Lipid Subclasses Differentiate Insulin Resistance by Triglyceride–Glucose Index
Abstract
1. Introduction
2. Methods
2.1. Data Source and Study Participants
2.2. Metabolomics
2.3. Statistical Analysis
3. Results
3.1. General Characteristics of Participants
3.2. Multivariate Analysis
3.3. Univariate Analysis
3.4. Functional Enrichment Analysis
3.5. Association Between Metabolites Associated with Insulin Sensitivity and Clinical Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, X.; An, X.; Yang, C.; Sun, W.; Ji, H.; Lian, F. The crucial role and mechanism of insulin resistance in metabolic disease. Front. Endocrinol. 2023, 14, 1149239. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, S.Y.; Choi, C.S. Insulin Resistance: From Mechanisms to Therapeutic Strategies. Diabetes Metab. J. 2022, 46, 15–37. [Google Scholar] [CrossRef]
- Adeva-Andany, M.M.; Martínez-Rodríguez, J.; González-Lucán, M.; Fernández-Fernández, C.; Castro-Quintela, E. Insulin resistance is a cardiovascular risk factor in humans. Diabetes Metab. Syndr. 2019, 13, 1449–1455. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ke, Y.; Nianogo, R. Trends in hyperinsulinemia and insulin resistance among nondiabetic US adults, NHANES, 1999–2018. Res. Sq. 2024, rs.3.rs-5279795. [Google Scholar] [CrossRef]
- Elrayess, M.A.; Rizk, N.M.; Fadel, A.S.; Kerkadi, A. Prevalence and Predictors of Insulin Resistance in Non-Obese Healthy Young Females in Qatar. Int. J. Environ. Res. Public Health 2020, 17, 5088. [Google Scholar] [CrossRef]
- Rooney, M.R.; Fang, M.; Ogurtsova, K.; Ozkan, B.; Echouffo-Tcheugui, J.B.; Boyko, E.J.; Magliano, D.J.; Selvin, E. Global Prevalence of Prediabetes. Diabetes Care 2023, 46, 1388–1394. [Google Scholar] [CrossRef]
- Goh, L.P.; Sani, S.A.; Sabullah, M.K.; Gansau, J.A. The Prevalence of Insulin Resistance in Malaysia and Indonesia: An Updated Systematic Review and Meta-Analysis. Medicina 2022, 58, 826. [Google Scholar] [CrossRef]
- Tahapary, D.L.; Pratisthita, L.B.; Fitri, N.A.; Marcella, C.; Wafa, S.; Kurniawan, F.; Rizka, A.; Tarigan, T.J.E.; Harbuwono, D.S.; Purnamasari, D.; et al. Challenges in the diagnosis of insulin resistance: Focusing on the role of HOMA-IR and Tryglyceride/glucose index. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102581. [Google Scholar] [CrossRef] [PubMed]
- Che, B.; Zhong, C.; Zhang, R.; Pu, L.; Zhao, T.; Zhang, Y.; Han, L. Triglyceride-glucose index and triglyceride to high-density lipoprotein cholesterol ratio as potential cardiovascular disease risk factors: An analysis of UK biobank data. Cardiovasc. Diabetol. 2023, 22, 34. [Google Scholar] [CrossRef]
