Metabolite Profile and Metabolic Network Analysis of Walnuts (Juglans regia L.) in Response to Chilling Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. REC Determination
- (1)
- 15-min boiling with subsequent sealing using parafilm
- (2)
- Continuous extraction in a temperature-controlled shaker (25 °C, 15 h)
- (3)
- Initial conductivity measurement (A1) using a DDSJ-307F conductivity meter (Shanghai Yidian Science Instrument Co., Ltd., Shanghai, China)
- (4)
- Thermal treatment in an 85 °C water bath for 20 min to achieve complete cellular disruption
- (5)
- Post-extraction conductivity measurement at 25 °C (A2)
2.3. Metabolites’ Extraction
2.3.1. Tissue Sample Processing
- (1)
- Homogenization: Approximately 100 mg of tissue specimen was cryogenically pulverized in liquid nitrogen using a mortar and pestle.
- (2)
- Extraction: The resulting homogenate was vortex-resuspended in 1 mL of pre-chilled 80% (v/v) methanol aqueous solution and maintained at 4 °C for 5 min with ice-bath incubation.
- (3)
- Primary Centrifugation: Cellular debris was removed by centrifugation at 15,000× g (4 °C, 20 min) using a refrigerated centrifuge.
- (4)
- Dilution: The methanolic supernatant was diluted with LC-MS grade water to achieve a final methanol concentration of 53% (v/v).
- (5)
- Secondary Clarification: The diluted solution underwent additional centrifugation under identical parameters (15,000× g, 4 °C, 20 min) in sterile microcentrifuge tubes.
- (6)
- LC-MS/MS Analysis: The clarified supernatant was directly injected into the LC-MS/MS system for metabolite profiling [38].
2.3.2. Quality Control (QC) Samples
2.3.3. Method Blank Preparation
2.4. Instrument Parameters
2.4.1. Chromatographic Conditions (Vanquish UHPLC, Thermo Fisher, Dreieich, Germany)
| Time | A% | B% |
| 0 | 98 | 2 |
| 1.5 | 98 | 2 |
| 12 | 0 | 100 |
| 14 | 0 | 100 |
| 14.1 | 98 | 2 |
| 17 | 98 | 2 |
2.4.2. Mass Spectrometry Conditions (Q Exactive™ HF, Thermo Fisher, Dreieich, Germany)
