Retrospective Urine Metabolomics of Clinical Toxicology Samples Reveals Features Associated with Cocaine Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. LC-qToF-MS Conditions/Settings
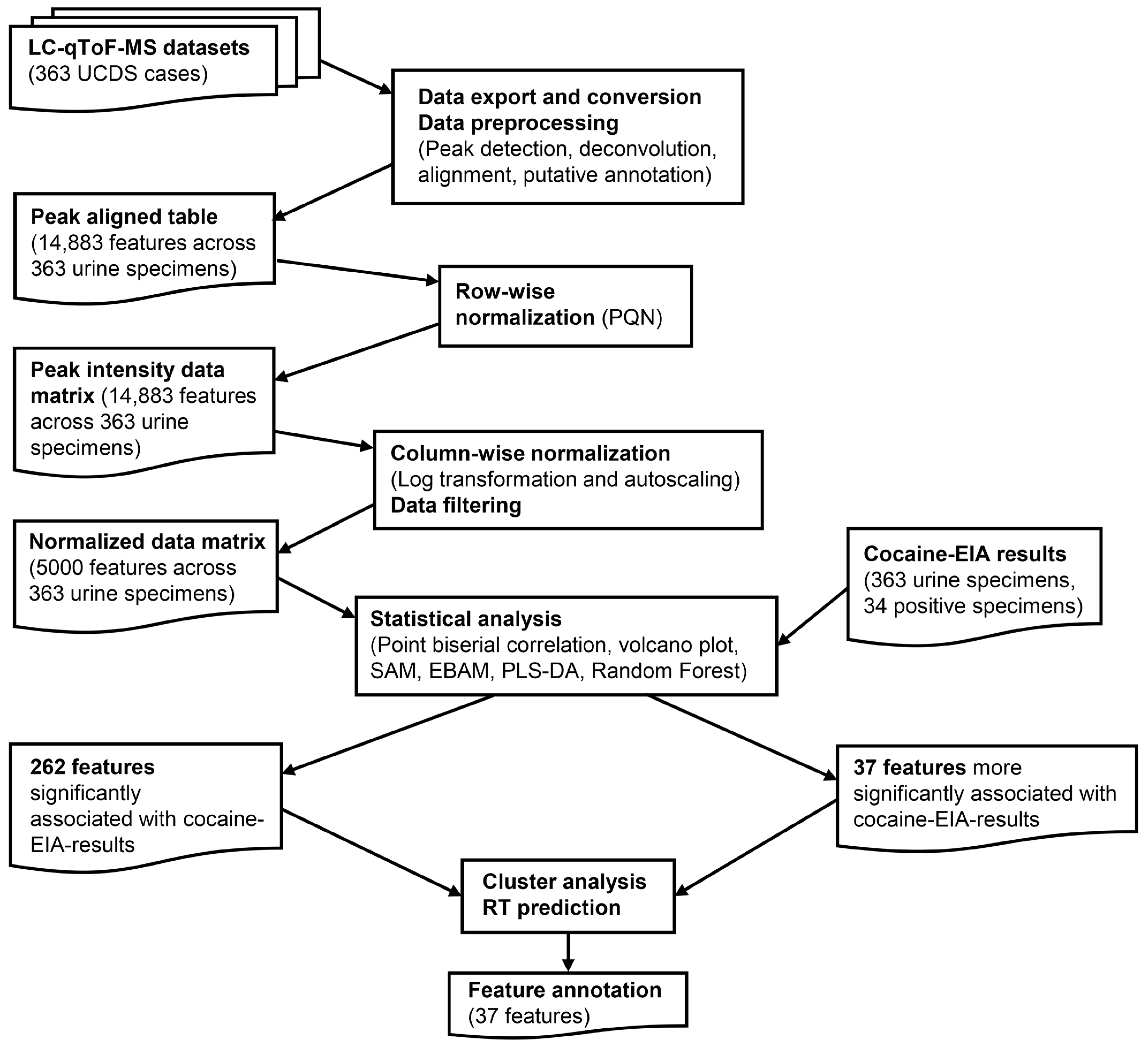
2.3. Data Export and Preprocessing
2.4. Data Analysis
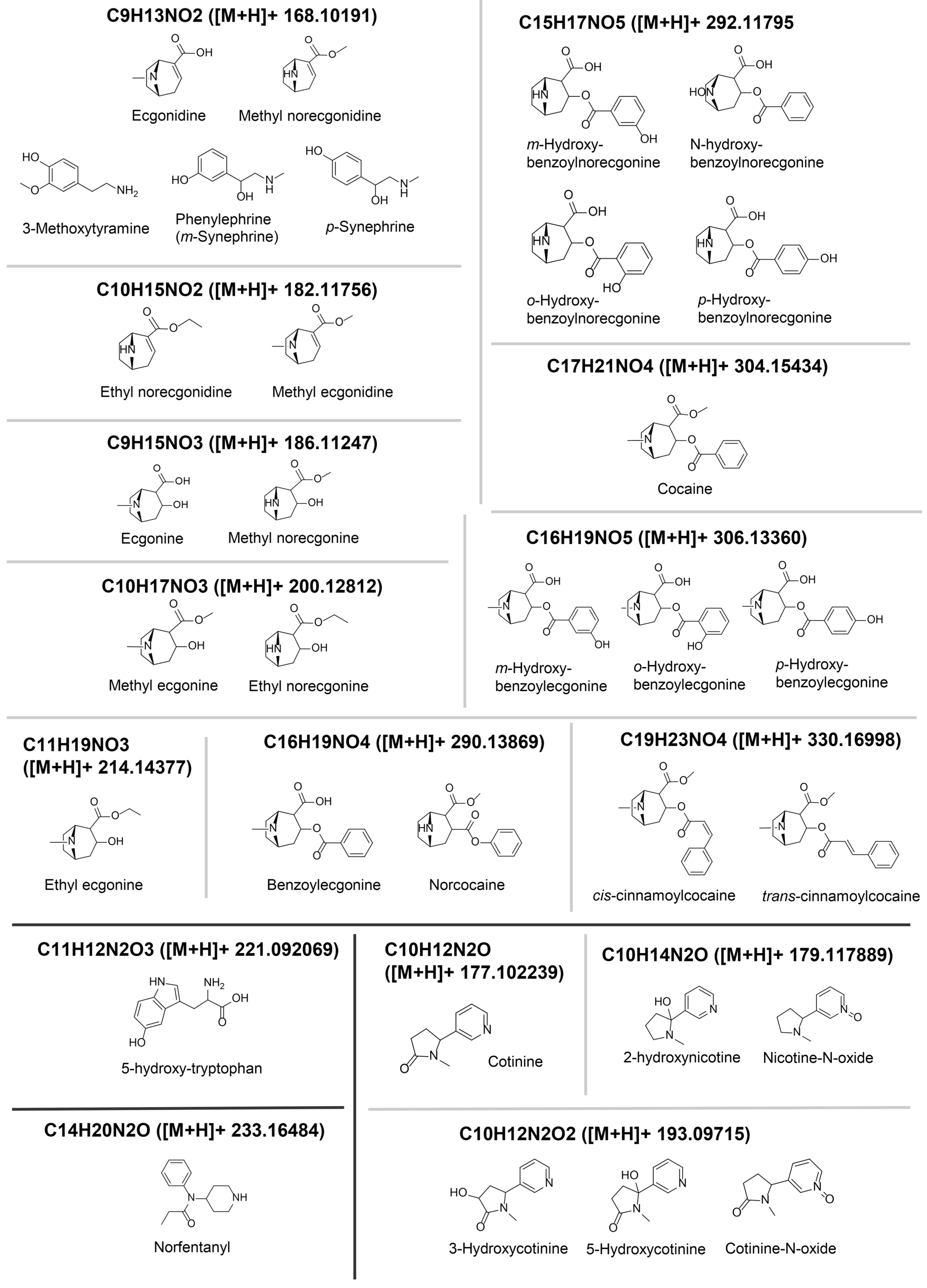
2.5. Feature Annotations
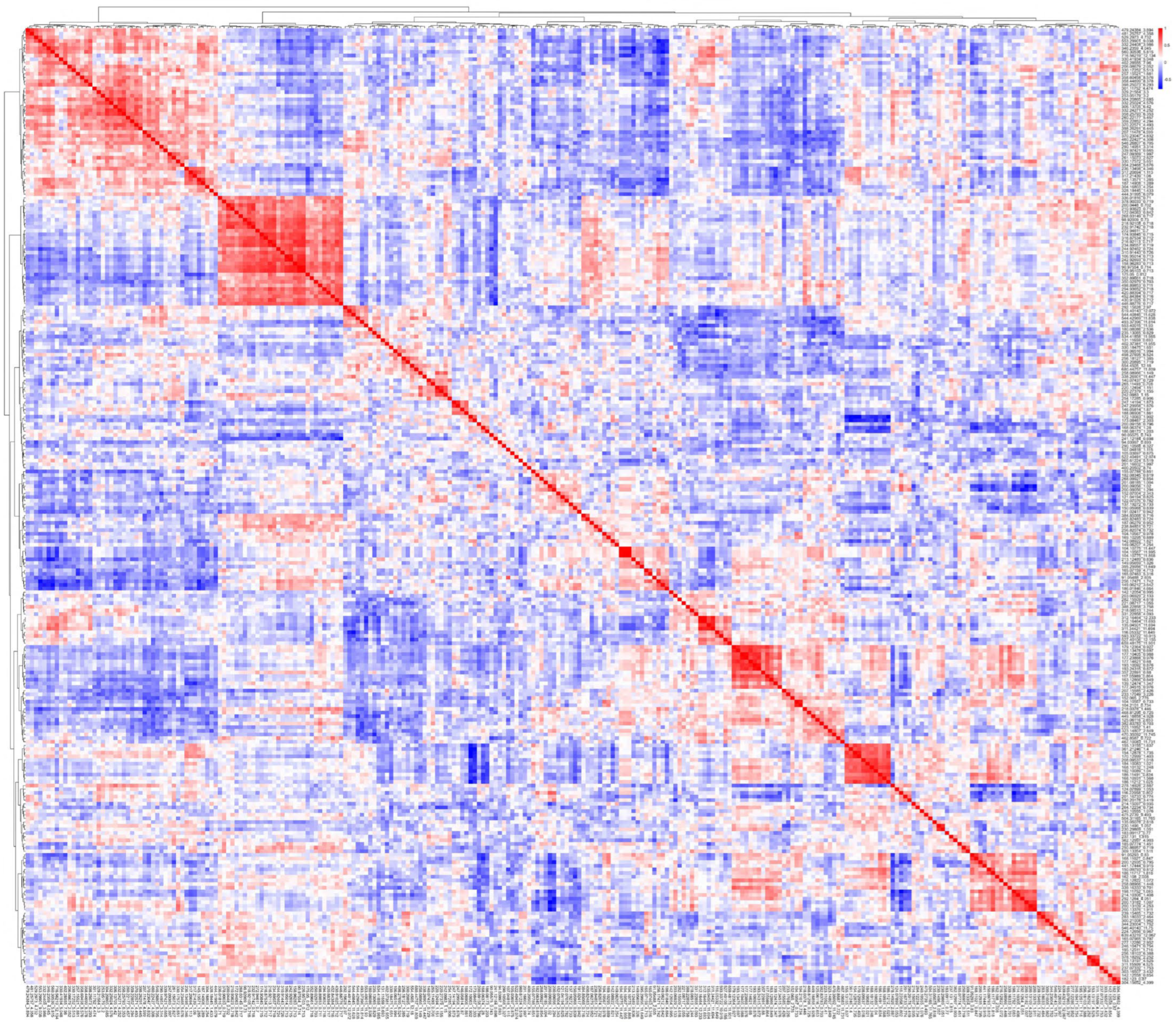
3. Results
3.1. Features 200.12935_0.795, 200.13182_1.007 and 200.13109_4.253
3.2. Features 304.15652_4.399 and 304.16803_4.254
3.3. Feature 182.123_0.87
3.4. Features 186.11491_0.834, 186.11212_1.025, and 186.11717_1.818
3.5. Features 330.17252_5.513 and 330.17572_5.651
3.6. Features 168.11021_0.847, 168.10132_1.248, and 168.10931_1.568
3.7. Feature 214.15097_0.935
3.8. Feature 233.17049_3.226
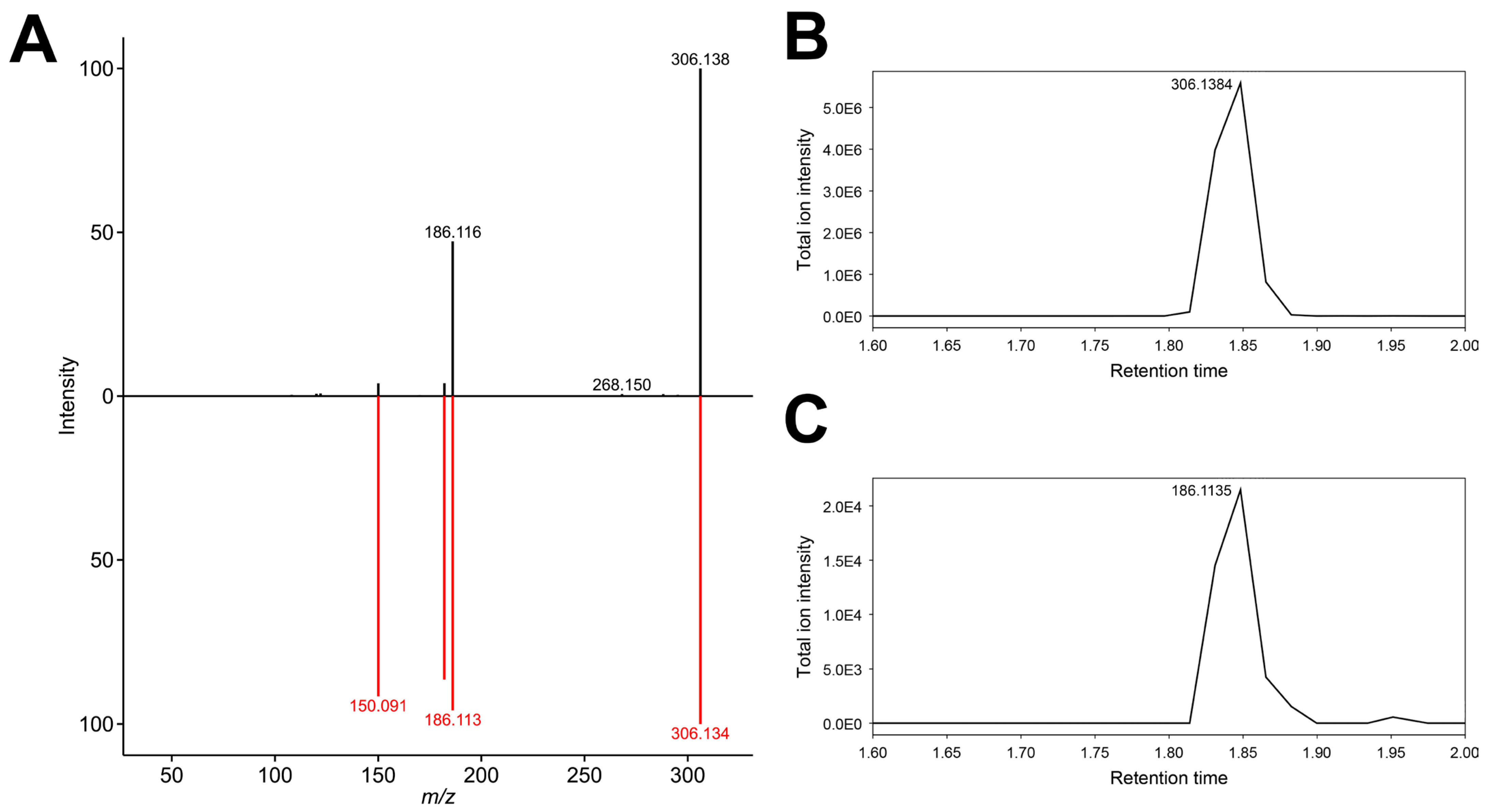
3.9. Feature 292.1264_4.051
3.10. Feature 290.14951_3.314
3.11. Feature 221.08717_1.055
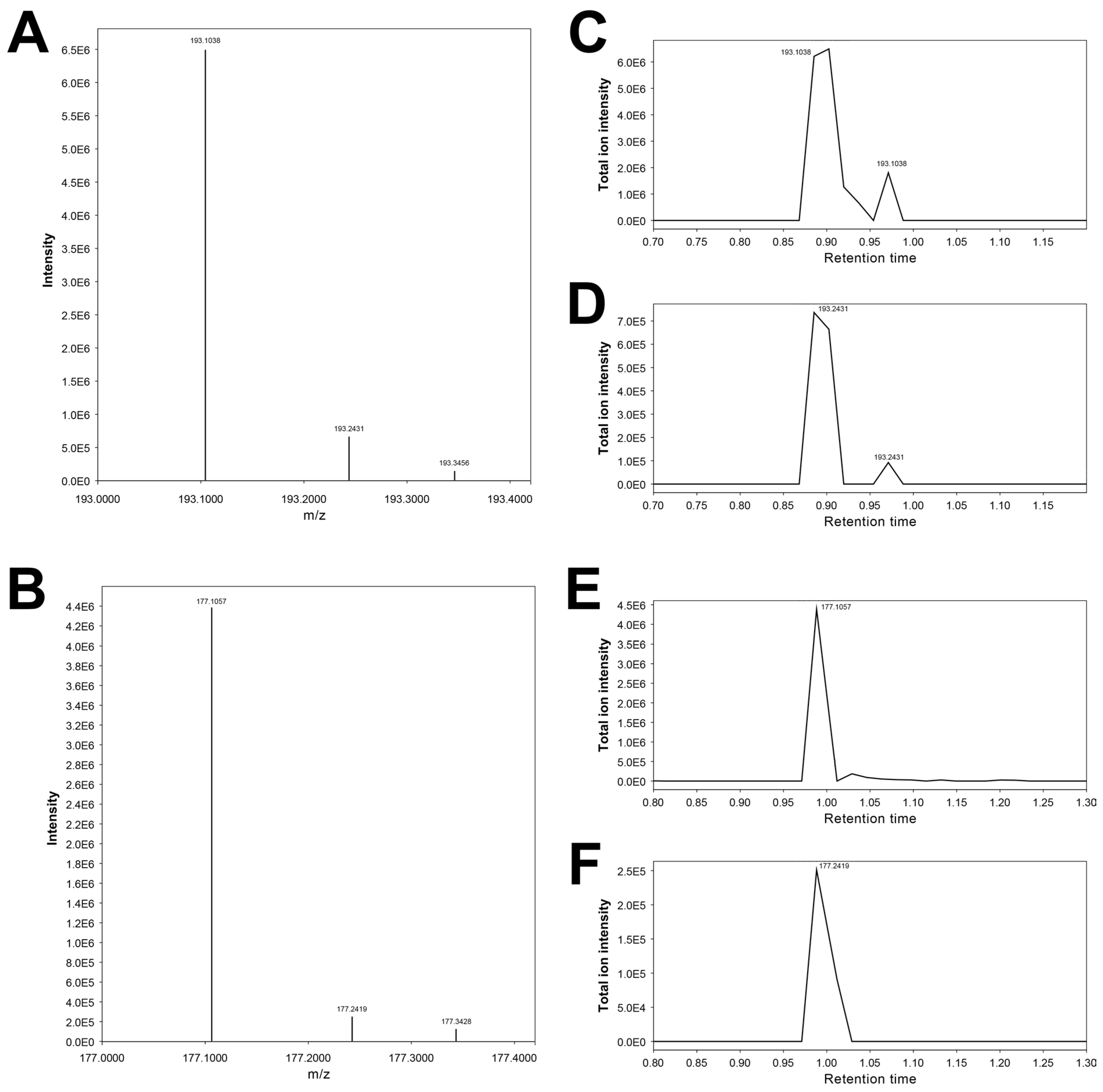
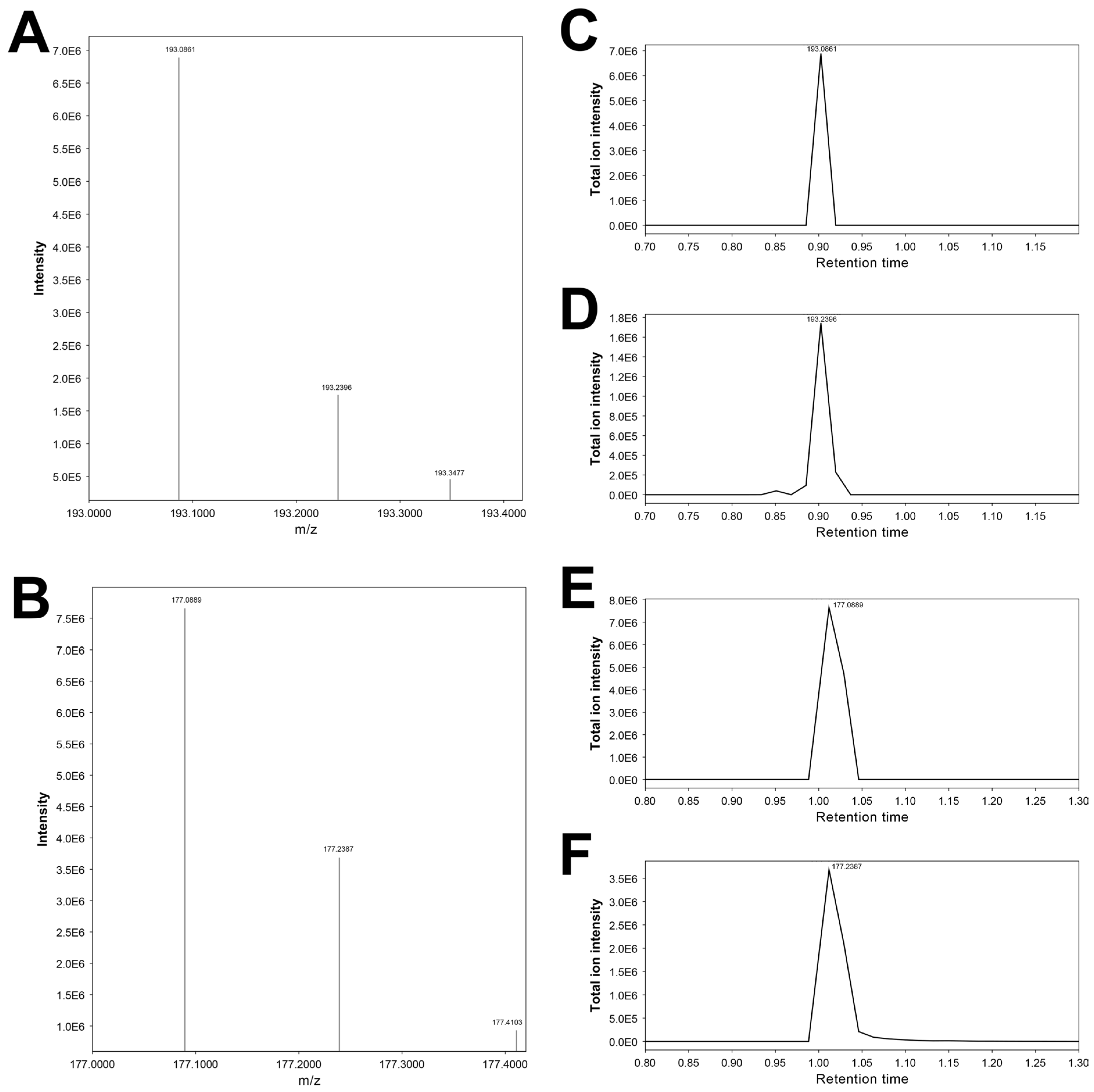
3.12. Features 193.10092_0.876 and 193.24315_0.872
3.13. Feature 179.12364_0.927
3.14. Features 177.10405_0.988 and 177.23868_0.976
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MS | Mass spectrometry |
| LC-qToF-MS | Liquid chromatography–quadrupole time-of-flight mass spectrometry |
| EIA | Enzyme immunoassay |
| LC-HRMS | Liquid chromatography–high-resolution mass spectrometry |
| UCDS | Urine comprehensive drug screening |
| UPMC | University of Pittsburgh Medical Center |
| ESI | Electron spray ionization |
| PQN | Probabilistic quotient normalization |
| SAM | Significance analysis of microarrays and metabolites |
| EBAM | Empirical Bayesian analysis of microarrays and metabolites |
| FDR | False discovery rate |
| PLS-DA | Partial least squares discriminant analysis |
| RT | Retention time |
| xgb | XGBoost |
| rf | Random Forest |
| brnn | Bayesian Neural Network |
References
- Goldstein, R.A.; DesLauriers, C.; Burda, A.M. Cocaine: History, social implications, and toxicity—A review. Dis. Mon. 2009, 55, 6–38. [Google Scholar] [CrossRef] [PubMed]
- Substance Abuse and Mental Health Services Administration (SAMHSA). Key Substance Use and Mental Health Indicators in the United States: Results from the 2023 National Survey on Drug Use and Health; Substance Abuse and Mental Health Services Administration: Rockville, MD, USA, 2024. [Google Scholar]
- Drug Enforcement Administration. National Drug Threat Assessment 2024; Drug Enforcement Administration: Arlington, VA, USA, 2024. [Google Scholar]
- European Monitoring Centre for Drugs and Drug Addiction (EMCDDA). European Drug Report 2024: Trends and Developments; European Monitoring Centre for Drugs and Drug Addiction: Luxembourg, 2024. [Google Scholar]
- Tamama, K.; Lynch, M.J. Newly Emerging Drugs of Abuse. In Substance Use Disorders; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2020; Volume 258, pp. 463–502. [Google Scholar] [CrossRef]
- Tamama, K. Advances in drugs of abuse testing. Clin. Chim. Acta 2021, 514, 40–47. [Google Scholar] [CrossRef]
- Caspani, G.; Sebok, V.; Sultana, N.; Swann, J.R.; Bailey, A. Metabolic phenotyping of opioid and psychostimulant addiction: A novel approach for biomarker discovery and biochemical understanding of the disorder. Br. J. Pharmacol. 2022, 179, 1578–1606. [Google Scholar] [CrossRef]
- Dinis-Oliveira, R.J. Metabolomics of cocaine: Implications in toxicity. Toxicol. Mech. Methods 2015, 25, 494–500. [Google Scholar] [PubMed]
