Early Gestational Hepatic Lipidomic Profiles Are Modulated by One-Carbon Metabolite Supplementation and Nutrient Restriction in Beef Heifers and Fetuses
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Ethics Statement
2.2. Sample Collection, Storage and Preparation
2.3. Quality Control
2.4. Ultrahigh-Performance Liquid Chromatography–Tandem Mass Spectroscopy (UPLC-MS/MS)
2.5. Data Extraction and Compound Identification
2.6. Statistical Analysis
3. Results
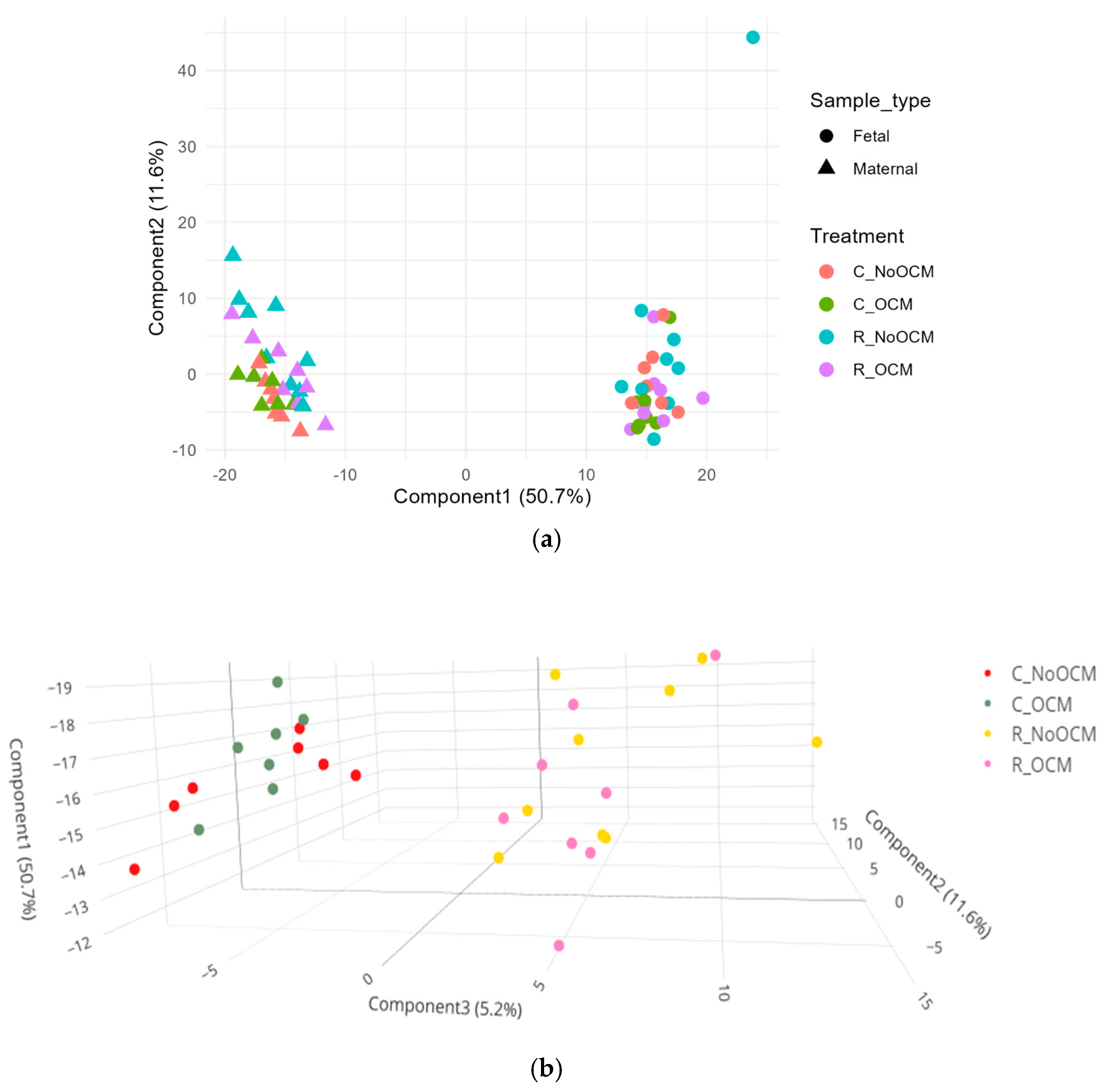
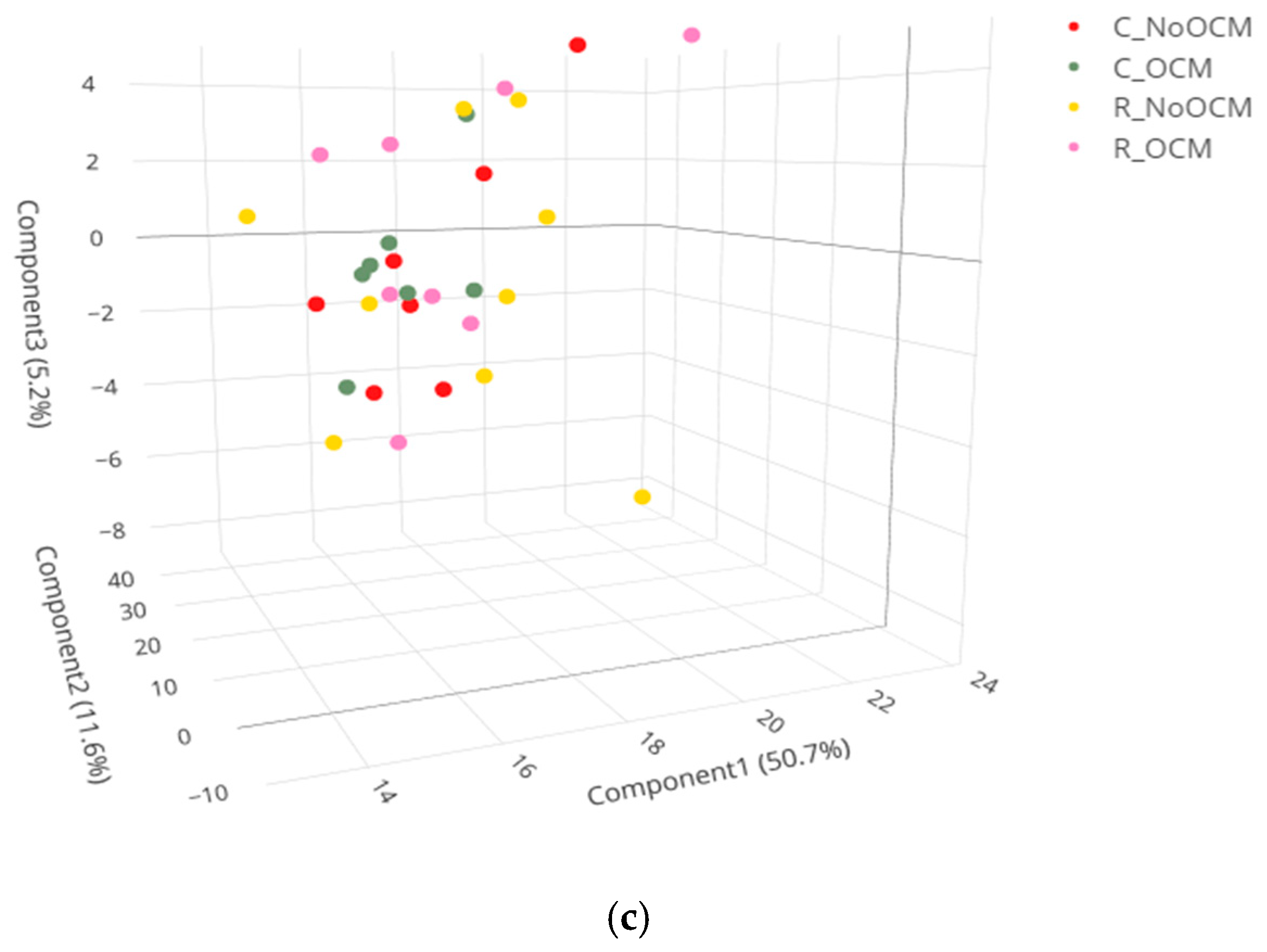
3.1. Principal Component Analysis (PCA) of Lipid Profiles
3.2. Metabolic Pathway Enrichment Analysis
3.3. Statistical Analysis of Main Effects and Interactions in Fetal Liver
3.4. Statistical Analysis of Main Effects and Interactions in Maternal Liver
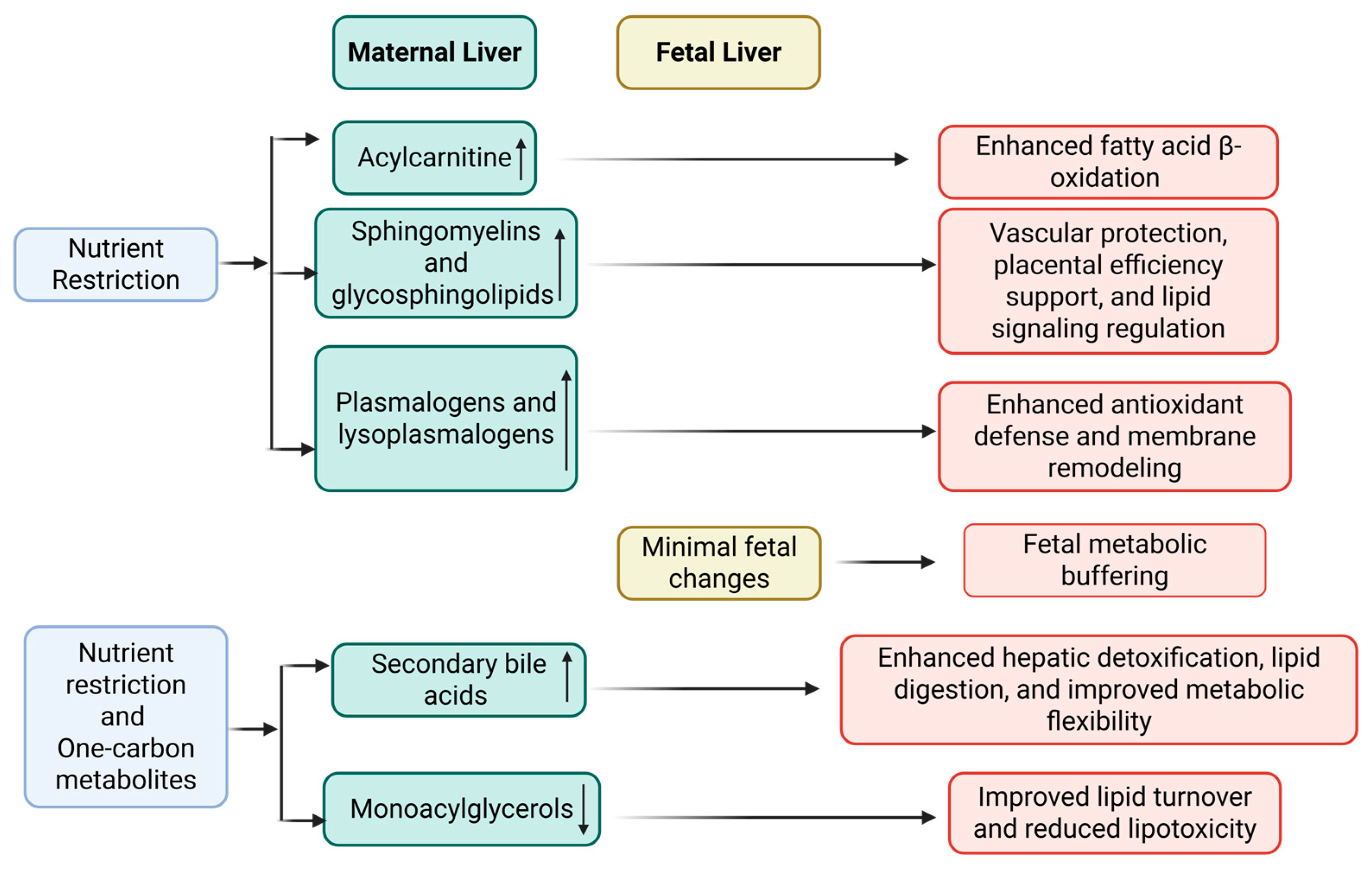
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADG | Average daily gain |
| CON | Control |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| OCM | One-carbon metabolism |
| PCA | Principal component analysis |
| MAG | Monoacylglycerol |
| S1P | Sphingosine-1-phosphate |
| FAO | Fatty acid β-oxidation |
| LCER | Lactosylceramides |
| UPLC | Ultra-performance liquid chromatography |
| VLDL | Very-low-density lipoprotein |
| SAM | S-Adenosylmethionine |
| HCER | Hexosylceramides |
| RES | Restricted |
References
