J.; Roch, L.; Baroukh, N.; Mavel, S.; Lefèvre, A.; Montigny, F.; Schibler, L.; Emond, P.; Nadal-Desbarats, L. Analytical Methodology for a Metabolome Atlas of Goat’s Plasma, Milk and Feces Using1h-Nmr and Uhplc-Hrms. Metabolites 2021, 11, 681. [Google Scholar] [CrossRef]
- Blanco-Pérez, F.; Steigerwald, H.; Schülke, S.; Vieths, S.; Toda, M.; Scheurer, S. The Dietary Fiber Pectin: Health Benefits and Potential for the Treatment of Allergies by Modulation of Gut Microbiota. Curr. Allergy Asthma Rep. 2021, 21, 43. [Google Scholar] [CrossRef]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic Distribution of Three Pathways for Propionate Production within the Human Gut Microbiota. ISME J. 2014, 8, 1323–1335. [Google Scholar] [CrossRef]
- Fang, S.; Chen, X.; Ye, X.; Zhou, L.; Xue, S.; Gan, Q. Effects of Gut Microbiome and Short-Chain Fatty Acids (SCFAs) on Finishing Weight of Meat Rabbits. Front. Microbiol. 2020, 11, 1835. [Google Scholar] [CrossRef]
- Zhao, J.; Bai, Y.; Tao, S.; Zhang, G.; Wang, J.; Liu, L.; Zhang, S. Fiber-Rich Foods Affected Gut Bacterial Community and Short-Chain Fatty Acids Production in Pig Model. J. Funct. Foods 2019, 57, 266–274. [Google Scholar] [CrossRef]
- Kelly, C.J.; Taylor, T.; Colgan Correspondence, S.P. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, Metabolism and Microbial Ecology of Butyrate-Producing Bacteria from the Human Large Intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Miranda, P.M.; De Palma, G.; Serkis, V.; Lu, J.; Louis-Auguste, M.P.; McCarville, J.L.; Verdu, E.F.; Collins, S.M.; Bercik, P. High Salt Diet Exacerbates Colitis in Mice by Decreasing Lactobacillus Levels and Butyrate Production. Microbiome 2018, 6, 57. [Google Scholar] [CrossRef]
- Fei, Y.; Wang, Y.; Pang, Y.; Wang, W.; Zhu, D.; Xie, M.; Lan, S.; Wang, Z. Xylooligosaccharide Modulates Gut Microbiota and Alleviates Colonic Inflammation Caused by High Fat Diet Induced Obesity. Front. Physiol. 2020, 10, 1601. [Google Scholar] [CrossRef]
- Berger, K.; Burleigh, S.; Lindahl, M.; Bhattacharya, A.; Patil, P.; Stålbrand, H.; Nordberg Karlsson, E.; Hållenius, F.; Nyman, M.; Adlercreutz, P. Xylooligosaccharides Increase Bifidobacteria and Lachnospiraceae in Mice on a High-Fat Diet, with a Concomitant Increase in Short-Chain Fatty Acids, Especially Butyric Acid. J. Agric. Food Chem. 2021, 69, 3617–3625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, Y.; Zha, X.; Ma, Y.; Liu, X.; Elsabagh, M.; Wang, H.; Wang, M. Dietary L-Arginine or N-Carbamylglutamate Alleviates Colonic Barrier Injury, Oxidative Stress, and Inflammation by Modulation of Intestinal Microbiota in Intrauterine Growth-Retarded Suckling Lambs. Antioxidants 2022, 11, 2251. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Yuan, M.; Chang, S.; Wang, C. N-Carbamylglutamate Supplementation Regulates Hindgut Microbiota Composition and Short-Chain Fatty Acid Contents in Charollais and Small Tail Han Crossbred Sheep. Front. Vet. Sci. 2023, 10, 1230190. [Google Scholar] [CrossRef]
- Ahn, I.S.; Yoon, J.; Diamante, G.; Cohn, P.; Jang, C.; Yang, X. Disparate Metabolomic Responses to Fructose Consumption between Different Mouse Strains and the Role of Gut Microbiota. Metabolites 2021, 11, 342. [Google Scholar] [CrossRef]
- Neis, E.P.J.G.; Dejong, C.H.C.; Rensen, S.S. The Role of Microbial Amino Acid Metabolism in Host Metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [PubMed]
- Denton, R.M.; Halestrap, A.P. Regulation of Pyruvate Metabolism in Mammalian Tissues. Essays Biochem. 1979, 15, 33–37. [Google Scholar]
