Secondary Metabolites Predict Diazotrophic Cyanobacteria: A Model-Based Cheminformatic Approach
Abstract
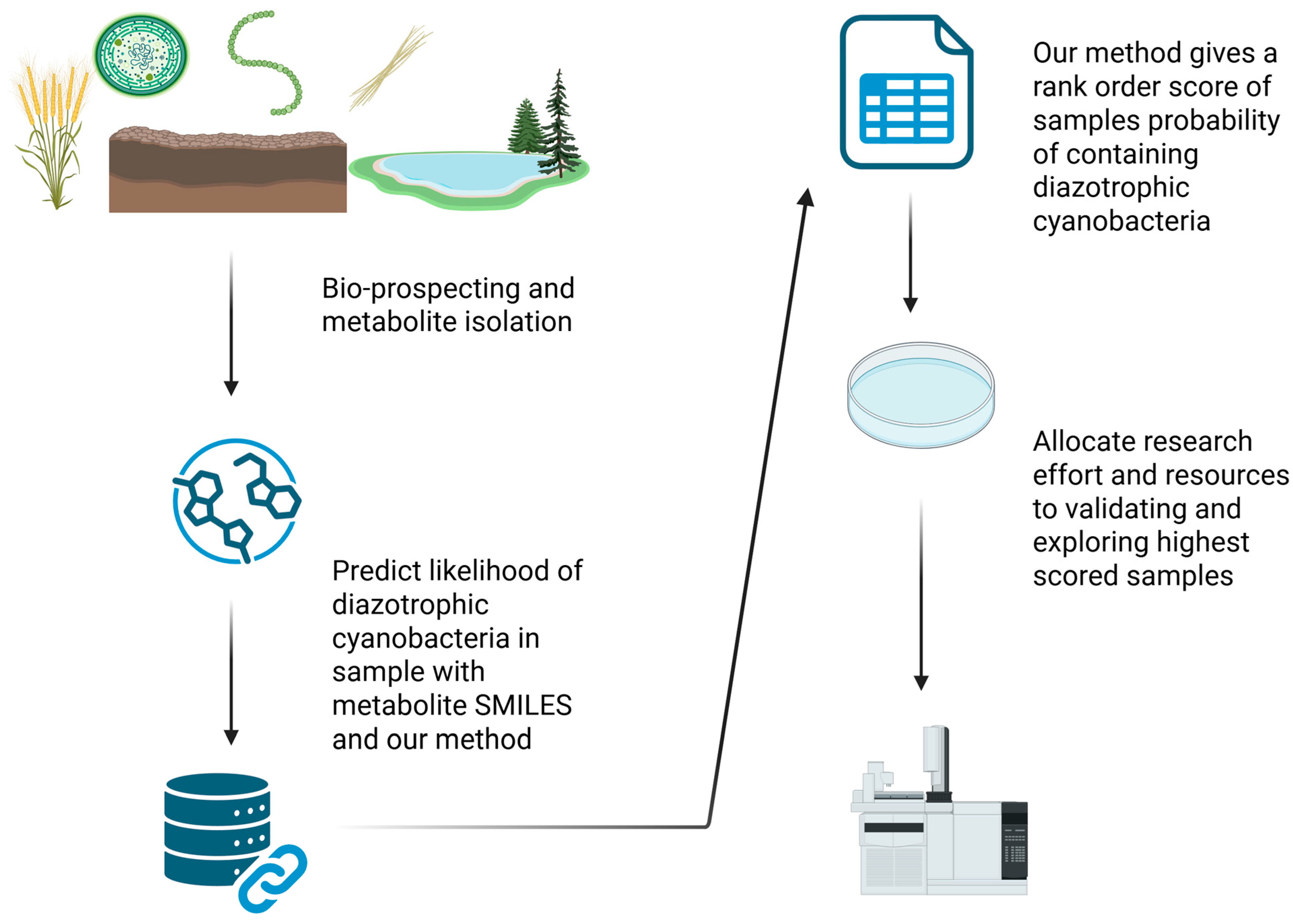
1. Introduction
2. Materials and Methods
3. Results
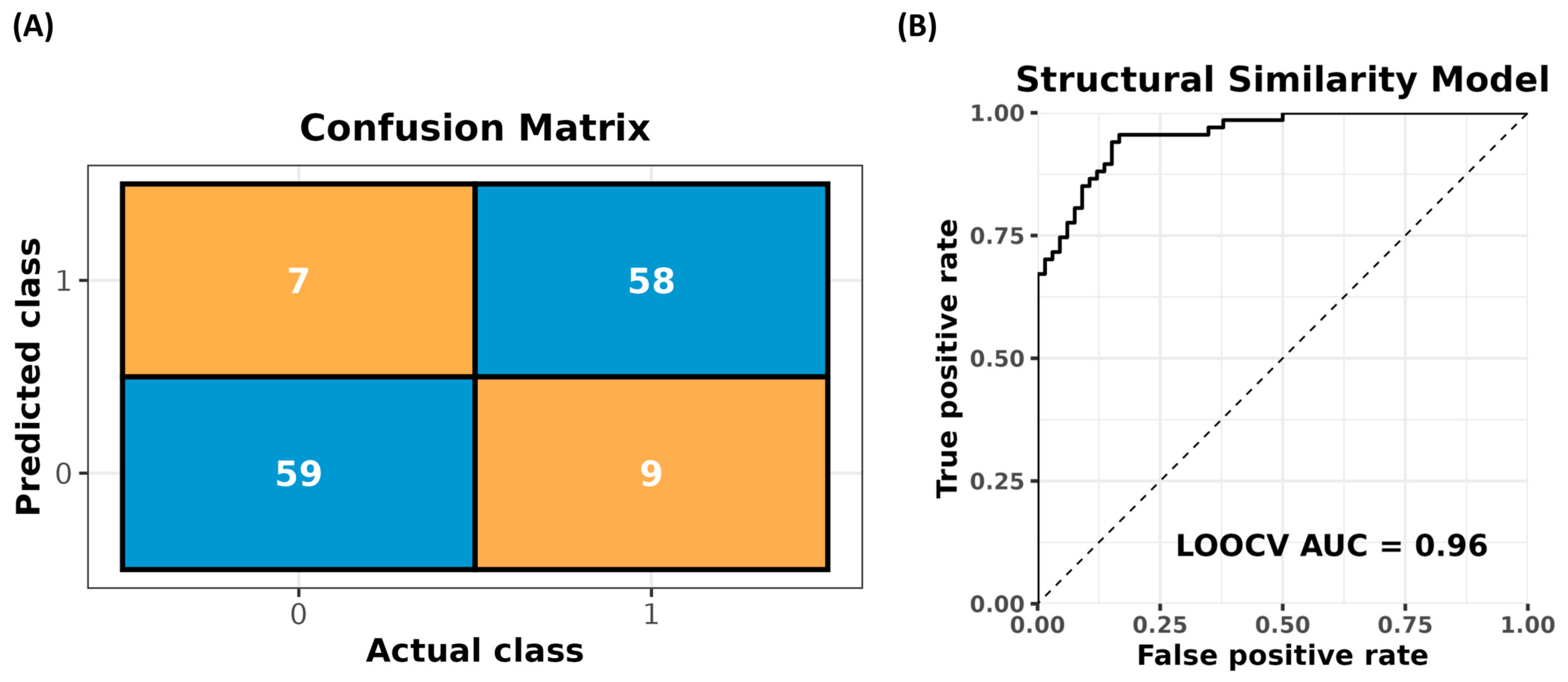
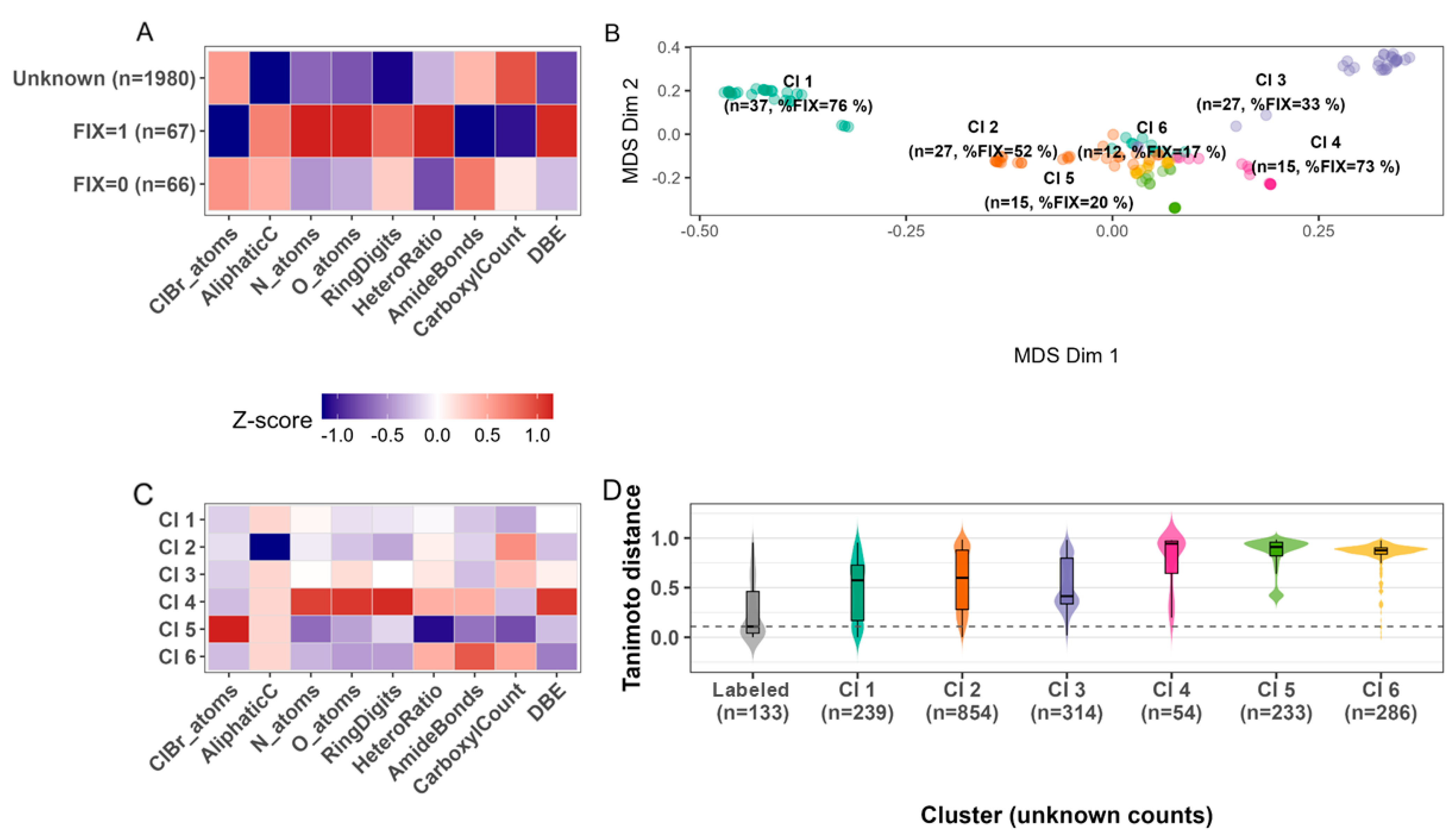
3.1. Design and Performance
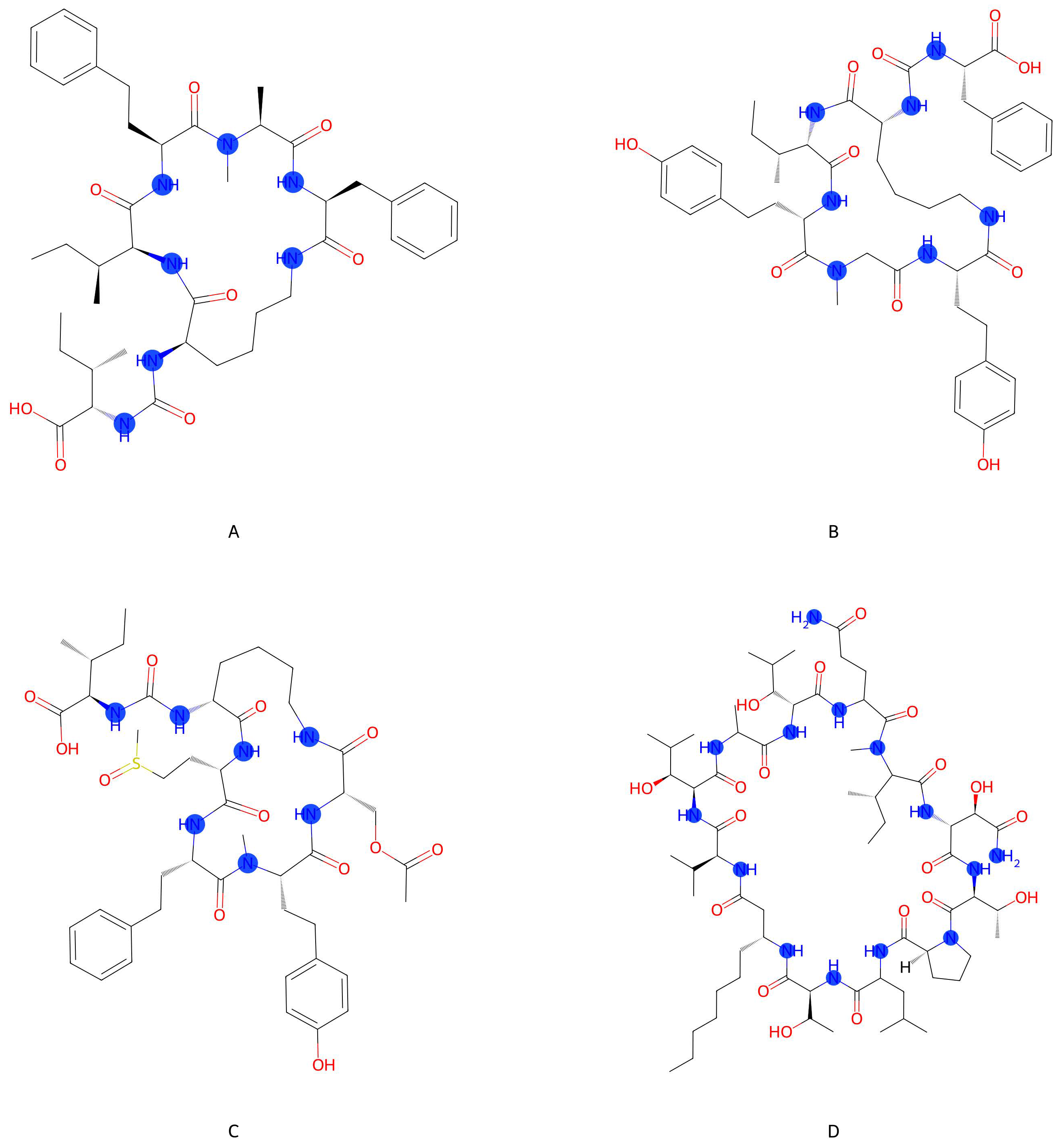
3.1.1. Metabolite Diversity Represented
3.1.2. The Similarity-Based Diazotrophic Prediction Algorithm
3.2. Top Unknown Strains and Metabolites for Exploration
| Strain | Max Probability | Metabolite Count |
|---|---|---|
| IL-208-2-2 | 0.999 | 1 |
| CCNP1411 | 0.999 | 6 |
| CENA352 | 0.994 | 2 |
| TAU NZ-3-1 | 0.986 | 3 |
| GSV 224 | 0.984 | 39 |
| AV1 | 0.978 | 17 |
| KAC 11 | 0.975 | 4 |
| NIES-81 | 0.975 | 2 |
| PCC7310 | 0.970 | 1 |
| ITEP-24 | 0.966 | 2 |
| Compound Name | Predicted Probability |
|---|---|
| Schizopeptin 791 | 0.999 |
| Anabaenopeptin NZ857 | 0.994 |
| Nodulapeptin B | 0.978 |
| Nodulapeptin 863 | 0.977 |
| Nodulapeptin 915a | 0.976 |
| Nodulapeptin 855b | 0.976 |
| Laxaphycin B | 0.974 |
| Laxaphycin B2 | 0.974 |
| Trichormamide C | 0.965 |
| Laxaphycin B3 | 0.963 |
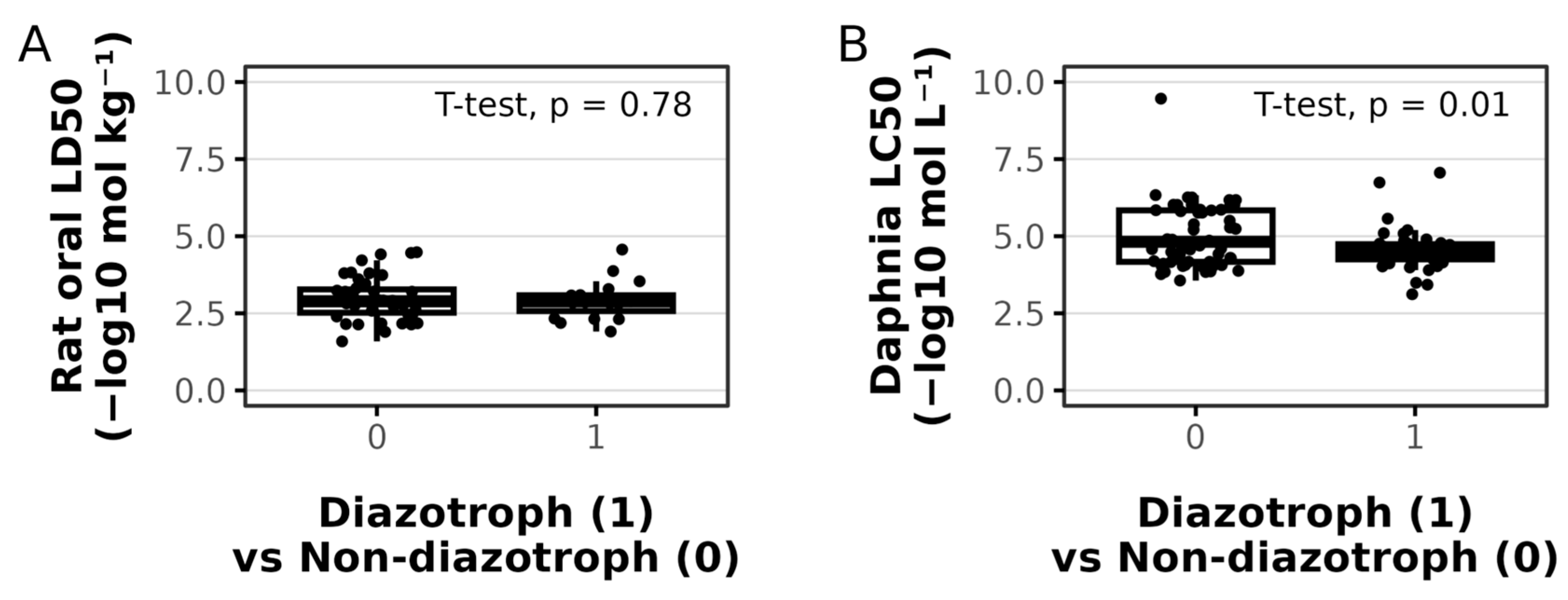