- Malarkey, D.E.; Johnson, K.; Ryan, L.; Boorman, G.; Maronpot, R.R. New insights into functional aspects of liver morphology. Toxicol. Pathol. 2005, 33, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Parrettini, S.; Caroli, A.; Torlone, E. Nutrition and metabolic adaptations in physiological and complicated pregnancy: Focus on obesity and gestational diabetes. Front. Endocrinol. 2020, 11, 611929. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, I.; Galan, X.; Peinado-Onsurbc, J.; Llobera, M. Feta! and Neonatal Physiology; Polin, R.A., Fox, W.W., Eds.; WB Saunder Co.: Philadelphia, PA, USA, 1998; pp. 535–541. [Google Scholar]
- Herrera, E. Implications of dietary fatty acids during pregnancy on placental, fetal and postnatal development—A review. Placenta 2002, 23, S9–S19. [Google Scholar] [CrossRef] [PubMed]
- Okala, S.G.; Sise, E.A.; Sosseh, F.; Prentice, A.M.; Woollett, L.A.; Moore, S.E. Maternal plasma lipid levels across pregnancy and the risks of small-for-gestational age and low birth weight: A cohort study from rural Gambia. BMC Pregnancy Childbirth 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Kramer, A.C.; Jansson, T.; Bale, T.L.; Powell, T.L. Maternal-fetal cross-talk via the placenta: Influence on offspring development and metabolism. Development 2023, 150, dev202088. [Google Scholar] [CrossRef]
- Reynolds, L.; Dahlen, C.; Ward, A.; Crouse, M.; Borowicz, P.; Davila-Ruiz, B.; Kanjanaruch, C.; Bochantin, K.; McLean, K.; McCarthy, K. Role of the placenta in developmental programming: Observations from models using large animals. Anim. Reprod. Sci. 2023, 257, 107322. [Google Scholar] [CrossRef]