2.5. Methods of Qualitative and Quantitative Analysis of Metabolites
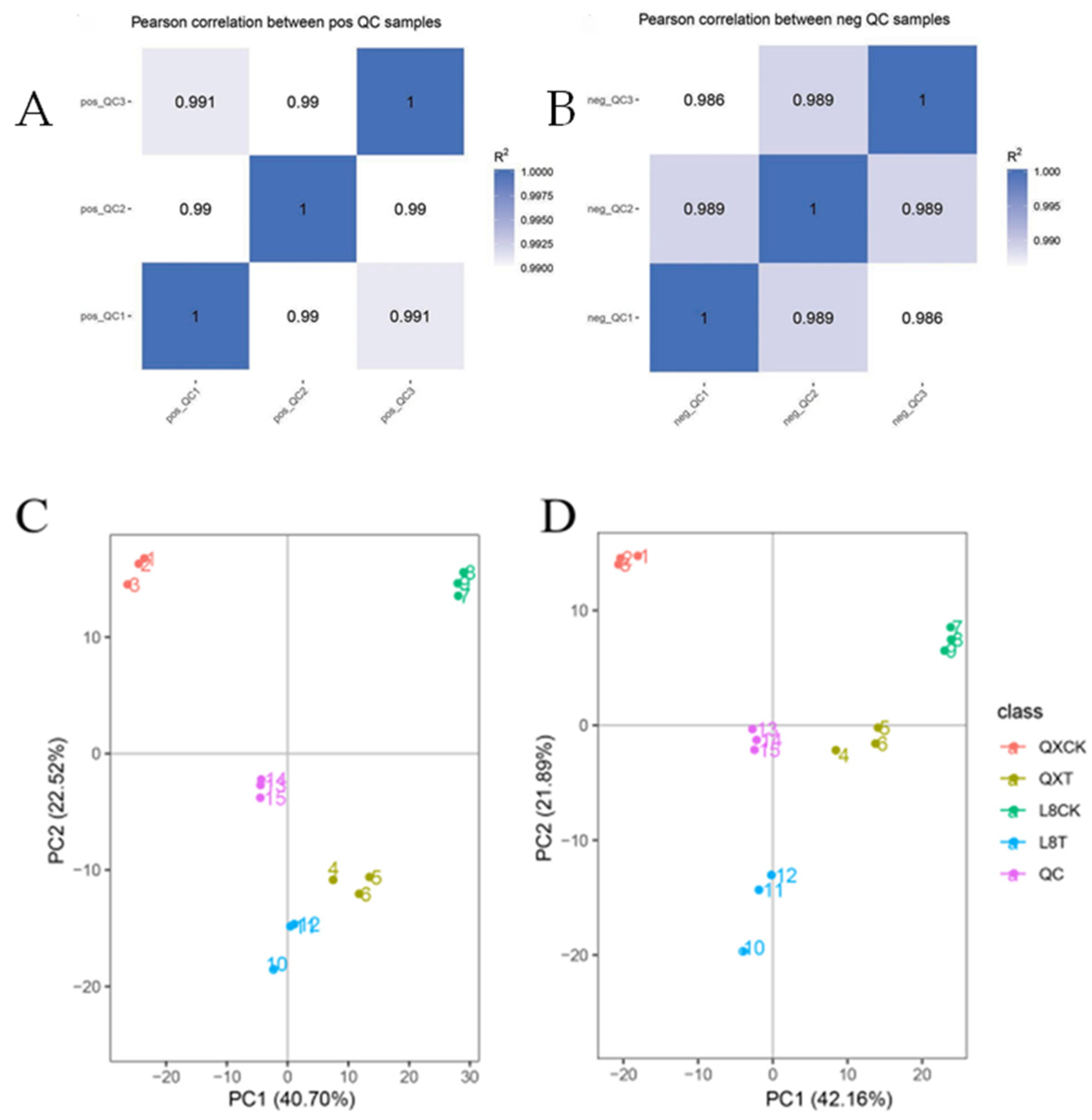
2.6. Data Quality Assessment
2.7. OPLS-DA Analysis
2.8. Differential Metabolite Analysis
2.9. Statistical Analysis of Data
3. Results
3.1. Relative Electrical Conductivity Changes in Walnut in Response to Chilling Stress
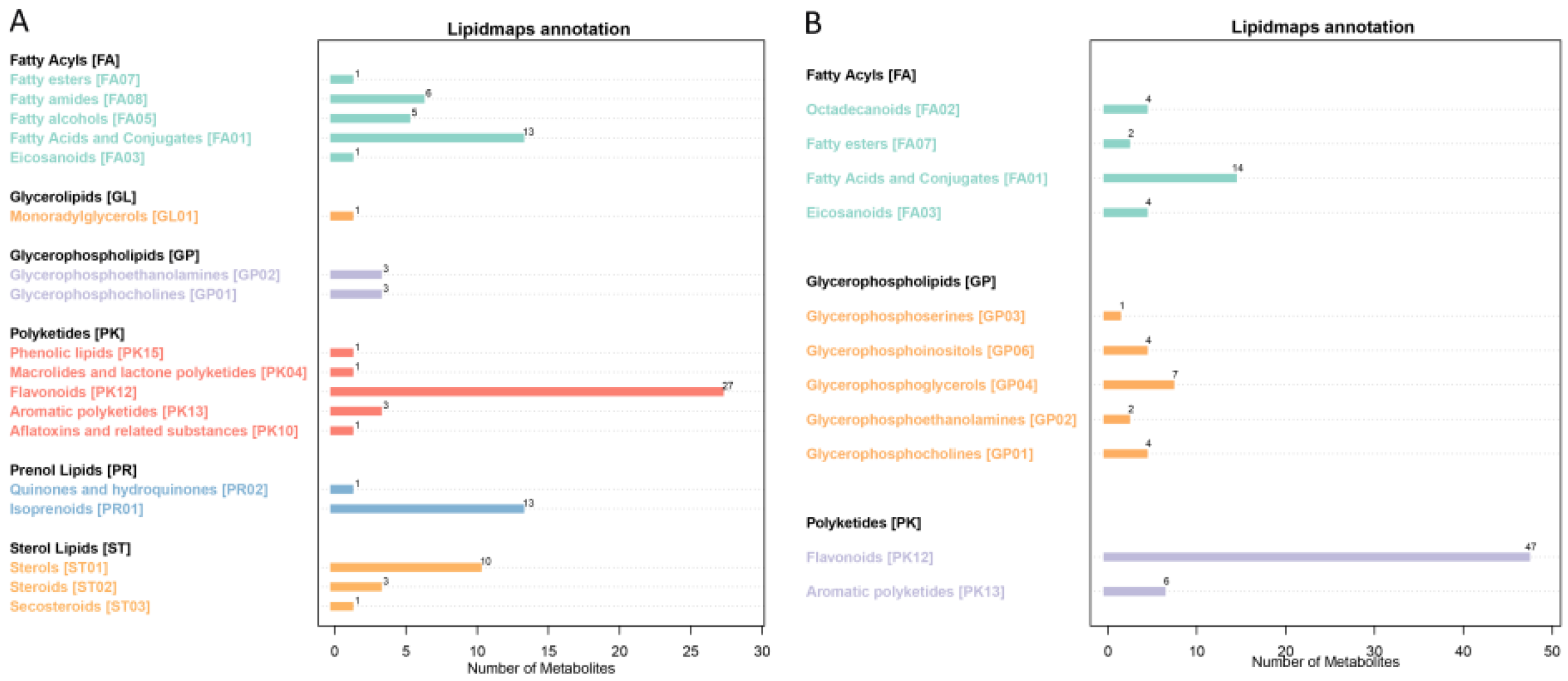
3.2. Qualitative and Quantitative Analysis of Metabolites
3.3. PLS-DA Analysis
3.4. Differential Metabolites’ Screening
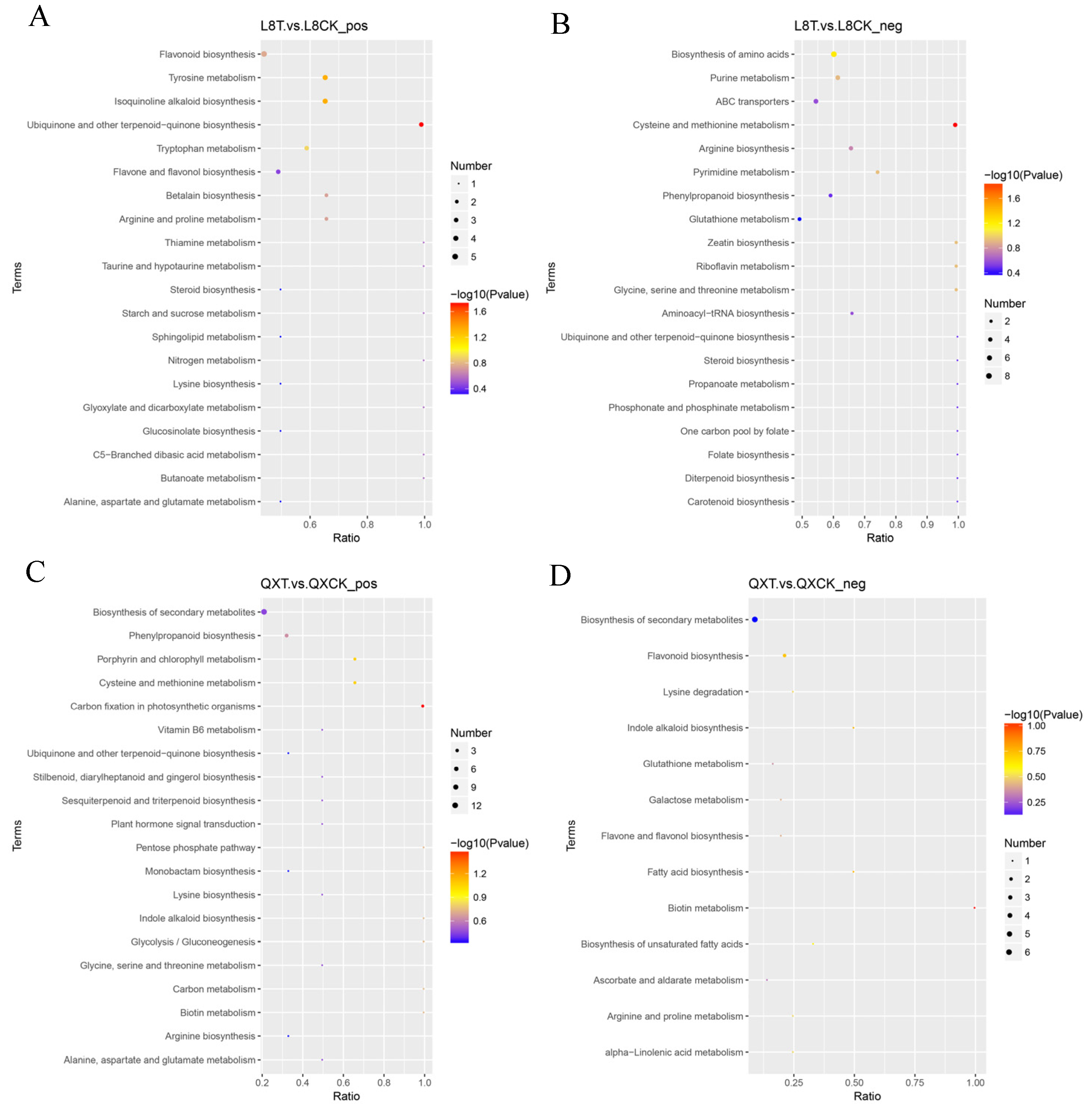
3.5. KEGG Enrichment Result Analysis
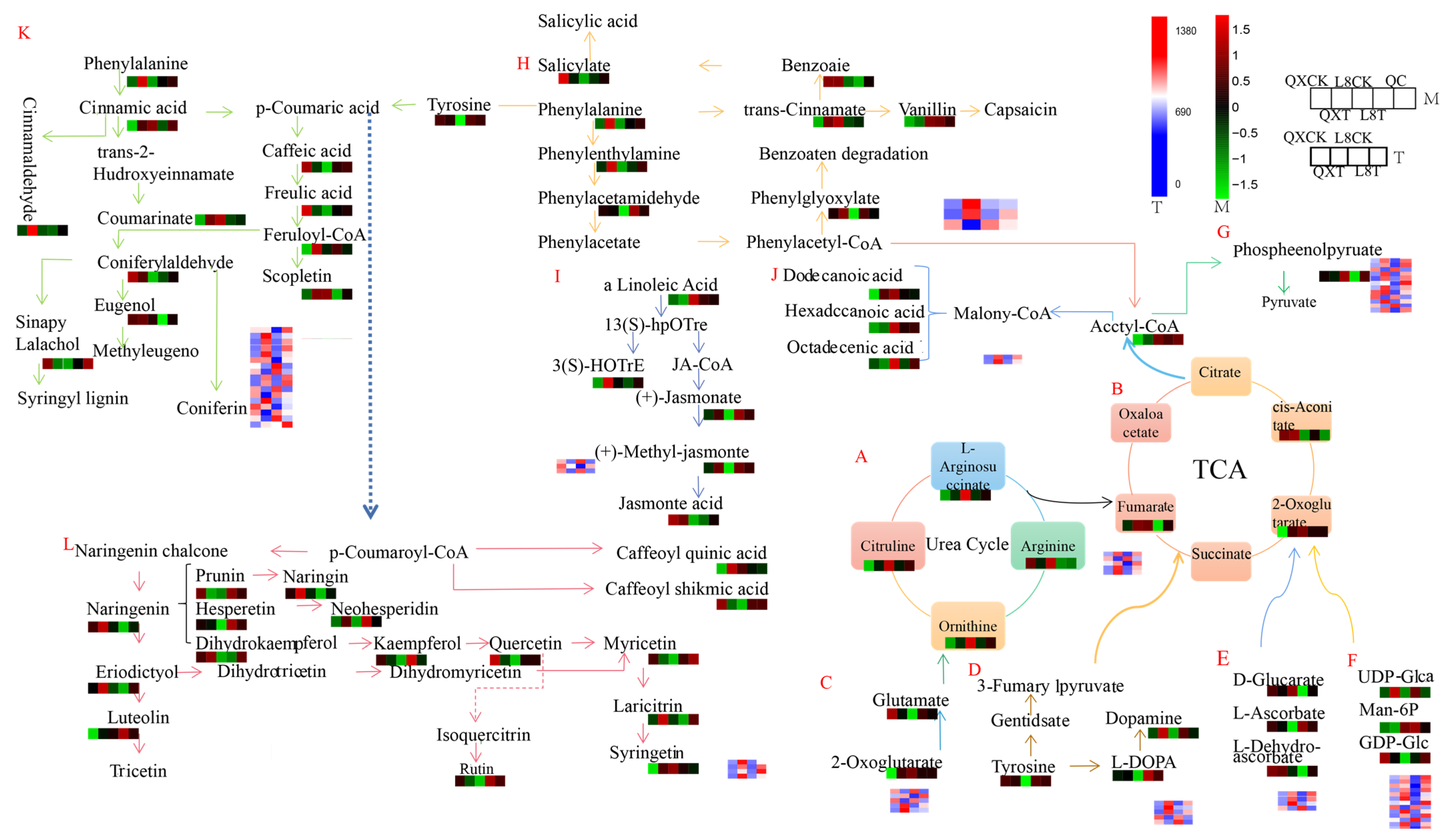
3.6. Analysis of Comprehensive Metabolomics and Transcriptomics Networks Under Chilling Stress
- -
- 109013393 was significantly negatively correlated with methyl dihydrojasmonate and methyl jasmonate (Table 2).
- -
- 109018148 was significantly positively correlated with L-phenylalanine and phenethylamine (Table 3).
- -
- 109010746 was significantly positively correlated with laricitrin (Table 4).
- -
- 108993196 was significantly positively correlated with neohesperidin and ferulaldehyde (Table 4).
- -
- 108989769 was positively correlated with neohesperidin, rutin, and hesperetin and significantly negatively correlated with eugenol (Table 4).
- -
- 109020389 was significantly positively correlated with L-phenylalanine and ferulaldehyde (Table 5).
- -
- 109020701 was highly significantly positively correlated with L-phenylalanine and significantly positively correlated with ferulaldehyde (Table 5).
- -
- 109003193 was significantly positively correlated with coniferin (Table 5).
- -
- 108999621 and 109004171 were significantly negatively correlated with ferulaldehyde (Table 5).
- -
- 109009576 and 108983554 were significantly positively correlated with ferulaldehyde, with the latter also showing a significant positive correlation with L-phenylalanine (Table 5).
- -
- 109005199 was significantly positively correlated with coniferin (Table 5).
- -