3.3. Toxicity Is Not Positively Associated with Diazotrophic Secondary Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bahl, J.; Lau, M.C.; Smith, G.J.; Vijaykrishna, D.; Cary, S.C.; Lacap, D.C.; Lee, C.K.; Papke, R.T.; Warren-Rhodes, K.A.; Wong, F.K.; et al. Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat. Commun. 2011, 2, 163. [Google Scholar] [CrossRef] [PubMed]
- Fiore, C.L.; Jarett, J.K.; Olson, N.D.; Lesser, M.P. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 2010, 18, 455–463. [Google Scholar] [CrossRef]
- Allen, R.S.; Tilbrook, K.; Warden, A.C.; Campbell, P.C.; Rolland, V.; Singh, S.P.; Wood, C.C. Expression of 16 Nitrogenase Proteins within the Plant Mitochondrial Matrix. Front. Plant Sci. 2017, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liberton, M.; Yu, J.; Pakrasi, H.B.; Bhattacharyya-Pakrasi, M. Engineering Nitrogen Fixation Activity in an Oxygenic Phototroph. mBio 2018, 9, e01029-18. [Google Scholar] [CrossRef]
- Tsujimoto, R.; Kotani, H.; Yokomizo, K.; Yamakawa, H.; Nonaka, A.; Fujita, Y. Functional expression of an oxygen-labile nitrogenase in an oxygenic photosynthetic organism. Sci. Rep. 2018, 8, 7380. [Google Scholar] [CrossRef]