- Avagimyan, A.; Pogosova, N.; Fogacci, F.; Aghajanova, E.; Djndoyan, Z.; Patoulias, D.; Sasso, L.L.; Bernardi, M.; Faggiano, A.; Mohammadifard, N.; et al. Triglyceride-glucose index (TyG) as a novel biomarker in the era of cardiometabolic medicine. Int. J. Cardiol. 2025, 418, 132663. [Google Scholar] [CrossRef]
- Wan, H.; Cao, H.; Ning, P. Superiority of the triglyceride glucose index over the homeostasis model in predicting metabolic syndrome based on NHANES data analysis. Sci. Rep. 2024, 14, 15499. [Google Scholar] [CrossRef]
- Kurniawan, L.B. Triglyceride-Glucose Index as A Biomarker of Insulin Resistance, Diabetes Mellitus, Metabolic Syndrome, and Cardiovascular Disease: A Review. Ejifcc 2024, 35, 44–51. [Google Scholar] [PubMed]
- Palmer, N.D.; Stevens, R.D.; Antinozzi, P.A.; Anderson, A.; Bergman, R.N.; Wagenknecht, L.E.; Newgard, C.B.; Bowden, D.W. Metabolomic profile associated with insulin resistance and conversion to diabetes in the Insulin Resistance Atherosclerosis Study. J. Clin. Endocrinol. Metab. 2015, 100, E463–E468. [Google Scholar] [CrossRef] [PubMed]
- Diboun, I.; Al-Mansoori, L.; Al-Jaber, H.; Albagha, O.; Elrayess, M.A. Metabolomics of Lean/Overweight Insulin-Resistant Females Reveals Alterations in Steroids and Fatty Acids. J. Clin. Endocrinol. Metab. 2021, 106, e638–e649. [Google Scholar] [CrossRef]
- Al Thani, A.; Fthenou, E.; Paparrodopoulos, S.; Al Marri, A.; Shi, Z.; Qafoud, F.; Afifi, N. Qatar Biobank Cohort Study: Study Design and First Results. Am. J. Epidemiol. 2019, 188, 1420–1433. [Google Scholar] [CrossRef]
- Thareja, G.; Al-Sarraj, Y.; Belkadi, A.; Almotawa, M.; Suhre, K.; Albagha, O.M.E. Whole genome sequencing in the Middle Eastern Qatari population identifies genetic associations with 45 clinically relevant traits. Nat. Commun. 2021, 12, 1250. [Google Scholar] [CrossRef]
- Suhre, K.; Stephan, N.; Zaghlool, S.; Triggle, C.R.; Robinson, R.J.; Evans, A.M.; Halama, A. Matching Drug Metabolites from Non-Targeted Metabolomics to Self-Reported Medication in the Qatar Biobank Study. Metabolites 2022, 12, 249. [Google Scholar] [CrossRef] [PubMed]
- Zaghlool, S.B.; Halama, A.; Stephan, N.; Gudmundsdottir, V.; Gudnason, V.; Jennings, L.L.; Thangam, M.; Ahlqvist, E.; Malik, R.A.; Albagha, O.M.E.; et al. Metabolic and proteomic signatures of type 2 diabetes subtypes in an Arab population. Nat. Commun. 2022, 13, 7121. [Google Scholar] [CrossRef]