- Dinis-Oliveira, R.J. Metabolomics of drugs of abuse: A more realistic view of the toxicological complexity. Bioanalysis 2014, 6, 3155–3159. [Google Scholar] [CrossRef]
- Doke, M.; McLaughlin, J.P.; Baniasadi, H.; Samikkannu, T. Sleep Disorder and Cocaine Abuse Impact Purine and Pyrimidine Nucleotide Metabolic Signatures. Metabolites 2022, 12, 869. [Google Scholar] [CrossRef]
- Zaitsu, K.; Hayashi, Y.; Kusano, M.; Tsuchihashi, H.; Ishii, A. Application of metabolomics to toxicology of drugs of abuse: A mini review of metabolomics approach to acute and chronic toxicity studies. Drug Metab. Pharmacokinet. 2016, 31, 21–26. [Google Scholar] [CrossRef]
- Zaitsu, K.; Miyawaki, I.; Bando, K.; Horie, H.; Shima, N.; Katagi, M.; Tatsuno, M.; Bamba, T.; Sato, T.; Ishii, A.; et al. Metabolic profiling of urine and blood plasma in rat models of drug addiction on the basis of morphine, methamphetamine, and cocaine-induced conditioned place preference. Anal. Bioanal. Chem. 2014, 406, 1339–1354. [Google Scholar] [CrossRef]
- Bouhifd, M.; Hartung, T.; Hogberg, H.T.; Kleensang, A.; Zhao, L. Review: Toxicometabolomics. J. Appl. Toxicol. 2013, 33, 1365–1383. [Google Scholar] [CrossRef] [PubMed]
- Tamama, K. Dilute and shoot approach for toxicology testing. Front. Chem. 2023, 11, 1278313. [Google Scholar] [CrossRef]
- Guo, J.; Shen, S.; Xing, S.; Yu, H.; Huan, T. ISFrag: De Novo Recognition of In-Source Fragments for Liquid Chromatography-Mass Spectrometry Data. Anal. Chem. 2021, 93, 10243–10250. [Google Scholar] [CrossRef]
- Nash, W.J.; Ngere, J.B.; Najdekr, L.; Dunn, W.B. Characterization of Electrospray Ionization Complexity in Untargeted Metabolomic Studies. Anal. Chem. 2024, 96, 10935–10942. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Lu, W.; Rabinowitz, J.D. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal. Chem. 2015, 87, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Benton, H.P.; Siuzdak, G. Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem. 2018, 90, 480–489. [Google Scholar] [CrossRef]
- Tsugawa, H. Advances in computational metabolomics and databases deepen the understanding of metabolisms. Curr. Opin. Biotechnol. 2018, 54, 10–17. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R.; Storey, J.D.; Tusher, V. Empirical Bayes Analysis of a Microarray Experiment. J. Am. Stat. Assoc. 2001, 96, 1151–1160. [Google Scholar] [CrossRef]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef]
- Katajamaa, M.; Miettinen, J.; Oresic, M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 2006, 22, 634–636. [Google Scholar] [CrossRef]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffmann, N.; Lu, M.; et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef]
- Bonini, P.; Kind, T.; Tsugawa, H.; Barupal, D.K.; Fiehn, O. Retip: Retention Time Prediction for Compound Annotation in Untargeted Metabolomics. Anal. Chem. 2020, 92, 7515–7522. [Google Scholar] [CrossRef]
- Chen, J.Y.; Sutaria, S.R.; Xie, Z.; Kulkarni, M.; Keith, R.J.; Bhatnagar, A.; Sears, C.G.; Lorkiewicz, P.; Srivastava, S. Simultaneous profiling of mercapturic acids, glucuronic acids, and sulfates in human urine. Environ. Int. 2025, 199, 109516. [Google Scholar] [CrossRef]
- Muñoz-Muñoz, A.C.; Pekol, T.; Schubring, D.; Johnson, C.; Andrade, L. Identification of Novel Opioid Interferences using High-Resolution Mass Spectrometry. J. Anal. Toxicol. 2018, 42, 6–16. [Google Scholar] [CrossRef]
- Yao, D.; Shi, X.; Wang, L.; Gosnell, B.A.; Chen, C. Characterization of differential cocaine metabolism in mouse and rat through metabolomics-guided metabolite profiling. Drug Metab. Dispos. 2013, 41, 79–88. [Google Scholar] [CrossRef]