- Young, J.; Gu, L.; Gibbons, W.; Zhou, R. Harnessing Solar-Powered Oxic N 2-fixing Cyanobacteria for the BioNitrogen Economy. In Cyanobacteria Biotechnology; Wiley: Hoboken, NJ, USA, 2021; pp. 407–439. [Google Scholar]
- Young, J.; Gu, L.; Hildreth, M.; Zhou, R. Unicellular Cyanobacteria Exhibit Light-Driven, Oxygen-Tolerant, Constitutive Nitrogenase Activity Under Continuous Illumination. bioRxiv 2019. preprint. [Google Scholar] [CrossRef]
- Nunnery, J.K.; Mevers, E.; Gerwick, W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotechnol. 2010, 21, 787–793. [Google Scholar] [CrossRef]
- Singh, R.K.; Tiwari, S.P.; Rai, A.K.; Mohapatra, T.M. Cyanobacteria: An emerging source for drug discovery. J. Antibiot. 2011, 64, 401–412. [Google Scholar] [CrossRef]
- Gademann, K.; Portmann, C. Secondary Metabolites from Cyanobacteria: Complex Structures and Powerful Bioactivities. Curr. Org. Chem. 2008, 12, 326–341. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Sinha, R.P. Biotechnological and industrial significance of cyanobacterial secondary metabolites. Biotechnol. Adv. 2009, 27, 521–539. [Google Scholar] [CrossRef] [PubMed]