- Ramdas Nayak, V.K.; Satheesh, P.; Shenoy, M.T.; Kalra, S. Triglyceride Glucose (TyG) Index: A surrogate biomarker of insulin resistance. J. Pak. Med. Assoc. 2022, 72, 986–988. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med. Open 2018, 4, 2. [Google Scholar] [CrossRef]
- Evans, A.; Bridgewater, B.; Liu, Q.; Mitchell, M.; Robinson, R.; Dai, H.; Stewart, S.; Dehaven, C.; Miller, L. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics 2014, 4, 2. [Google Scholar] [CrossRef]
- Torres, M.; Parets, S.; Fernández-Díaz, J.; Beteta-Göbel, R.; Rodríguez-Lorca, R.; Román, R.; Lladó, V.; Rosselló, C.A.; Fernández-García, P.; Escribá, P.V. Lipids in Pathophysiology and Development of the Membrane Lipid Therapy: New Bioactive Lipids. Membranes 2021, 11, 919. [Google Scholar] [CrossRef]
- Chang, W.; Hatch, G.M.; Wang, Y.; Yu, F.; Wang, M. The relationship between phospholipids and insulin resistance: From clinical to experimental studies. J. Cell. Mol. Med. 2019, 23, 702–710. [Google Scholar] [CrossRef]
- Wentworth, J.M.; Naselli, G.; Ngui, K.; Smyth, G.K.; Liu, R.; O′Brien, P.E.; Bruce, C.; Weir, J.; Cinel, M.; Meikle, P.J.; et al. GM3 ganglioside and phosphatidylethanolamine-containing lipids are adipose tissue markers of insulin resistance in obese women. Int. J. Obes. 2016, 40, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.J.; Liu, T.T.; Pan, J.C.; Man, Q.Q.; Song, S.; Zhang, J. The Association between the Plasma Phospholipid Profile and Insulin Resistance: A Population-Based Cross-Section Study from the China Adult Chronic Disease and Nutrition Surveillance. Nutrients 2024, 16, 1205. [Google Scholar] [CrossRef] [PubMed]
- Al-Sulaiti, H.; Anwardeen, N.; Bashraheel, S.S.; Naja, K.; Elrayess, M.A. Alterations in Choline Metabolism in Non-Obese Individuals with Insulin Resistance and Type 2 Diabetes Mellitus. Metabolites 2024, 14, 457. [Google Scholar] [CrossRef] [PubMed]
- Razquin, C.; Toledo, E.; Clish, C.B.; Ruiz-Canela, M.; Dennis, C.; Corella, D.; Papandreou, C.; Ros, E.; Estruch, R.; Guasch-Ferré, M.; et al. Plasma Lipidomic Profiling and Risk of Type 2 Diabetes in the PREDIMED Trial. Diabetes Care 2018, 41, 2617–2624. [Google Scholar] [CrossRef]
- Yang, J.; Wang, M.; Yang, D.; Yan, H.; Wang, Z.; Yan, D.; Guo, N. Integrated lipids biomarker of the prediabetes and type 2 diabetes mellitus Chinese patients. Front. Endocrinol. 2023, 13, 1065665. [Google Scholar] [CrossRef]
- Hirano, T. Pathophysiology of Diabetic Dyslipidemia. J. Atheroscler. Thromb. 2018, 25, 771–782. [Google Scholar] [CrossRef]