- Jeyanathan, J.; Martin, C.; Morgavi, D.P. The Use of Direct-Fed Microbials for Mitigation of Ruminant Methane Emissions: A Review. Animal 2014, 8, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Regassa, A.; Kiarie, E.; Sands, J.S.; Walsh, M.C.; Kim, W.K.; Nyachoti, C.M. Nutritional and Metabolic Implications of Replacing Cornstarch with D-Xylose in Broiler Chickens Fed Corn and Soybean Meal-Based Diet. Poult. Sci. 2017, 96, 388–396. [Google Scholar] [CrossRef]
- Huntley, N.F.; Patience, J.F. Xylose: Absorption, Fermentation, and Post-Absorptive Metabolism in the Pig. J. Anim. Sci. Biotechnol. 2018, 9, 4. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Zhang, J.; Liu, W.; Jing, W.; Lyu, B.; Yu, H.; Zhang, Z. Insoluble Dietary Fiber from Okara Combined with Intermittent Fasting Treatment Synergistically Confers Antiobesity Effects by Regulating Gut Microbiota and Its Metabolites. J. Agric. Food Chem. 2023, 71, 13346–13362. [Google Scholar] [CrossRef]
- Simpson, H.L.; Campbell, B.J. Review Article: Dietary Fibre-Microbiota Interactions. Aliment. Pharmacol. Ther. 2015, 42, 158–179. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Ou, Y.; Guo, Y.; Chen, M.; Lu, X.; Guo, Z.; Zheng, B. Gut Microbiome–Serum Metabolic Profiles: Insight into the Hypoglycemic Effect of Porphyra Haitanensis Glycoprotein on Hyperglycemic Mice. Food Funct. 2023, 14, 7977–7991. [Google Scholar] [CrossRef]
- Zhang, J.; Song, L.; Wang, Y.; Liu, C.; Zhang, L.; Zhu, S.; Liu, S.; Duan, L. Beneficial Effect of Butyrate-Producing Lachnospiraceae on Stress-Induced Visceral Hypersensitivity in Rats. J. Gastroenterol. Hepatol. 2019, 34, 1368–1376. [Google Scholar] [CrossRef] [PubMed]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef] [PubMed]
- Hoa, T.T.; Duc, L.H.; Isticato, R.; Baccigalupi, L.; Ricca, E.; Van, P.H.; Cutting, S.M. Fate and Dissemination of Bacillus Subtilis Spores in a Murine Model. Appl. Environ. Microbiol. 2001, 67, 3819–3823. [Google Scholar] [CrossRef] [PubMed]
- Casula, G.; Cutting, S.M. Bacillus Probiotics: Spore Germination in the Gastrointestinal Tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, X.; Zheng, L.; Duan, L.; Gao, Z.; Hu, D.; Li, J.; Li, X.; Shen, X.; Xiao, H. Ban-Lan-Gen Granule Alleviates Dextran Sulfate Sodium-Induced Chronic Relapsing Colitis in Mice via Regulating Gut Microbiota and Restoring Gut SCFA Derived-GLP-1 Production. J. Inflamm. Res. 2022, 15, 1457–1470. [Google Scholar] [CrossRef]
- Fu, R.; Niu, R.; Li, R.; Yue, B.; Zhang, X.; Cao, Q.; Wang, J.; Sun, Z. Fluoride-Induced Alteration in the Diversity and Composition of Bacterial Microbiota in Mice Colon. Biol. Trace Elem. Res. 2020, 196, 537–544. [Google Scholar] [CrossRef]
- Zhu, T.; Xue, Q.; Liu, Y.; Xu, Y.; Xiong, C.; Lu, J.; Yang, H.; Zhang, Q.; Huang, Y. Analysis of Intestinal Microflora and Metabolites From Mice With DSS-Induced IBD Treated With Schistosoma Soluble Egg Antigen. Front. Cell Dev. Biol. 2021, 9, 777218. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, Y.; Nie, X.; Zhu, C.; Luo, Q.; Laghi, L.; Picone, G. Metabolomic Analysis of Feces vs. Cecum Content in Animals: A Comparative Study Investigated by 1H-NMR. Metabolites 2025, 15, 565. https://doi.org/10.3390/metabo15090565
Li X, Li Y, Nie X, Zhu C, Luo Q, Laghi L, Picone G. Metabolomic Analysis of Feces vs. Cecum Content in Animals: A Comparative Study Investigated by 1H-NMR. Metabolites. 2025; 15(9):565. https://doi.org/10.3390/metabo15090565
Chicago/Turabian StyleLi, Xiexin, Yang Li, Xin Nie, Chenglin Zhu, Qiqi Luo, Luca Laghi, and Gianfranco Picone. 2025. "Metabolomic Analysis of Feces vs. Cecum Content in Animals: A Comparative Study Investigated by 1H-NMR" Metabolites 15, no. 9: 565. https://doi.org/10.3390/metabo15090565
APA StyleLi, X., Li, Y., Nie, X., Zhu, C., Luo, Q., Laghi, L., & Picone, G. (2025). Metabolomic Analysis of Feces vs. Cecum Content in Animals: A Comparative Study Investigated by 1H-NMR. Metabolites, 15(9), 565. https://doi.org/10.3390/metabo15090565