- 108996382 was significantly negatively correlated with L-phenylalanine and ferulaldehyde (Table 5).
| id | 109013393 | 109002743 | 108997763 | |
|---|---|---|---|---|
| Linoleic acid | 0.483 | −0.341 | 0.264 | |
| 13(S)-HOTrE | −0.236 | −0.035 | −0.273 | |
| Jasmonic acid | −0.15 | 0.251 | 0.069 | |
| Methyl dihydrojasmonate | −0.683 * | 0.13 | −0.568 | |
| Methyl jasmonate | −0.582 * | 0.175 | −0.545 | |
| id | 109018148 | 108987596 | 109009407 | |
|---|---|---|---|---|
| Salicylic acid | −0.268 | −0.227 | 0.042 | |
| L-phenylalanine | 0.690 * | 0.544 | −0.356 | |
| Phenethylamine | 0.756 ** | 0.52 | −0.347 | |
| Phenylacetaldehyde | 0.059 | 0.559 | −0.225 | |
| Phenylglyoxylic acid | 0.307 | 0.24 | −0.026 | |
| Trans-cinnamic acid | 0.357 | 0.209 | −0.131 | |
| Vanillin | −0.243 | 0.036 | 0.182 | |
| Benzoic acid | 0.35 | 0.078 | −0.17 | |
| id | 109010746 | 108993196 | 108989769 | 108997708 | |
|---|---|---|---|---|---|
| 1,3-Dicaffeoylquinic acid | 0.529 | 0.202 | −0.032 | 0.432 | |
| Myricetin | −0.125 | 0.198 | 0.446 | −0.105 | |
| Laricitrin | 0.619 * | 0.498 | −0.061 | 0.377 | |
| Syringetin | 0.138 | −0.018 | 0.07 | −0.002 | |
| Naringin | 0.463 | 0.026 | −0.163 | 0.29 | |
| Naringenin | 0.492 | −0.013 | −0.264 | 0.283 | |
| Luteolin | 0.079 | 0.332 | 0.559 | 0.229 | |
| 5-O-Caffeoylshikimic acid | −0.213 | −0.047 | −0.012 | −0.164 | |
| Neohesperidin | 0.381 | 0.806 ** | 0.695 * | 0.512 | |
| Kaempferol | 0.133 | 0.437 | 0.43 | −0.058 | |
| Quercetin | −0.124 | −0.106 | −0.002 | −0.182 | |
| Rutin | −0.103 | 0.511 | 0.616 * | −0.054 | |
| Prunin | −0.349 | 0.014 | 0.423 | −0.301 | |
| Hesperetin | 0.055 | 0.535 | 0.629 * | 0.179 | |
| Dihydrokaempferol | 0.412 | −0.132 | 0.195 | 0.19 | |
| Eriodictyol | 0.432 | −0.028 | −0.174 | 0.306 | |
| id | 109020389 | 108993196 | 109020701 | 109003193 | 108999621 | 109009576 | 109004171 | 108983554 | 109005199 | 108996382 | 108989769 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L-phenylalanine | 0.890 ** | 0.466 | 0.712 ** | 0.155 | −0.414 | 0.37 | −0.487 | 0.802 ** | 0.124 | −0.696 * | 0.282 | |