- Zhao, J.; Zhu, Y.; Hyun, N.; Zeng, D.; Uppal, K.; Tran, V.T.; Yu, T.; Jones, D.; He, J.; Lee, E.T.; et al. Novel metabolic markers for the risk of diabetes development in American Indians. Diabetes Care 2015, 38, 220–227. [Google Scholar] [CrossRef]
- Jacobs, R.L.; Devlin, C.; Tabas, I.; Vance, D.E. Targeted deletion of hepatic CTP: Phosphocholine cytidylyltransferase alpha in mice decreases plasma high density and very low density lipoproteins. J. Biol. Chem. 2004, 279, 47402–47410. [Google Scholar] [CrossRef]
- Funai, K.; Lodhi, I.J.; Spears, L.D.; Yin, L.; Song, H.; Klein, S.; Semenkovich, C.F. Skeletal Muscle Phospholipid Metabolism Regulates Insulin Sensitivity and Contractile Function. Diabetes 2016, 65, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Jiang, X.C. Sphingomyelin and its role in cellular signaling. Adv. Exp. Med. Biol. 2013, 991, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Green, C.D.; Maceyka, M.; Cowart, L.A.; Spiegel, S. Sphingolipids in metabolic disease: The good, the bad, and the unknown. Cell. Metab. 2021, 33, 1293–1306. [Google Scholar] [CrossRef]
- Griess, K.; Rieck, M.; Müller, N.; Karsai, G.; Hartwig, S.; Pelligra, A.; Hardt, R.; Schlegel, C.; Kuboth, J.; Uhlemeyer, C.; et al. Sphingolipid subtypes differentially control proinsulin processing and systemic glucose homeostasis. Nat. Cell. Biol. 2023, 25, 20–29. [Google Scholar] [CrossRef]
- Straczkowski, M.; Kowalska, I.; Nikolajuk, A.; Dzienis-Straczkowska, S.; Kinalska, I.; Baranowski, M.; Zendzian-Piotrowska, M.; Brzezinska, Z.; Gorski, J. Relationship between insulin sensitivity and sphingomyelin signaling pathway in human skeletal muscle. Diabetes 2004, 53, 1215–1221. [Google Scholar] [CrossRef]
- Semba, R.D.; Gonzalez-Freire, M.; Moaddel, R.; Sun, K.; Fabbri, E.; Zhang, P.; Carlson, O.D.; Khadeer, M.; Chia, C.W.; Salem, N., Jr.; et al. Altered Plasma Amino Acids and Lipids Associated with Abnormal Glucose Metabolism and Insulin Resistance in Older Adults. J. Clin. Endocrinol. Metab. 2018, 103, 3331–3339. [Google Scholar] [CrossRef]
- Hanamatsu, H.; Ohnishi, S.; Sakai, S.; Yuyama, K.; Mitsutake, S.; Takeda, H.; Hashino, S.; Igarashi, Y. Altered levels of serum sphingomyelin and ceramide containing distinct acyl chains in young obese adults. Nutr. Diabetes 2014, 4, e141. [Google Scholar] [CrossRef]
- Sokolowska, E.; Blachnio-Zabielska, A. The Role of Ceramides in Insulin Resistance. Front. Endocrinol. 2019, 10, 577. [Google Scholar] [CrossRef] [PubMed]