- Chen, L.; Pan, H.; Zhai, G.; Luo, Q.; Li, Y.; Fang, C.; Shi, F. Widespread occurrence of in-source fragmentation in the analysis of natural compounds by liquid chromatography-electrospray ionization mass spectrometry. Rapid Commun. Mass. Spectrom. 2023, 37, e9519. [Google Scholar] [CrossRef]
- Buré, C.; Le Falher, G.; Lange, C.; Delmas, A. Fragmentation study of peptide acetals and aldehydes using in-source collision-induced dissociation. J. Mass. Spectrom. 2004, 39, 817–823. [Google Scholar] [CrossRef]
- Gabelica, V.; De Pauw, E. Internal energy and fragmentation of ions produced in electrospray sources. Mass. Spectrom. Rev. 2005, 24, 566–587. [Google Scholar] [CrossRef]
- Moore, J.M.; Casale, J.F. In-depth chromatographic analyses of illicit cocaine and its precursor, coca leaves. J. Chromatogr. A 1994, 674, 165–205. [Google Scholar] [CrossRef]
- Moore, J.M.; Moore, J.F.; Fodor, G.; Jones, A.B. Detection and Characterization of Cocaine and Related Tropane Alkaloids in Coca Leaf, Cocaine, and Biological Specimens. Forensic Sci. Rev. 1995, 7, 77–101. [Google Scholar] [PubMed]
- Choe, A.J.; Ellison, R.; Ramaswamy, S.R.; Schult, R.F.; Gerona, R.; Nacca, N. Profound Hyperthermia Associated With Fentanyl and Cocaine Use With Suspected Synephrine Adulteration. J. Emerg. Med. 2023, 64, 259–262. [Google Scholar] [CrossRef]
- Wyatt, R.J.; Angrist, B.; Karoum, F. Urinary Dopamine Metabolites During Cocaine Abstinence. Am. J. Addict. 1995, 4, 133–140. [Google Scholar] [CrossRef]
- Ciccarone, D. The rise of illicit fentanyls, stimulants and the fourth wave of the opioid overdose crisis. Curr. Opin. Psychiatry 2021, 34, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Park, J.N.; Rashidi, E.; Foti, K.; Zoorob, M.; Sherman, S.; Alexander, G.C. Fentanyl and fentanyl analogs in the illicit stimulant supply: Results from U.S. drug seizure data, 2011–2016. Drug Alcohol. Depend. 2021, 218, 108416. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huan, T. Mechanistic Understanding of the Discrepancies between Common Peak Picking Algorithms in Liquid Chromatography-Mass Spectrometry-Based Metabolomics. Anal. Chem. 2023, 95, 5894–5902. [Google Scholar] [CrossRef] [PubMed]
- Pirttila, K.; Balgoma, D.; Rainer, J.; Pettersson, C.; Hedeland, M.; Brunius, C. Comprehensive Peak Characterization (CPC) in Untargeted LC-MS Analysis. Metabolites 2022, 12I, 137. [Google Scholar] [CrossRef]
- Davidson, M.; Rashidi, N.; Hossain, M.K.; Raza, A.; Nurgali, K.; Apostolopoulos, V. Tryptophan and Substance Abuse: Mechanisms and Impact. Int. J. Mol. Sci. 2023, 24, 2737. [Google Scholar] [CrossRef]
- Patkar, A.A.; Rozen, S.; Mannelli, P.; Matson, W.; Pae, C.U.; Krishnan, K.R.; Kaddurah-Daouk, R. Alterations in tryptophan and purine metabolism in cocaine addiction: A metabolomic study. Psychopharmacology 2009, 206, 479–489. [Google Scholar] [CrossRef]
- Araos, P.; Vidal, R.; O’Shea, E.; Pedraz, M.; Garcia-Marchena, N.; Serrano, A.; Suarez, J.; Castilla-Ortega, E.; Ruiz, J.J.; Campos-Cloute, R.; et al. Serotonin is the main tryptophan metabolite associated with psychiatric comorbidity in abstinent cocaine-addicted patients. Sci. Rep. 2019, 9, 16842. [Google Scholar] [CrossRef]
- Neurath, G.B.; Dunger, M.; Krenz, O.; Orth, D.; Pein, F.G. Trans-3′-hydroxycotinine—A main metabolite in smokers. Klin. Wochenschr. 1988, 66 (Suppl. 11), 2–4. [Google Scholar] [CrossRef]