| Trans-cinnamic acid | 0.36 | −0.137 | 0.231 | −0.432 | 0.15 | −0.164 | 0.215 | 0.24 | −0.417 | −0.266 | −0.231 | |
| Cinnamaldehyde | 0.566 | 0.434 | 0.536 | 0.18 | −0.357 | 0.483 | −0.174 | 0.366 | 0.177 | −0.255 | 0.068 | |
| Ferulaldehyde | 0.811 ** | 0.638 * | 0.639 * | 0.139 | −0.599 * | 0.615 * | −0.721 ** | 0.796 ** | 0.036 | −0.631 * | 0.549 | |
| Eugenol | 0.164 | −0.428 | 0.038 | 0.158 | −0.095 | −0.433 | 0.305 | 0.209 | 0.224 | −0.102 | −0.607 * | |
| Coniferin | 0.158 | −0.046 | 0.166 | 0.614 * | −0.243 | −0.003 | −0.165 | 0.223 | 0.659 * | −0.18 | −0.173 | |
| Caffeic acid | −0.331 | −0.183 | −0.233 | 0.499 | −0.058 | −0.12 | −0.057 | −0.157 | 0.477 | 0.16 | −0.074 | |
| Ferulic acid | −0.42 | −0.308 | −0.318 | 0.507 | −0.063 | −0.248 | 0.076 | −0.236 | 0.557 | 0.244 | −0.246 | |
| Sinapyl alcohol | −0.094 | −0.236 | 0.155 | 0.16 | −0.176 | −0.098 | 0.087 | −0.222 | 0.197 | 0.256 | 0.131 | |
| Scopoletin | 0.469 | −0.162 | 0.226 | −0.258 | 0.031 | −0.274 | 0.102 | 0.406 | −0.196 | −0.37 | −0.277 | |
| Scopolin | −0.112 | −0.244 | −0.216 | 0.457 | −0.149 | −0.352 | 0.033 | 0.047 | 0.548 | 0.058 | −0.283 | |
| Cinnamic acid | 0.428 | −0.014 | 0.233 | −0.372 | −0.006 | −0.165 | 0.067 | 0.332 | −0.305 | −0.295 | −0.023 | |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zeb, U.; Rahim, F.; Azizullah, A.; Saleh, I.A.; Wali, S.; Khan, A.A.; Khan, H.; Fiaz, S.; AbdElgawad, H.; Iqbal, B.; et al. Effects of copper sulphate stress on the morphological and biochemical characteristics of Spinacia oleracea and Avena sativa. BMC Plant Biol. 2024, 24, 889. [Google Scholar] [CrossRef]
- Dong, C.; Li, L.; Cao, N.; Shang, Q.; Zhang, Z. Roles of phenylalanine ammonia-lyase in low temperature tolerance in cucumber seedlings. Chin. J. Appl. Ecol. 2015, 26, 2041–2049. [Google Scholar]
- Li, C.; Xu, W.; Liu, L.; Yang, J.; Xin, K.; Guo, W. Changes of endogenous hormone contents and antioxidative enzyme activities in wheat leaves under low temperature stress at jointing stage. Chin. J. Appl. Ecol. 2015, 26, 2015–2022. [Google Scholar]