- Reali, F.; Morine, M.J.; Kahramanoğulları, O.; Raichur, S.; Schneider, H.-C.; Crowther, D.; Priami, C. Mechanistic interplay between ceramide and insulin resistance. Sci. Rep. 2017, 7, 41231. [Google Scholar] [CrossRef]
- Bandet, C.L.; Mahfouz, R.; Véret, J.; Sotiropoulos, A.; Poirier, M.; Giussani, P.; Campana, M.; Philippe, E.; Blachnio-Zabielska, A.; Ballaire, R.; et al. Ceramide Transporter CERT Is Involved in Muscle Insulin Signaling Defects Under Lipotoxic Conditions. Diabetes 2018, 67, 1258–1271. [Google Scholar] [CrossRef]
- Kolak, M.; Gertow, J.; Westerbacka, J.; Summers, S.A.; Liska, J.; Franco-Cereceda, A.; Orešič, M.; Yki-Järvinen, H.; Eriksson, P.; Fisher, R.M. Expression of ceramide-metabolising enzymes in subcutaneous and intra-abdominal human adipose tissue. Lipids Health Dis. 2012, 11, 115. [Google Scholar] [CrossRef]
- Zemski Berry, K.A.; Garfield, A.; Jambal, P.; Zarini, S.; Perreault, L.; Bergman, B.C. Oxidised phosphatidylcholine induces sarcolemmal ceramide accumulation and insulin resistance in skeletal muscle. Diabetologia 2024, 67, 2819–2832. [Google Scholar] [CrossRef] [PubMed]
- Bozelli, J.C.; Azher, S.; Epand, R.M. Plasmalogens and Chronic Inflammatory Diseases. Front. Physiol. 2021, 12, 730829. [Google Scholar] [CrossRef] [PubMed]
- Almsherqi, Z.A. Potential Role of Plasmalogens in the Modulation of Biomembrane Morphology. Front. Cell. Dev. Biol. 2021, 9, 673917. [Google Scholar] [CrossRef] [PubMed]
- Beyene, H.B.; Huynh, K.; Wang, T.; Paul, S.; Cinel, M.; Mellett, N.A.; Olshansky, G.; Meikle, T.G.; Watts, G.F.; Hung, J.; et al. Development and validation of a plasmalogen score as an independent modifiable marker of metabolic health: Population based observational studies and a placebo-controlled cross-over study. eBioMedicine 2024, 105, 105187. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Di, J.; Zhang, X.; Liu, Y.; Zhang, Y.; Li, B.; Qi, S.; Cao, X.; Liu, L.; et al. High coverage of targeted lipidomics revealed lipid changes in the follicular fluid of patients with insulin-resistant polycystic ovary syndrome and a positive correlation between plasmalogens and oocyte quality. Front. Endocrinol. 2024, 15, 1414289. [Google Scholar] [CrossRef]
- Tonks, K.T.; Coster, A.C.; Christopher, M.J.; Chaudhuri, R.; Xu, A.; Gagnon-Bartsch, J.; Chisholm, D.J.; James, D.E.; Meikle, P.J.; Greenfield, J.R.; et al. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity 2016, 24, 908–916. [Google Scholar] [CrossRef]