- Varvarigou, A.A. Intrauterine growth restriction as a potential risk factor for disease onset in adulthood. J. Pediatr. Endocrinol. Metab. 2010, 23, 215–224. [Google Scholar] [CrossRef]
- Caton, J.S.; Crouse, M.S.; McLean, K.J.; Dahlen, C.R.; Ward, A.K.; Cushman, R.A.; Grazul-Bilska, A.T.; Neville, B.W.; Borowicz, P.P.; Reynolds, L.P. Maternal periconceptual nutrition, early pregnancy, and developmental outcomes in beef cattle. J. Anim. Sci. 2020, 98, skaa358. [Google Scholar] [CrossRef]
- Roche, J.F. The effect of nutritional management of the dairy cow on reproductive efficiency. Anim. Reprod. Sci. 2006, 96, 282–296. [Google Scholar] [CrossRef]
- Clare, C.E.; Brassington, A.H.; Kwong, W.Y.; Sinclair, K.D. One-carbon metabolism: Linking nutritional biochemistry to epigenetic programming of long-term development. Annu. Rev. Anim. Biosci. 2019, 7, 263–287. [Google Scholar] [CrossRef]
- Clare, C.E. One-Carbon Metabolism and Epigenetic Programming of Mammalian Development. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2021. [Google Scholar]
- Safain, K.S.; Crouse, M.S.; Syring, J.G.; Entzie, Y.L.; King, L.E.; Hirchert, M.R.; Ward, A.K.; Reynolds, L.P.; Borowicz, P.P.; Dahlen, C.R. One-carbon metabolites supplementation and nutrient restriction alter the fetal liver metabolomic profile during early gestation in beef heifers. J. Anim. Sci. 2024, 