- Liu, K.; Li, J.; Wang, H.; Li, L.; Zhao, S.; Zhang, J.; Ma, Q.; Zhang, Z. Identification and transcriptional expression analysis of walnut JrJAZ gene family. J. Hebei Agric. Univ. 2021, 44, 41–50. [Google Scholar]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef]
- Liu, F.; Wang, H.; Hu, C. Current situation of main fruit tree industry in China and it’s development countermeasure during the “14th five-year plan” period. Chin. Acad. Agric. Sci. 2021, 1, 1–5. [Google Scholar]
- Xi, S. Gene Resources of juglans and genetic improvement of Juglas regia in China. Sci. Silvae Sin. 1987, 23, 342–350. [Google Scholar]
- Ma, T.; Ning, D. Analysis of international competitiveness of walnut Industry in China. Firest Sci. Technol. 2021, 64, 3–7. [Google Scholar]
- Khan, K.; Zeb, U.; Gul, S.; Aziziullah, A.; Ali, N.; Fiaz, S.; Abushady, A.M.; Ullah, S.; Muhammad, S.; Attia, K.A.; et al. Genome wide identification and expression analysis of U-box gene family in Juglans regia L. Genet. Resour. Crop Evol. 2023, 70, 2337–2352. [Google Scholar]
- Zhou, Q.; Han, Y.; Pan, J.; Yuan, H.; Li, X.; Qin, M.; Peng, B.; Zhu, Y. Research progress in plant cold resistance mechanism. J. Xinyang Norm. Univ. 2019, 32, 511–516. [Google Scholar]
- Zhao, X.; Chen, S.; Cheng, J.; Guo, H.; Qu, X.; An, D.; Li, Z. Analysis of cold resistance on three peach cultivars. J. Fruit Resour. 2020, 1, 14–19. [Google Scholar]
- Xiao, C.; Peng, T. Research progress on abiotic stress response mechanism of fruit tree. South China Fruits 2019, 48, 133–144. [Google Scholar]
- Liu, J.; Qiu, X.; Li, Y.; Li, Y.; Zhang, J.; Bai, Z. The present situation, problems and development countermeasures of walnut production in China. North. Fruit Trees 2020, 6, 38–41. [Google Scholar]
- The Food and Agriculture Organization Database. Available online: http://www.fao.org/faostat/en/#data (accessed on 24 January 2021).