- Apantaku-Olajide, T.; Darker, C.D.; Smyth, B.P. Onset of cocaine use: Associated alcohol intoxication and psychosocial characteristics among adolescents in substance abuse treatment. J. Addict. Med. 2013, 7, 183–188. [Google Scholar] [CrossRef]
- Stinson, F.S.; Grant, B.F.; Dawson, D.A.; Ruan, W.J.; Huang, B.; Saha, T. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol. Depend. 2005, 80, 105–116. [Google Scholar] [CrossRef]
- Pennings, E.J.; Leccese, A.P.; Wolff, F.A. Effects of concurrent use of alcohol and cocaine. Addiction 2002, 97, 773–783. [Google Scholar] [CrossRef]
- McCance, E.F.; Price, L.H.; Kosten, T.R.; Jatlow, P.I. Cocaethylene: Pharmacology, physiology and behavioral effects in humans. J. Pharmacol. Exp. Ther. 1995, 274, 215–223. [Google Scholar] [CrossRef]
- Benowitz, N.L. Pharmacology of nicotine: Addiction, smoking-induced disease, and therapeutics. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 57–71. [Google Scholar] [CrossRef]
- Estadt, A.T.; White, B.N.; Ricks, J.M.; Lancaster, K.E.; Hepler, S.; Miller, W.C.; Kline, D. The impact of fentanyl on state- and county-level psychostimulant and cocaine overdose death rates by race in Ohio from 2010 to 2020: A time series and spatiotemporal analysis. Harm Reduct. J. 2024, 21, 13. [Google Scholar] [CrossRef]
- Jones, C.M.; Bekheet, F.; Park, J.N.; Alexander, G.C. The Evolving Overdose Epidemic: Synthetic Opioids and Rising Stimulant-Related Harms. Epidemiol. Rev. 2020, 42, 154–166. [Google Scholar] [CrossRef]
- Park, J.N.; Schneider, K.E.; Fowler, D.; Sherman, S.G.; Mojtabai, R.; Nestadt, P.S. Polysubstance Overdose Deaths in the Fentanyl Era: A Latent Class Analysis. J. Addict. Med. 2022, 16, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Spencer, M.R.; Minino, A.M.; Warner, M. Drug Overdose Deaths in the United States, 2001–2021. NCHS Data Brief. 2022, 457, 1–8. [Google Scholar]
- Liu, X.; Singer, M.E. Intentional use of both opioids and cocaine in the United States. Prev. Med. Rep. 2023, 33, 102227. [Google Scholar] [CrossRef] [PubMed]
- Nolan, M.L.; Shamasunder, S.; Colon-Berezin, C.; Kunins, H.V.; Paone, D. Increased Presence of Fentanyl in Cocaine-Involved Fatal Overdoses: Implications for Prevention. J. Urban. Health 2019, 96, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Rakusanova, S.; Cajka, T. Tips and tricks for LC–MS-based metabolomics and lipidomics analysis. Trends Anal. Chem. 2024, 180, 117940. [Google Scholar] [CrossRef]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef]
- Arbouche, N.; de Lestrange, A.; Raul, J.S.; Kintz, P. Mariani wine: What’s really in it? Analysis of the most popular tonic drink of the 19th century after 100 years of storage. J. Pharm. Biomed. Anal. 2024, 238, 115804. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Maher, N.; Torres, R.; Cotto, C.; Hastings, B.; Dasgupta, M.; Hyman, R.; Huebert, N.; Caldwell, G.W. Isobaric metabolite interferences and the requirement for close examination of raw data in addition to stringent chromatographic separations in liquid chromatography/tandem mass spectrometric analysis of drugs in biological matrix. Rapid Commun. Mass. Spectrom. 2008, 22, 2021–2028. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Song, Q.; Liu, W.; Li, J.; Tu, P. High-confidence structural identification of metabolites relying on tandem mass spectrometry through isomeric identification: A tutorial. Trends Anal. Chem. 2023, 160, 116982. [Google Scholar] [CrossRef]
- Mahieu, N.G.; Patti, G.J. Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites. Anal. Chem. 2017, 89, 10397–10406. [Google Scholar] [CrossRef]