102, skae258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Garrow, T.A.; Dong, X.; Luchini, D.N.; Loor, J.J. Hepatic activity and transcription of betaine-homocysteine methyltransferase, methionine synthase, and cystathionine synthase in periparturient dairy cows are altered to different extents by supply of methionine and choline. J. Nutr. 2017, 147, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Młodzik-Czyżewska, M.A.; Szwengiel, A.; Malinowska, A.M.; Chmurzynska, A. Comparison of associations between one-carbon metabolism, lipid metabolism, and fatty liver markers in normal-weight and overweight people aged 20–40 years. Ann. Nutr. Metab. 2021, 77, 221–230. [Google Scholar] [CrossRef]
- Syring, J.G.; Crouse, M.S.; Entzie, Y.L.; King, L.E.; Hirchert, M.R.; Ward, A.K.; Reynolds, L.P.; Borowicz, P.P.; Dahlen, C.R.; Caton, J.S. One-carbon metabolite supplementation increases vitamin B12, folate, and methionine cycle metabolites in beef heifers and fetuses in an energy dependent manner at day 63 of gestation. J. Anim. Sci. 2024, 102, skae202. [Google Scholar] [CrossRef]
- Daneshi, M.; Borowicz, P.P.; Entzie, Y.L.; Syring, J.G.; King, L.E.; Safain, K.S.; Anas, M.; Reynolds, L.P.; Ward, A.K.; Dahlen, C.R. Influence of Maternal Nutrition and One-Carbon Metabolites Supplementation during Early Pregnancy on Bovine Fetal Small Intestine Vascularity and Cell Proliferation. Vet. Sci. 2024, 11, 146. [Google Scholar] [CrossRef]