- May, D.S.; Crnkovic, C.M.; Krunic, A.; Wilson, T.A.; Fuchs, J.R.; Orjala, J.E. 15N Stable Isotope Labeling and Comparative Metabolomics Facilitates Genome Mining in Cultured Cyanobacteria. ACS Chem. Biol. 2020, 15, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Calteau, A.; Fewer, D.P.; Latifi, A.; Coursin, T.; Laurent, T.; Jokela, J.; Kerfeld, C.A.; Sivonen, K.; Piel, J.; Gugger, M. Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genom. 2014, 15, 977. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Huang, G.; Lechno-Yossef, S.; Wolk, C.P.; Kaneko, T.; Tabata, S. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 2005, 58, 227–243. [Google Scholar] [CrossRef]
- Dagan, T.; Roettger, M.; Stucken, K.; Landan, G.; Koch, R.; Major, P.; Gould, S.B.; Goremykin, V.V.; Rippka, R.; Tandeau de Marsac, N.; et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 2013, 5, 31–44. [Google Scholar] [CrossRef]
- Liaimer, A.; Helfrich, E.J.; Hinrichs, K.; Guljamow, A.; Ishida, K.; Hertweck, C.; Dittmann, E. Nostopeptolide plays a governing role during cellular differentiation of the symbiotic cyanobacterium Nostoc punctiforme. Proc. Natl. Acad. Sci. USA 2015, 112, 1862–1867. [Google Scholar] [CrossRef]