- Cech, N.B.; Enke, C.G. Practical implications of some recent studies in electrospray ionization fundamentals. Mass. Spectrom. Rev. 2001, 20, 362–387. [Google Scholar] [CrossRef]
- Holcapek, M.; Kolárová, L.; Nobilis, M. High-performance liquid chromatography-tandem mass spectrometry in the identification and determination of phase I and phase II drug metabolites. Anal. Bioanal. Chem. 2008, 391, 59–78. [Google Scholar] [CrossRef]
- Pitt, J.J.; Eggington, M.; Kahler, S.G. Comprehensive screening of urine samples for inborn errors of metabolism by electrospray tandem mass spectrometry. Clin. Chem. 2002, 48, 1970–1980. [Google Scholar] [CrossRef]
| Male | Female | |||
|---|---|---|---|---|
| Age (Years) | Outpatient | Non-Outpatient | Outpatient | Non-Outpatient |
| 0–9 | 0 | 38 (2) | 0 | 33 |
| 10–19 | 1 | 23 | 3 | 29 |
| 20–29 | 7 | 10 (1) | 10 | 7 (3) |
| 30–39 | 12 (4) | 11 (2) | 19 (3) | 10 (1) |
| 40–49 | 23 (4) | 8 (2) | 21 (2) | 6 (3) |
| 50–59 | 20 | 13 (1) | 15 | 6 (1) |
| 60–69 | 12 (1) | 3 (2) | 6 | 5 |
| 70- | 5 (2) | 4 | 2 | 2 |
| Cluster A (Table 2A) | Cluster B (Table 2B) | ||||||||||||||
| A1 | A2 | A3 | B1 | ||||||||||||
| A1-1 | A2-1 | A3-1 | B1-1 | ||||||||||||
| 90.05575_0.743 | 145.13571_1.285 | 154.12878_1.735 | 90.97554_0.714 | ||||||||||||
| 241.12184_0.698 | 187.14908_1.289 | 155.13155_1.637 | 106.95014_0.713 | ||||||||||||
| A1-2 | 206.08679_2.052 | 170.12999_1.403 | 158.96283_0.713 | ||||||||||||
| 142.12558_0.854 | 304.16803_4.254 | 361.21246_1.4 | 174.93845_0.715 | ||||||||||||
| 201.16602_1.997 | 328.18445_1.433 | A3-2 | 175.05_0.912 | ||||||||||||
| 400.20502_8.74 | 330.17252_5.513 | 236.13498_4.346 | 216.92113_0.717 | ||||||||||||
| A1-3 | 332.24408_3.986 | 317.20694_1.113 | 226.95103_0.713 | ||||||||||||
| 330.41934_5.048 | 346.2359_4.045 | 317.21439_1.06 | 234.89557_0.719 | ||||||||||||
| 402.28555_7.96 | 560.30536_5.815 | 242.92693_0.715 | |||||||||||||
| 716.56219_12.134 | A2-2 | 244.92462_0.724 | |||||||||||||
| 960.61224_5.519 | 207.11478_4.599 | 272.94611_0.7 | |||||||||||||
| 260.22177_5.457 | 294.93652_0.718 | ||||||||||||||
| 290.14951_3.314 | 310.91443_0.726 | ||||||||||||||
| 304.20865_2.685 | 316.87534_0.712 | ||||||||||||||
| 306.13705_6.42 | 352.89661_0.718 | ||||||||||||||
| 332.24271_4.252 | 420.88394_0.717 | ||||||||||||||
| 339.97421_0.665 | 430.91025_0.712 | ||||||||||||||
| 358.25793_6.353 | 446.88776_0.717 | ||||||||||||||
| 359.22852_4.294 | 452.84384_0.716 | ||||||||||||||
| 370.22571_4.493 | B1-2 | ||||||||||||||
| 370.23047_4.832 | 98.92009_0.73 | ||||||||||||||
| 398.26291_6.445 | 172.04063_0.842 | ||||||||||||||
| 444.31995_6.079 | 200.0448_0.702 | ||||||||||||||
| 460.22427_4.308 | 210.93623_0.718 | ||||||||||||||
| 546.26807_6.705 | 218.92108_0.718 | ||||||||||||||
| A2-3 | 232.91742_0.718 | ||||||||||||||
| 257.15521_1.881 | 268.03149_0.717 | ||||||||||||||
| 358.44699_8.378 | 336.01816_0.71 | ||||||||||||||
| 358.60458_8.576 | 378.90033_0.719 | ||||||||||||||
| 398.25073_6.293 | |||||||||||||||
| A2-4 | |||||||||||||||
| 479.24384_3.894 | |||||||||||||||
| 481.25757_4.358 | |||||||||||||||
| 529.2973_4.732 | |||||||||||||||
| 553.29901_9.038 | |||||||||||||||
| Cluster C (Table 2C) | Cluster D (Table 2D) | ||||||||||||||
| C1 | C2 | D1 | D2 | D3 | |||||||||||
| C1-1 | C2-1 | D1-1 | D2-1 | D3-1 | |||||||||||
| 91.05293_0.83 | 135.05078_2.818 | 91.05488_2.605 | 94.03997_0.693 | 116.22058_0.802 | |||||||||||
| 168.11021_0.847 | 230.1496_1.057 | 149.06212_3.642 | 105.03697_0.875 | 200.09056_1.02 | |||||||||||