| Test | Variable | Insulin-Sensitive Group (N = 620) | Insulin-Resistant Group (N = 635) | p-Value |
|---|---|---|---|---|
| General characteristics | Sex (M/F) | 322/298 | 349/286 | 0.3 |
| Age | 35 (29–43.25) | 47 (41–54.5) | <0.0001 | |
| BMI (kg/m2) | 27.74 (24.7–31.76) | 30.48 (27.26–34.12) | <0.0001 | |
| Waist to hip ratio | 0.8 (0.75–0.86) | 0.9 (0.83–0.96) | <0.0001 | |
| Systolic blood pressure (mmHg) | 109 (102–118) | 121 (112–131) | <0.0001 | |
| Diastolic blood pressure (mmHg) | 71 (66–77.5) | 78 (72–85) | <0.0001 | |
| Insulin resistance | TyG index | 8 (7.83–8.16) | 9.21 (9.01–9.52) | <0.0001 |
| Blood sugar | Fasting blood glucose (mmol/L) | 4.9 (4.6–5.16) | 5.9 (5.1–8.1) | <0.0001 |
| HbA1C (%) | 5.3 (5.1–5.6) | 5.9 (5.5–6.8) | <0.0001 | |
| C-peptide (ng/mL) | 1.88 (1.39–2.52) | 3.39 (2.49–4.84) | <0.0001 | |
| Insulin (uU/mL) | 7.2 (5–11) | 17 (10.1–30) | <0.0001 | |
| Lipid profile | Total cholesterol (mmol/L) | 4.6 (4.14–5.2) | 5.23 (4.56–5.9) | <0.0001 |
| HDL-cholesterol (mmol/L) | 1.46 (1.23–1.73) | 1.11 (0.96–1.31) | <0.0001 | |
| LDL-cholesterol (mmol/L) | 2.9 (2.19–3.24) | 3 (2.46–3.83) | <0.0001 | |
| Triglyceride (mmol/L) | 0.77 (0.63–0.9) | 2.04 (1.68–2.6) | <0.0001 | |
| Cardiac function | NT-proBNP (pg/mL) | 27 (14.78–44) | 20.6 (11.83–38.15) | 0.0003 |
| Homocysteine (µmol/L) | 8.4 (7–10.25) | 8.3 (6.8–10.1) | 0.3261 | |
| Kidney function | Chloride (mmol/L) | 101 (100–103) | 101 (99–102) | <0.0001 |
| Urea (mmol/L) | 4.2 (3.5–5) | 4.5 (3.7–5.3) | 0.0001 | |
| Bicarbonate (mmol/L) | 27 (25–28) | 27 (25–28) | 0.6 | |
| Total protein (g/L) | 73 (70–75) | 73 (70–75) | 0.7 | |
| Liver function | Albumin (g/L) | 45 (43–47) | 45 (43–47) | 0.0034 |
| Bilirubin (µmol/L) | 6.6 (5–9) | 5.9 (4–8) | <0.0001 | |
| ALT (U/L) | 16 (12–23) | 22 (16–32) | <0.0001 | |
| AST (U/L) | 17 (14–21) | 18 (15–23) | 0.0092 |
| Metabolite | Superpathway | Subpathway | Estimate | SE | p-Value | FDR |
|---|---|---|---|---|---|---|
| 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) | Lipid | Phosphatidylethanolamine | −0.72 | 0.033 | 5.5 × 10−84 | 1.6 × 10−81 |
| 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4) | Lipid | Phosphatidylethanolamine | −0.57 | 0.026 | 7.2 × 10−82 | 1.6 × 10−79 |
| 1-palmitoyl-2-linoleoyl-GPE (16:0/18:2) | Lipid | Phosphatidylethanolamine | −0.66 | 0.031 | 3.6 × 10−77 | 5.2 × 10−75 |
| 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) | Lipid | Phosphatidylethanolamine | −0.64 | 0.030 | 9.0 × 10−77 | 1.1 × 10−74 |