- Schoffelen, N.J.; Mohr, W.; Ferdelman, T.G.; Duerschlag, J.; Littmann, S.; Ploug, H.; Kuypers, M.M.M. Phosphate availability affects fixed nitrogen transfer from diazotrophs to their epibionts. ISME J. 2019, 13, 2701–2713. [Google Scholar] [CrossRef]
- Guha, R. Chemical Informatics Functionality in R. J. Stat. Softw. 2007, 18, 1–16. [Google Scholar] [CrossRef]
- Mullowney, M.W.; Duncan, K.R.; Elsayed, S.S.; Garg, N.; van der Hooft, J.J.J.; Martin, N.I.; Meijer, D.; Terlouw, B.R.; Biermann, F.; Blin, K.; et al. Artificial intelligence for natural product drug discovery. Nat. Rev. Drug Discov. 2023, 22, 895–916. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, G.; Cheng, X.; Cao, C.; Cai, Z.; Zhou, J. Screening for Potential Antiviral Compounds from Cyanobacterial Secondary Metabolites Using Machine Learning. Mar. Drugs 2024, 22, 501. [Google Scholar] [CrossRef]
- Hardy, R.W.F.; Havelka, U.D. Nitrogen Fixation Research: A Key to World Food? Science 1975, 188, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Stal, L.J.; Krumbein, W.E. Temporal separation of nitrogen fixation and photosynthesis in the filamentous, non-heterocystous cyanobacterium Oscillatoria sp. Arch. Microbiol. 1987, 149, 76–80. [Google Scholar] [CrossRef]