- Wu, Y.; Tian, J.; Qiao, Y.; Shi, M.; Tian, X.; Liu, Z.; Ru, H.; Liang, Q.; Cheng, P. Analysis of physiological and biochemical indexes of cold resistance of walnut leaves under low temperature stress. China Agric. Circ. 2016, 32, 101–106. [Google Scholar]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolic networks of cucurbita maxima phloem. Phytochemistry 2002, 62, 875–886. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the fruit metabolome in tomato breeding. Cell 2018, 172, 249–261. [Google Scholar] [CrossRef]
- Cong, R.; Zhang, J.; Wang, S.; Yu, H.; Yan, X.; Pei, X.; He, S. Research progress of plant cold resistance identification and its physiological and ecological mechanism. J. Temp. For. Res. 2020, 3, 27–33. [Google Scholar]
- Dong, Y.; Silbermann, M.; Speiser, A.; Forieri, I.; Linster, E.; Poschet, G.; Samami, A.A.; Wanatabe, M.; Sticht, C.; Aurelio, A.; et al. Sulfur availability regulates plant growth via glucose-TOR signaling. Nat. Commun. 2020, 11, 1174. [Google Scholar]
- Giraud, E.; Van Aken, O.; Ho, L.H.; Whelan, J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009, 150, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Plant Cell Environ. 2012, 35, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, R.; Niu, K.; Zhu, R.; Wang, Y.; Ma, X.; Ma, H. Metabonomics study of qinghai wild Poa pratensis in response to low temperature stress. Acta Agrestia Sin. 2020, 28, 904–914. [Google Scholar]
- Zhao, M.; Ren, Y.; Wei, W.; Yang, J.; Zhong, Q.; Li, Z. Metabolite analysis of jerusalem artichoke (Helianthus tuberosus L.) seedlings in response to polyethylene glycol-simulated drought stress. Int. J. Mol. Sci. 2021, 22, 3294. [Google Scholar] [CrossRef]
- Cook, D.; Fowler, S.; Fiehn, O.; Thomashow, M.F. A prominent role for the CBF cold response pathway in configuring the low-temperature netabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 15243–15248. [Google Scholar] [CrossRef]
- Zhou, C.; Di, Y.; Qiu, Q.; Cai, J.; Liu, Y.; Xu, H.; Li, K. Combined analysis of transcriptome and metabolome on the effects of low temperature stress on the growth and development of Gastrodia elata. Mol. Plant Breed. 2023, 21, 110–122. [Google Scholar]
- Zhou, F.; Tang, F.; Zhang, Q.; Song, W.; Ning, m.; Cai, W.; Shan, C. Metabolomic and transcriptome analysis of phytohormone metabolism in Hami Melon during low temperature storage. Food Sci. 2022, 43, 181–191. [Google Scholar]
- Maria, B.; Toshihiro, O.; Nikolay, M.; Ivan, I.; Veselin, P.; Valentina, T.; Fernie, A.R.; Gechev, T.S. Comparative metabolic profiling of Haberlea rhodopensis, Thellungiella halophyla, and Arabidopsis thaliana exposed to low temperature. Front. Plant Sci. 2013, 4, 499. [Google Scholar]
- Li, C.; Qiao, J.; Huang, L.; Zhang, M.; Zhang, P.; Niu, J.; Liu, J. Transcriptome and metabolome analysis to reveal the mechanisms responding to high temperature stress in anthesis stage of maize. Acta Agric. Boreali-Sin. 2020, 35, 8–21. [Google Scholar]
- Ma, W.; Zou, L.; Zhao, H.; Ge, F. Analysis of the main metabolic pathway of drought resistance of Dontostemon elegans Maxim based on transcriptome sequencing. Mol. Plant Breed. 2022, 20, 3548–3561. [Google Scholar]
- Du, W.; Chen, Z.; Xu, D. Physiological response and differentially expressed genes analysis of transcriptome in Momordica charantia L. leaf under cold stress. J. Nucl. Agric. Sci. 2021, 35, 338–348. [Google Scholar]
- Zhou, L.; Li, Y.; Wang, W.; S, Z. Research progress in the metabolomics for plants response to temperature stress. J. Shanxi Agric. Sci. 2017, 45, 317–320. [Google Scholar]