| 1-stearoyl-2-oleoyl-GPE (18:0/18:1) | Lipid | Phosphatidylethanolamine | −0.67 | 0.032 | 3.0 × 10−76 | 3.3 × 10−74 |
| 1-stearoyl-2-arachidonoyl-GPE (18:0/20:4) | Lipid | Phosphatidylethanolamine | −0.51 | 0.026 | 3.5 × 10−68 | 2.8 × 10−66 |
| 1-(1-enyl-palmitoyl)-2-oleoyl-GPC (P-16:0/18:1) | Lipid | Plasmalogen | 0.36 | 0.020 | 1.1 × 10−59 | 7.9 × 10−58 |
| 1-palmitoyl-2-docosahexaenoyl-GPE (16:0/22:6) | Lipid | Phosphatidylethanolamine | −0.60 | 0.035 | 1.1 × 10−54 | 6.8 × 10−53 |
| 1-(1-enyl-palmitoyl)-2-palmitoleoyl-GPC (P-16:0/16:1) | Lipid | Plasmalogen | 0.34 | 0.021 | 4.7 × 10−47 | 2.6 × 10−45 |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPC (P-16:0/18:2) | Lipid | Plasmalogen | 0.30 | 0.019 | 3.2 × 10−46 | 1.7 × 10−44 |
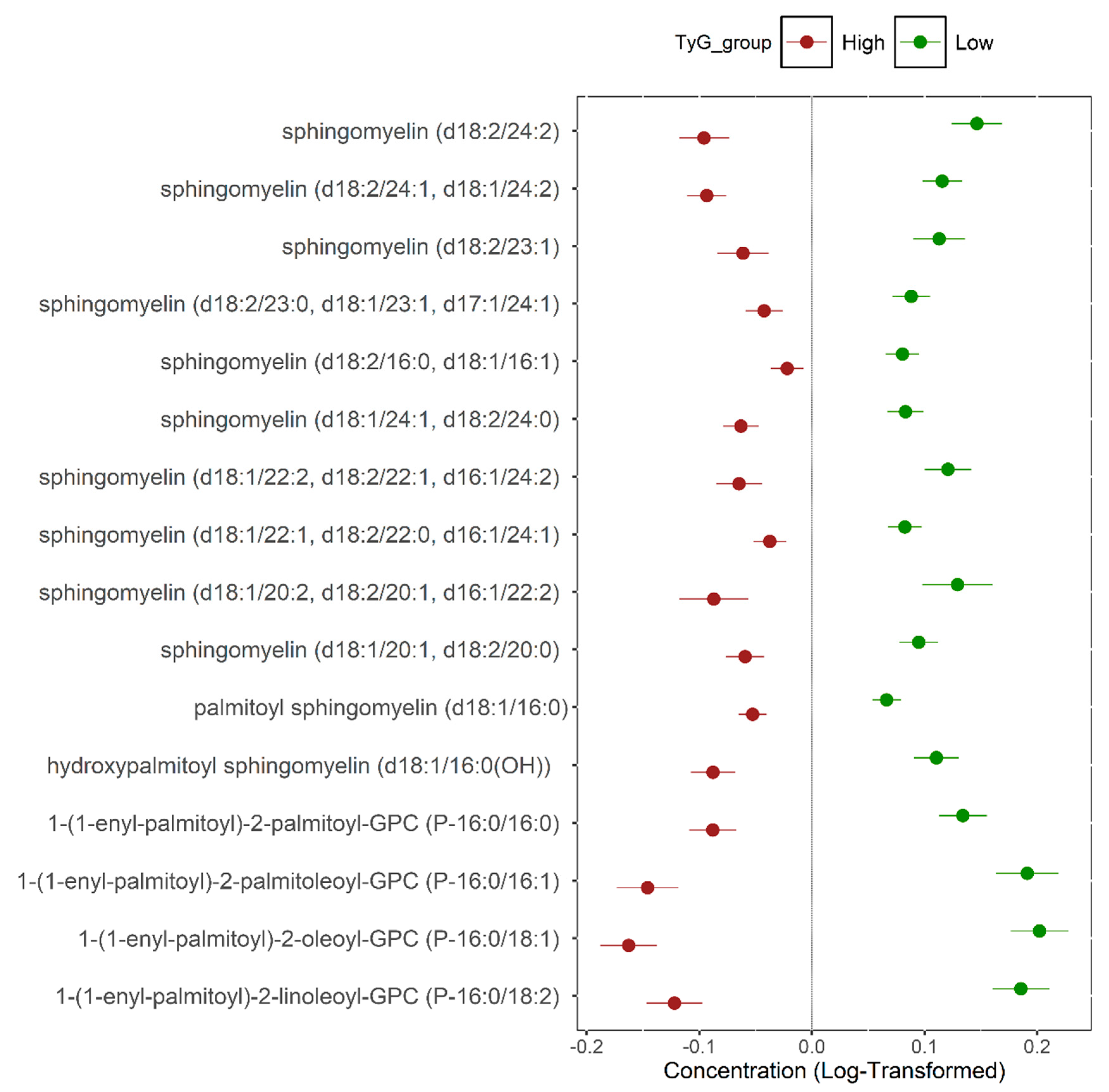
| Sphingomyelin (d18:2/24:1, d18:1/24:2) | Lipid | Sphingomyelins | 0.21 | 0.013 | 5.9 × 10−46 | 2.9 × 10−44 |
| Sphingomyelin (d18:2/24:2) | Lipid | Sphingomyelins | 0.25 | 0.017 | 9.8 × 10−41 | 4.1 × 10−39 |
| 1-palmitoyl-2-palmitoleoyl-GPC (16:0/16:1) | Lipid | Phosphatidylcholine | −0.39 | 0.029 | 6.0 × 10−36 | 2.2 × 10−34 |
| 1-(1-enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0) | Lipid | Plasmalogen | 0.22 | 0.016 | 1.9 × 10−35 | 6.6 × 10−34 |
| 1-palmitoyl-2-arachidonoyl-GPI (16:0/20:4) | Lipid | Phosphatidylinositol | −0.37 | 0.028 | 1.3 × 10−34 | 4.3 × 10−33 |
| Hydroxypalmitoyl sphingomyelin (d18:1/16:0(OH)) | Lipid | Sphingomyelins | 0.20 | 0.015 | 2.5 × 10−32 | 7.5 × 10−31 |
| 1-myristoyl-2-arachidonoyl-GPC (14:0/20:4) | Lipid | Phosphatidylcholine | −0.43 | 0.036 | 5.9 × 10−32 | 1.7 × 10−30 |
| Sphingomyelin (d18:1/22:2, d18:2/22:1, d16:1/24:2) | Lipid | Sphingomyelins | 0.19 | 0.017 | 4.7 × 10−29 | 1.3 × 10−27 |
| Sphingomyelin (d18:1/20:1, d18:2/20:0) | Lipid | Sphingomyelins | 0.16 | 0.014 | 4.7 × 10−28 | 1.3 × 10−27 |
| Sphingomyelin (d18:1/24:1, d18:2/24:0) | Lipid | Sphingomyelins | 0.15 | 0.013 | 8.7 × 10−29 | 2.3 × 10−27 |
| Palmitoyl sphingomyelin (d18:1/16:0) | Lipid | Sphingomyelins | 0.12 | 0.010 | 3.0 × 10−28 | 7.7 × 10−27 |
| Glucose | Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | −0.22 | 0.020 | 8.2 × 10−28 | 2.1 × 10−26 |
| 1-palmitoyl-2-dihomo-linolenoyl-GPC (16:0/20:3n3 or 6) | Lipid | Phosphatidylcholine | −0.28 | 0.025 | 1.3 × 10−27 | 3.1 × 10−26 |
| 1-stearoyl-2-arachidonoyl-GPI (18:0/20:4) | Lipid | Phosphatidylinositol | −0.21 | 0.019 | 1.4 × 10−27 | 3.4 × 10−26 |
| 1-palmitoyl-2-arachidonoyl-GPC (16:0/20:4n6) | Lipid | Phosphatidylcholine | −0.19 | 0.018 | 9.9 × 10−26 | 2.1 × 10−24 |