| 200.12935_0.795 | 230.29869_1.051 | 186.01846_4.664 | 290.10568_6.327 | 200.09056_1.294 | |||||||||||
| 441.17444_0.915 | 168.10132_1.248 | D1-2 | 522.40491_12.074 | D3-2 | |||||||||||
| C1-2 | 168.10931_1.568 | 121.04194_0.825 | D2-2 | 168.06374_1.28 | |||||||||||
| 117.05989_0.864 | 184.10083_1.021 | 122.07076_0.782 | 107.04818_1.115 | 186.08173_1.203 | |||||||||||
| 139.12474_1.347 | 186.11212_1.025 | 137.19272_0.733 | 546.40143_11.75 | 200.09158_0.796 | |||||||||||
| 163.12866_0.849 | 186.11491_0.834 | 150.05968_0.839 | D2-3 | ||||||||||||
| 177.34515_0.976 | 192.10089_1.04 | 152.07004_2.343 | 131.11659_0.693 | ||||||||||||
| 198.11752_1.083 | 208.09537_1.018 | 191.02417_0.942 | 402.37381_11.955 | ||||||||||||
| 214.10306_1.408 | C2-2 | D1-3 | 534.41858_11.965 | ||||||||||||
| 216.12822_1.072 | 177.10405_0.988 | 140.07437_0.729 | D2-4 | ||||||||||||
| C1-3 | 177.14621_0.68 | 185.07774_1.451 | 172.10063_1.902 | ||||||||||||
| 150.09793_0.812 | 177.23868_0.976 | 201.08185_1.004 | 173.08467_2.303 | ||||||||||||
| 162.108_2.008 | 179.12364_0.927 | 265.11493_0.706 | D2-5 | ||||||||||||
| 186.11717_1.818 | 193.10092_0.876 | D1-4 | 187.06279_0.952 | ||||||||||||
| 258.08966_1.448 | 193.13478_0.697 | 155.07788_0.851 | 238.84851_0.721 | ||||||||||||
| 339.16333_0.701 | 193.24315_0.872 | 182.08345_0.819 | 256.82074_0.732 | ||||||||||||
| 357.22891_0.88 | 268.09927_0.854 | 384.85068_0.716 | |||||||||||||
| C2-3 | 400.82483_0.724 | ||||||||||||||
| 214.15097_0.935 | |||||||||||||||
| 240.10555_1.076 | |||||||||||||||
| 264.12234_0.734 | |||||||||||||||
| 475.2739_9.493 | |||||||||||||||
| 504.31165_11.765 | |||||||||||||||
| C2-4 | |||||||||||||||
| 218.08513_1.244 | |||||||||||||||
| 221.08717_1.055 | |||||||||||||||
| 282.15509_4.618 | |||||||||||||||
| 309.13354_1.511 | |||||||||||||||
| 331.22858_4.093 | |||||||||||||||
| 388.22858_3.758 | |||||||||||||||
| Cluster E (Table 2E) | Cluster F (Table 2F) | Cluster G (Table 2G) | |||||||||||||
| E1 | E2 | F1 | F2 | G1 | |||||||||||
| E1-1 | E2-1 | F1-1 | F2-1 | G1-1 | |||||||||||
| 104.10567_0.733 | 104.10567_11.995 | 106.06516_1.994 | 153.13197_4.525 | 116.05332_11.649 | |||||||||||
| 104.2101_0.734 | 104.10775_11.868 | 256.18127_1.385 | 311.15509_4.525 | 135.04501_11.694 | |||||||||||
| 152.065_2.776 | 104.10775_11.447 | 258.08966_1.149 | 378.18292_2.252 | 311.34421_11.694 | |||||||||||
| 207.15585_2.426 | 149.05659_1.326 | 300.20895_1.719 | F2-2 | 312.16464_12.233 | |||||||||||
| 233.17049_3.226 | 213.12489_0.836 | 330.18475_1.651 | 165.07965_0.797 | 312.16464_11.693 | |||||||||||
| E1-2 | 395.29956_11.649 | 338.26901_11.447 | 224.12656_0.967 | 527.40106_12.155 | |||||||||||
| 125.06116_2.653 | E2-2 | 498.27695_6.524 | 277.12286_2.952 | 593.33722_10.015 | |||||||||||
| 223.11052_1.41 | 104.10567_0.978 | 554.4505_12.18 | 292.15628_2.97 | 659.48175_11.901 | |||||||||||
| 323.14807_2.849 | 142.08922_1.821 | 680.44757_11.809 | 519.40143_12.072 | G1-2 | |||||||||||
| 382.83783_0.703 | 149.06207_4.254 | F1-2 | 544.40845_11.628 | 142.12054_0.995 | |||||||||||
| 449.16858_4.928 | 169.10295_0.889 | 124.07899_1.053 | 639.43219_12.067 | 203.06929_2.133 | |||||||||||
| 470.35092_11.745 | E2-3 | 253.05179_3.2 | F2-3 | 237.07332_1.753 | |||||||||||
| E1-3 | 165.07159_4.713 | 278.14926_2.887 | 180.08086_2.536 | 303.16507_3.432 | |||||||||||
| 462.8587_0.722 | 165.07463_5.316 | 301.11752_4.474 | 235.13065_0.829 | G1-3 | |||||||||||
| 467.10083_11.731 | 218.0378_1.446 | 329.21564_3.3 | F2-4 | 146.05814_1.87 | |||||||||||