- Murry, M.A.; Wolk, C.P. Evidence that the barrier to the penetration of oxygen into heterocysts depends upon two layers of the cell envelope. Arch. Microbiol. 1989, 151, 469–474. [Google Scholar] [CrossRef]
- Zhou, R.; Wolk, C.P. A Two-component System Mediates Developmental Regulation of Biosynthesis of a Heterocyst Polysaccharide. J. Biol. Chem. 2003, 278, 19939–19946. [Google Scholar] [CrossRef]
- Harding, K.; Turk-Kubo, K.A.; Sipler, R.E.; Mills, M.M.; Bronk, D.A.; Zehr, J.P. Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic Ocean. Proc. Natl. Acad. Sci. USA 2018, 115, 13371–13375. [Google Scholar] [CrossRef] [PubMed]
- Baunach, M.; Guljamow, A.; Miguel-Gordo, M.; Dittmann, E. Harnessing the potential: Advances in cyanobacterial natural product research and biotechnology. Nat. Prod. Rep. 2024, 41, 347–369. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, T.; Fahad, S.; Gu, L.; Xu, L.; Zhou, R. Harnessing Nitrogen-Fixing Cyanobacteria for Sustainable Agriculture: Opportunities, Challenges, and Implications for Food Security. Nitrogen. 2025, 6, 16. [Google Scholar] [CrossRef]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a comprehensive public database of secondary metabolites from cyanobacteria. Water Res. 2021, 196, 117017. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531. [Google Scholar] [CrossRef]