| 1-palmitoyl-2-oleoyl-GPI (16:0/18:1) | Lipid | Phosphatidylinositol | −0.30 | 0.029 | 1.7 × 10−24 | 3.5 × 10−23 |
| 1-palmitoyl-2-linoleoyl-GPI (16:0/18:2) | Lipid | Phosphatidylinositol | −0.28 | 0.027 | 3.0 × 10−24 | 6.0 × 10−23 |
| N-Lactoyl phenylalanine | Amino Acid | Phenylalanine Metabolism | −0.28 | 0.028 | 7.7 × 10−23 | 1.4 × 10−21 |
| Sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1) | Lipid | Sphingomyelins | 0.12 | 0.012 | 2.5 × 10−22 | 4.5 × 10−21 |
| Sphingomyelin (d18:2/23:1) | Lipid | Sphingomyelins | 0.18 | 0.018 | 2.8 × 10−22 | 4.8 × 10−21 |
| Pyruvate | Carbohydrate | Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | −0.16 | 0.016 | 3.3 × 10−22 | 5.5 × 10−21 |
| Sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) | Lipid | Sphingomyelins | 0.13 | 0.013 | 3.8 × 10−22 | 6.4 × 10−21 |
| 1-palmitoyl-2-oleoyl-GPC (16:0/18:1) | Lipid | Phosphatidylcholine | −0.16 | 0.016 | 5.2 × 10−22 | 8.5 × 10−21 |
| Sphingomyelin (d18:1/20:2, d18:2/20:1, d16:1/22:2) | Lipid | Sphingomyelins | 0.22 | 0.025 | 4.3 × 10−18 | 5.5 × 10−17 |
| 1-myristoyl-2-palmitoyl-GPC (14:0/16:0) | Lipid | Phosphatidylcholine | −0.32 | 0.037 | 1.7 × 10−17 | 2.1 × 10−16 |
| Sphingomyelin (d18:2/16:0, d18:1/16:1) | Lipid | Sphingomyelins | 0.10 | 0.012 | 2.8 × 10−17 | 3.3 × 10−16 |
| Enriched Pathways | p-Value | FDR |
|---|---|---|
| Sphingomyelins | 2.8 × 10−7 | 2.8 × 10−5 |
| Phosphatidylethanolamine (PE) | 3.6 × 10−6 | 1.7 × 10−4 |
| Plasmalogen | 8.6 × 10−6 | 2.8 × 10−4 |
| Phosphatidylcholine (PC) | 7.1 × 10−5 | 1.1 × 10−3 |
| Phosphatidylinositol (PI) | 3.2 × 10−4 | 4.5 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naja, K.; Anwardeen, N.; Albagha, O.; Elrayess, M.A. Lipid Subclasses Differentiate Insulin Resistance by Triglyceride–Glucose Index. Metabolites 2025, 15, 342. https://doi.org/10.3390/metabo15050342
Naja K, Anwardeen N, Albagha O, Elrayess MA. Lipid Subclasses Differentiate Insulin Resistance by Triglyceride–Glucose Index. Metabolites. 2025; 15(5):342. https://doi.org/10.3390/metabo15050342
Chicago/Turabian StyleNaja, Khaled, Najeha Anwardeen, Omar Albagha, and Mohamed A. Elrayess. 2025. "Lipid Subclasses Differentiate Insulin Resistance by Triglyceride–Glucose Index" Metabolites 15, no. 5: 342. https://doi.org/10.3390/metabo15050342
APA StyleNaja, K., Anwardeen, N., Albagha, O., & Elrayess, M. A. (2025). Lipid Subclasses Differentiate Insulin Resistance by Triglyceride–Glucose Index. Metabolites, 15(5), 342. https://doi.org/10.3390/metabo15050342