| 250.86957_0.719 | 332.25024_4.576 | 195.12511_1.716 | 188.06906_1.861 | ||||||||||||
| 256.17471_1.742 | 183.09117_0.77 | 246.10471_0.754 | 247.14194_1.873 | ||||||||||||
| 350.02979_0.703 | 201.16733_0.774 | 256.18103_6.388 | 247.29958_1.876 | ||||||||||||
| 468.81296_0.725 | 237.131_1.815 | F2-5 | G1-4 | ||||||||||||
| 498.89853_0.711 | 290.20178_3.419 | 239.15465_1.732 | 182.123_0.87 | ||||||||||||
| 362.12997_4.903 | 283.18033_2.464 | 200.13109_4.253 | |||||||||||||
| F1-3 | 300.21008_1.962 | 200.13182_1.007 | |||||||||||||
| 220.12494_1.151 | 344.23004_1.752 | 200.13376_1.911 | |||||||||||||
| 220.27379_1.155 | 292.1264_4.051 | ||||||||||||||
| 242.0983_1.15 | 304.15652_4.399 | ||||||||||||||
| 247.09369_1.997 | |||||||||||||||
| 254.17285_0.906 | |||||||||||||||
| 261.15073_2.827 | |||||||||||||||
| 330.17572_5.651 | |||||||||||||||
| 354.23468_5.676 | |||||||||||||||
| F1-4 | |||||||||||||||
| 493.37396_11.814 | |||||||||||||||
| 544.42969_11.838 | |||||||||||||||
| 553.40015_11.93 | |||||||||||||||
| Feature | Correlation | Volcano | PLS | EBAM | SAM | RF | Annotation |
|---|---|---|---|---|---|---|---|
| 200.12935_0.795 | X | X | X | Methyl ecgonine * | |||
| 304.15652_4.399 | X | X | X | X | X | X | Cocaine * |
| 200.13182_1.007 | X | X | X | Ethyl norecgonine (putative) | |||
| 182.123_0.87 | X | X | X | X | X | X | ISF (-H2O) of methyl ecgonine |
| 186.11491_0.834 | X | X | X | X | X | X | Ecgonine * |
| 330.17252_5.513 | X | X | X | Cinnamoylcocaine (putative) | |||
| 168.11021_0.847 | X | X | X | X | X | X | Ecgonidine *# |
| 198.11752_1.083 | X | X | Unknown | ||||
| 277.12286_2.952 | X | X | X | Unknown | |||
| 330.17572_5.651 | X | X | X | Cinnamoylcocaine (putative) | |||
| 162.108_2.008 | X | X | X | Unknown | |||
| 186.11212_1.025 | X | X | X | X | X | X | Methyl norecgonine (putative) |
| 214.15097_0.935 | X | X | Ethyl ecgonine * | ||||
| 233.17049_3.226 | X | X | Norfentanyl * | ||||
| 186.11717_1.818 | X | X | X | ISF of Hydroxy-benzoylecgonine (putative) | |||
| 200.13109_4.253 | X | X | X | Unknown $ | |||
| 91.05293_0.83 | X | X | Unknown | ||||
| 292.1264_4.051 | X | X | N-Hydroxy-norbenzoylecgonine (putative) | ||||
| 168.10931_1.568 | X | X | Methyl norecgonidine (putative) | ||||
| 290.14951_3.314 | X | X | Benzoylecgonine ^ | ||||
| 216.12822_1.072 | X | X | Unknown | ||||
| 168.10132_1.248 | X | X | X | X | Methyl norecgonidine and ISF of methyl norecgonine # (putative) | ||
| 304.16803_4.254 | X | X | X | X | Cocaine * | ||
| 155.13155_1.637 | X | X | Unknown | ||||
| 560.30536_5.815 | X | X | Unknown | ||||
| 221.08717_1.055 | X | X | X | 5-Hydroxy-L-tryptophan * | |||
| 193.13478_0.697 | X | X | Unknown | ||||
| 206.08679_2.052 | X | X | Unknown | ||||
| 292.15628_2.97 | X | X | Unknown | ||||
| 104.2101_0.734 | X | X | Unknown | ||||
| 188.06906_1.861 | X | X | Unknown | ||||
| 193.10092_0.876 | X | X | X | 3-Hydroxycotinine # | |||
| 177.23868_0.976 | X | X | X | Cotinine artifact (putative) | |||
| 177.14621_0.68 | X | X | X | Unknown | |||
| 177.10405_0.988 | X | X | Cotinine * | ||||
| 179.12364_0.927 | X | X | Nicotine-N-oxide * | ||||
| 193.24315_0.872 | X | X | 3-Hydroxycotinine artifact (putative) |
| RT (Experimentally Determined) | xgb | Rf | brnn | XLogP | ALogP | nHBDon | nBase | |
|---|---|---|---|---|---|---|---|---|
| Cocaine-related metabolites | ||||||||
| Benzoylecgonine | 2.95 | 3.1 | 3.36 | 2.56 | 3.18 | 3.02 | 3.17 | 3.2 |
| cis-Cinnamoylcocaine | 6.94 | 7.02 | 4.77 | 6.38 | 5.99 | 5.93 | 5.99 | |