- Ni, J.; Yang, X.; Zhang, H.; Ni, Y.; Wu, H.; Qi, W. Metabolomics and its application to plant stress research. World For. Res. 2014, 27, 11–17. [Google Scholar]
- Liu, K.; Feng, Q.; Liu, X.; Wang, H.; An, X.; Zhang, Z. Study on cold resistance of two walnut varieties based on leaf microstructure. J. Hebei Agric. Univ. 2021, 44, 45–50. [Google Scholar]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal andhuman tissues via UPLC-MS. Nat. Protoc. 2012, 8, 17–32. [Google Scholar] [CrossRef]
- Dai, W.; Xie, D.; Lu, M.; Li, P.; Lv, H.; Yang, C.; Peng, Q.; Zhu, Y.; Guo, L.; Zhang, Y.; et al. Characterization of white tea metabolome: Comparison against green and black tea by a nontargeted metabolomics approach. Food Res. Int. 2017, 96, 40–45. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Heischmann, S.; Quinn, K.; Cruickshank-Quinn, C.; Liang, L.-P.; Reisdorph, R.; Reisdorph, N.; Patel, M. Exploratory Metabolomics Profiling in the Kainic Acid Rat Model Reveals Depletion of 25-Hydroxyvitamin D3 during Epileptogenesis. Sci. Rep. 2016, 6, 31424. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Clément, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Wang, X.; Fu, L.; Sun, J.; Guan, T.; Li, Z. Effects of low temperature stress on membrane fatty axid in tillering node of winter wheat. J. Triticeae Crops 2013, 33, 746–751. [Google Scholar]
- Wang, X.; Li, S.; Dong, H.; Gao, Z.; Dai, S. The effect of low temperature on free fatty acid in tomato deedlings and florescence. Acta Hortic. Sin. 1997, 24, 58–61. [Google Scholar]
- Zhang, J.; Yan, S.; Wang, H.; Si, L.; Ma, Z.; Yang, J. Study on the relationship between the contents of fatty acids and low temperature tolerance in cucumber seeds. J. Anhui Agric. 2008, 36, 4859–4861. [Google Scholar]
- Sun, B.; Ma, L.; Zeng, X.; Niu, Z.; Wang, W.; Hu, F.; Lu, X.; Qi, W.; Wu, J.; Liu, L. Fatty acid components and differential expression of FAD3 in winter Brassica rapa L. under low temperature stress. Agric. Res. Arid. Areas 2021, 39, 65–74. [Google Scholar]
- Li, D.; Wang, Q.; Hu, B.; Zhang, H.; Wu, W. Effects of low temperature on fatty acid composition and gene expression of fatty acid desaturase in safflower. Mol. Plant Breed. 2018, 16, 5223–5231. [Google Scholar]
- Mao, J.; Xiong, X.; Lu, Y. Advances in the regulation of plant stress response by Jasmonic Acid. Chin. J. Bioprocess Eng. 2021, 19, 413–419. [Google Scholar]
- Wei, X.; Wang, X.; Chen, J. Trancsriptome and jasmin metabolism gene analysis of Passiflora edulia Sims under low temperature stress. J. Nucl. Agric. Sci. 2021, 35, 815–825. [Google Scholar]
- Zhang, H.; Xing, X.; Niu, L.; Lu, Y.; Zhang, L.; Liu, Y. Effects of exogenous MeJA on cold tolerance of tobacco seedlings. Guizhou Agric. Sci. 2020, 48, 8–12. [Google Scholar]
- Wang, J.; Liang, Q.; Zhou, Y.; Li, R.; Xue, L.; Hao, X. Effects of exogenous Meja on the regulation of fatty acid synthesis and FAD3 gene expression in Perilla frutescens under low tempperature. J. Shanxi Agric. Univ. 2019, 39, 1–6. [Google Scholar]
- Taylor, L.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Janas, K.M.; Cvikrová, M.; Pałagiewicz, A.; Szafranska, K.; Posmyk, M.M. Constitutive elevated accumulation of phenylpropanoids in soybean roots at low temperature. Plant Sci. 2002, 163, 369–373. [Google Scholar] [CrossRef]
- Luo, L.; Chen, Z.; Ding, J.; Zhou, P. The influence of low latitude and high elevation regions UV-B radiation on flavonoids content of primula. J. Yunnan Agric. Univ. 2007, 2, 229–233. [Google Scholar]