- Latysheva, N.; Junker, V.L.; Palmer, W.J.; Codd, G.A.; Barker, D. The evolution of nitrogen fixation in cyanobacteria. Bioinformatics 2012, 28, 603–606. [Google Scholar] [CrossRef]
- Koirala, A.; Brözel, V.S. Phylogeny of Nitrogenase Structural and Assembly Components Reveals New Insights into the Origin and Distribution of Nitrogen Fixation across Bacteria and Archaea. Microorganisms 2021, 9, 1662. [Google Scholar] [CrossRef]
- Swamidass, S.J.; Azencott, C.A.; Lin, T.W.; Gramajo, H.; Tsai, S.C.; Baldi, P. Influence relevance voting: An accurate and interpretable virtual high throughput screening method. J. Chem. Inf. Model. 2009, 49, 756–766. [Google Scholar] [CrossRef]
- Li, M.; Cheng, L.; Tang, J.; Daroch, M. Molecular Components of Nitrogen Fixation Gene Cluster and Associated Enzymatic Activities of Non-Heterocystous Thermophilic Cyanobacterium Thermoleptolyngbya sp. Life 2021, 11, 640. Life 2021, 11, 640. [Google Scholar] [CrossRef]
- Ccte, E.P.A. Toxicity Estimation Software Tool (TEST); The United States Environmental Protection Agency’s Center for Computational Toxicology and Exposure: Washington, DC, USA, 2022.