| trans-Cinnamoylcocaine | 6.94 | 7.02 | 4.77 | 6.38 | 5.99 | 5.93 | 5.99 | |
| Cocaethylene | 5.55 | 5.79 | 5.58 | 5.39 | 5.56 | 5.63 | 5.16 | 5.84 |
| Cocaine | 4.46 | 4.26 | 4.32 | 4.11 | 4.37 | 4.61 | 4.52 | 4.53 |
| Ecgonidine | 0.84 | 1.24 | 1.73 | 1.05 | 1.18 | 0.91 | 1.69 | 0.91 |
| Ecgonine | 0.80 | 0.51 | 1.3 | 0.34 | 0.29 | 0.87 | 1.27 | 1.52 |
| Ethyl ecgonine | 0.93 | 1.95 | 3.05 | 1.85 | 2 | 1.9 | 1.79 | 1.9 |
| Ethyl norecgonidine | 3.08 | 3.46 | 3.52 | 2.16 | 2.03 | 1.66 | 2.03 | |
| Ethyl norecgonine | 1.84 | 2.93 | 2.18 | 1.83 | 1.58 | 1.56 | 1.58 | |
| Methyl ecgonidine | 1.10 | 1.39 | 1.69 | 1.62 | 1.32 | 1.16 | 1.5 | 1.71 |
| Methyl ecgonine | 0.80 | 0.88 | 1.19 | 1 | 0.7 | 0.74 | 0.91 | 0.83 |
| Methyl norecgonidine | 2.76 | 3.17 | 2.4 | 1.55 | 1.59 | 1.05 | 1.59 | |
| Methyl norecgonine | 0.87 | 1.32 | 1.25 | 1 | 1.17 | 1.1 | 1.17 | |
| m-Hydroxy-benzoylecgonine | 2.68 | 3.67 | 2.17 | 3.89 | 4.14 | 3.77 | 4.14 | |
| o-Hydroxy-benzoylecgonine | 2.94 | 4 | 2.12 | 2.97 | 3.28 | 2.63 | 3.28 | |
| p-Hydroxy-benzoylecgonine | 2.74 | 3.67 | 1.96 | 3.86 | 4 | 3.77 | 4 | |
| m-Hydroxy-benzoylnorecgonine | 2.66 | 3.11 | 2.6 | 2.45 | 2.44 | 2.63 | 3.23 | |
| N-Hydroxy-benzoylnorecgonine | 3.22 | 3.15 | 4.12 | 2.84 | 2.74 | 3.08 | 3.18 | |
| o-Hydroxy-benzoylnorecgonine | 2.81 | 3.61 | 2.82 | 2.84 | 2.79 | 3.29 | 3.84 | |
| p-Hydroxy-benzoylnorecgonine | 2.66 | 3.1 | 2.41 | 2.46 | 2.38 | 2.63 | 3.23 | |
| Norcocaine | 4.54 | 4.32 | 4.05 | 4.47 | 4.1 | 4.32 | 4.19 | 4.31 |
| Nicotine-related metabolites | ||||||||
| Cotinine N-oxide | 1.18 | 1.41 | 0.76 | 1.59 | 1.65 | 1.62 | 1.29 | |
| Cotinine | 1.06 | 1.12 | 1.23 | 0.65 | 1.18 | 1.2 | 1.38 | 1.39 |
| 2-Hydroxynicotine | 0.7 | 1.4 | -0.18 | 0.55 | 0.8 | 1.14 | 0.79 | |
| 3-Hydroxycotinine | 0.90 | 0.78 | 1.36 | 0.21 | 0.86 | 0.96 | 1.23 | 1.01 |
| 5-Hydroxycotinine | 1.11 | 1.37 | -0.03 | 1.39 | 1.07 | 1.78 | 2.05 | |
| Nicotine N-oxide | 0.92 | 1.07 | 1.4 | 0.12 | 1.2 | 1.29 | 1.28 | 1.42 |
| Other xenobiotics | ||||||||
| p-Synephrine | 0.83 | 0.92 | 1.5 | 0.55 | 0.89 | 0.81 | 1.35 | 1.21 |
| Phenylephrine (m-synephrine) | 0.83 | 0.92 | 1.47 | 0.62 | 0.81 | 0.83 | 1.31 | 1.06 |
| Norfentanyl | 3.25 | 3.24 | 3.49 | 2.78 | 3.32 | 3.26 | 3.42 | 3.16 |
| Endogenous metabolite | ||||||||
| 3-Methoxytyramine | 0.98 | 1.5 | 1.53 | 1.94 | 1.19 | 1.18 | 1.17 | 1.43 |
| 5-Hydroxytryptophan | 0.90 | 1.19 | 1.66 | 0.25 | 0.78 | 0.71 | 1.6 | 1.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanderschelden, R.K.; Kundu, R.; Morrow, D.; Patel, S.; Tamama, K. Retrospective Urine Metabolomics of Clinical Toxicology Samples Reveals Features Associated with Cocaine Exposure. Metabolites 2025, 15, 563. https://doi.org/10.3390/metabo15090563
Vanderschelden RK, Kundu R, Morrow D, Patel S, Tamama K. Retrospective Urine Metabolomics of Clinical Toxicology Samples Reveals Features Associated with Cocaine Exposure. Metabolites. 2025; 15(9):563. https://doi.org/10.3390/metabo15090563
Chicago/Turabian StyleVanderschelden, Rachel K., Reya Kundu, Delaney Morrow, Simmi Patel, and Kenichi Tamama. 2025. "Retrospective Urine Metabolomics of Clinical Toxicology Samples Reveals Features Associated with Cocaine Exposure" Metabolites 15, no. 9: 563. https://doi.org/10.3390/metabo15090563
APA StyleVanderschelden, R. K., Kundu, R., Morrow, D., Patel, S., & Tamama, K. (2025). Retrospective Urine Metabolomics of Clinical Toxicology Samples Reveals Features Associated with Cocaine Exposure. Metabolites, 15(9), 563. https://doi.org/10.3390/metabo15090563