- Leng, P.; Zhang, G.; Wu, X.; Qi, J. Change of phenols in persimmon during autumn and winter. J. China Agric. Univ. 2001, 6, 63–67. [Google Scholar]
- Hernández, I.; Alegre, L.; Munné-Bosch, S. Drought-induced changes in flavonoids and other low molecular weight antioxidants in Cistus clusii grown under Mediterranean field conditions. Tree Physiol. 2004, 24, 1303–1311. [Google Scholar] [CrossRef]
- Zhou, P.; Lin, Z.; Guo, R.; Yan, S.; Zhang, X.; Ma, X.; Jing, G. Effects of low temperature treatment on gene expression and flavonoids biosynthesis metabolism in peach (Prunus persica) Leaves. J. Agric. Biotechnol. 2021, 29, 1283–1294. [Google Scholar]
- Xu, Y.; Yang, S.; Jia, R.; Zhao, X.; He, L.; Meng, W.; Zhou, M.; Ge, H. Physiological and proteomics analysis on freezing tolerance of rosa beggeriana branches during overwintering. J. Plant Genet. Resour. 2020, 21, 1568–1576. [Google Scholar]
- Liu, W.; Zhang, R.; Xiang, C.; Zhang, R.; Wang, Q.; Wang, T.; Li, X.; Lu, X.; Gao, S.; Liu, Z.; et al. Transcriptomic and Physiological Analysis Reveal That α-Linolenic Acid Biosynthesis Responds to Early Chilling Tolerance in Pumpkin Rootstock Varieties. Front. Plant Sci. 2021, 12, 669565. [Google Scholar] [CrossRef]
- Cao, F.; Wang, H.; Yu, W.; Cheng, H. Research progress on fatty acid desaturases and coding genes in higher plants. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2012, 36, 125–132. [Google Scholar]
- Li, G.; Yang, D.; Hu, T. Changes in fatty acid content of walnuts from different production areas in sichuan province. For. Sci. 2007, 5, 36–41. [Google Scholar]
- Wang, L.; Fu, Z.; Gao, J.; Dong, X.; Zhang, J.; Yuan, X.; Jiang, H.; Wang, H.; Li, Y.; Shi, M.; et al. The biosynthesis and regulation of unsaturated fatty acids in plants. Genom. Appl. Biol. 2020, 39, 254–258. [Google Scholar]
- Xie, P.; Wu, Y.; Lee, Y.-Y.; Wang, Y.; Zhang, Z. Asterias Rolleston starfish gonad lipids: A novel source of Omega-3 fatty acids-assessment of major components and their antioxidant activities. Food Chem. 2024, 456, 140005. [Google Scholar] [CrossRef] [PubMed]




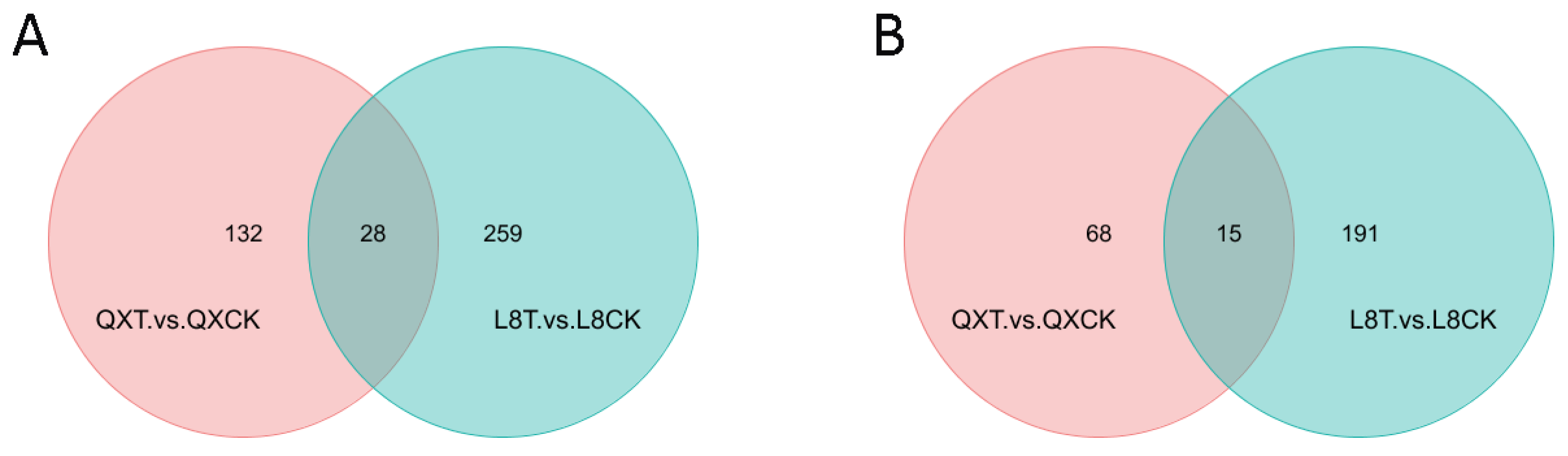

| Compared Samples | Total Identified | Total Significantly Changed | Significantly Upregulated | Significantly Downregulated |
|---|---|---|---|---|
| QXT vs. QXCK positive | 871 | 160 | 33 | 127 |
| L8T vs. L8CK positive | 871 | 287 | 233 | 54 |
| QXT vs. QXCK negative | 633 | 83 | 37 | 46 |
| L8T vs. L8CK negative | 633 | 206 | 114 | 92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Li, Y.; Sang, Y.; Zhang, Y.; An, X.; Wang, H.; Zhang, R. Metabolite Profile and Metabolic Network Analysis of Walnuts (Juglans regia L.) in Response to Chilling Stress. Metabolites 2025, 15, 394. https://doi.org/10.3390/metabo15060394
Liu K, Li Y, Sang Y, Zhang Y, An X, Wang H, Zhang R. Metabolite Profile and Metabolic Network Analysis of Walnuts (Juglans regia L.) in Response to Chilling Stress. Metabolites. 2025; 15(6):394. https://doi.org/10.3390/metabo15060394
Chicago/Turabian StyleLiu, Kai, Yang Li, Yaxin Sang, Yaru Zhang, Xiuhong An, Hongxia Wang, and Ruifen Zhang. 2025. "Metabolite Profile and Metabolic Network Analysis of Walnuts (Juglans regia L.) in Response to Chilling Stress" Metabolites 15, no. 6: 394. https://doi.org/10.3390/metabo15060394
APA StyleLiu, K., Li, Y., Sang, Y., Zhang, Y., An, X., Wang, H., & Zhang, R. (2025). Metabolite Profile and Metabolic Network Analysis of Walnuts (Juglans regia L.) in Response to Chilling Stress. Metabolites, 15(6), 394. https://doi.org/10.3390/metabo15060394