- Berrendero, E.; Valiente, E.F.; Perona, E.; Gómez, C.L.; Loza, V.; Muñoz-Martín, M.Á.; Mateo, P. Nitrogen fixation in a non-heterocystous cyanobacterial mat from a mountain river. Sci. Rep. 2016, 6, 30920. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sher, D.; Kelly, L.; Shi, Y.; Huang, K.; Knerr, P.J.; Joewono, I.; Rusch, D.; Chisholm, S.W.; van der Donk, W.A. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 10430–10435. [Google Scholar] [CrossRef] [PubMed]
- Rapp, J.; Rath, P.; Kilian, J.; Brilisauer, K.; Grond, S.; Forchhammer, K. A bioactive molecule made by unusual salvage of radical SAM enzyme byproduct 5-deoxyadenosine blurs the boundary of primary and secondary metabolism. J. Biol. Chem. 2021, 296, 100621. [Google Scholar] [CrossRef] [PubMed]
- Tooming-Klunderud, A.; Sogge, H.; Rounge, T.B.; Nederbragt, A.J.; Lagesen, K.; Glöckner, G.; Hayes, P.K.; Rohrlack, T.; Jakobsen, K.S. From green to red: Horizontal gene transfer of the phycoerythrin gene cluster between Planktothrix strains. Appl. Environ. Microbiol. 2013, 79, 6803–6812. [Google Scholar] [CrossRef] [PubMed]
- Popin, R.V.; Alvarenga, D.O.; Castelo-Branco, R.; Fewer, D.P.; Sivonen, K. Mining of Cyanobacterial Genomes Indicates Natural Product Biosynthetic Gene Clusters Located in Conjugative Plasmids. Front. Microbiol. 2021, 12, 684565. [Google Scholar] [CrossRef]
| Fingerprint | Depth | Bits | Accuracy | Precision | Recall | F1 | AUC | AUC > Random Chance p-Value |
|---|---|---|---|---|---|---|---|---|
| PubChem (881 bits) | NA | 881 | 0.905 (0.895–0.914) | 0.907 (0.895–0.920) | 0.903 (0.894–0.912) | 0.905 (0.896–0.914) | 0.962 (0.956–0.968) | 1.76 × 10−17 |
| MACCS 166 | NA | 166 | 0.886 (0.875–0.896) | 0.897 (0.886–0.908) | 0.873 (0.860–0.887) | 0.885 (0.874–0.896) | 0.957 (0.949–0.966) | 4.41 × 10−16 |
| Extended d15 262 k | 15 | 262,144 | 0.874 (0.862–0.887) | 0.868 (0.849–0.886) | 0.887 (0.875–0.898) | 0.877 (0.865–0.888) | 0.948 (0.942–0.954) | 1.45 × 10−17 |
| Path-based d30 262 k (paper) | 30 | 262,144 | 0.868 (0.854–0.883) | 0.874 (0.854–0.894) | 0.864 (0.848–0.880) | 0.869 (0.855–0.883) | 0.950 (0.944–0.956) | 1.75 × 10−17 |
| Graph 4 k | NA | 4096 | 0.820 (0.809–0.832) | 0.866 (0.851–0.882) | 0.761 (0.746–0.776) | 0.810 (0.797–0.823) | 0.864 (0.857–0.871) | 4.43 × 10−16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, J.; Nawaz, T.; Gu, L.; Zhou, R. Secondary Metabolites Predict Diazotrophic Cyanobacteria: A Model-Based Cheminformatic Approach. Metabolites 2025, 15, 562. https://doi.org/10.3390/metabo15090562
Young J, Nawaz T, Gu L, Zhou R. Secondary Metabolites Predict Diazotrophic Cyanobacteria: A Model-Based Cheminformatic Approach. Metabolites. 2025; 15(9):562. https://doi.org/10.3390/metabo15090562
Chicago/Turabian StyleYoung, James, Taufiq Nawaz, Liping Gu, and Ruanbao Zhou. 2025. "Secondary Metabolites Predict Diazotrophic Cyanobacteria: A Model-Based Cheminformatic Approach" Metabolites 15, no. 9: 562. https://doi.org/10.3390/metabo15090562
APA StyleYoung, J., Nawaz, T., Gu, L., & Zhou, R. (2025). Secondary Metabolites Predict Diazotrophic Cyanobacteria: A Model-Based Cheminformatic Approach. Metabolites, 15(9), 562. https://doi.org/10.3390/metabo15090562