- Crouse, M.S.; McCarthy, K.L.; Menezes, A.C.B.; Kassetas, C.J.; Baumgaertner, F.; Kirsch, J.D.; Dorsam, S.; Neville, T.L.; Ward, A.K.; Borowicz, P.P. Vitamin and mineral supplementation and rate of weight gain during the first trimester of gestation in beef heifers alters the fetal liver amino acid, carbohydrate, and energy profile at day 83 of gestation. Metabolites 2022, 12, 696. [Google Scholar] [CrossRef]
- Menezes, A.C.B.; Dahlen, C.R.; McCarthy, K.L.; Kassetas, C.J.; Baumgaertner, F.; Kirsch, J.D.; Dorsam, S.T.; Neville, T.L.; Ward, A.K.; Borowicz, P.P. Fetal Hepatic Lipidome Is More Greatly Affected by Maternal Rate of Gain Compared with Vitamin and Mineral Supplementation at Day 83 of Gestation. Metabolites 2023, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Mosley, J.D.; Schock, T.B.; Beecher, C.W.; Dunn, W.B.; Kuligowski, J.; Lewis, M.R.; Theodoridis, G.; Ulmer Holland, C.Z.; Vuckovic, D.; Wilson, I.D. Establishing a framework for best practices for quality assurance and quality control in untargeted metabolomics. Metabolomics 2024, 20, 20. [Google Scholar] [CrossRef]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef]
- Simintiras, C.A.; Sánchez, J.M.; McDonald, M.; Martins, T.; Binelli, M.; Lonergan, P. Biochemical characterization of progesterone-induced alterations in bovine uterine fluid amino acid and carbohydrate composition during the conceptus elongation window. Biol. Reprod. 2019, 100, 672–685. [Google Scholar] [CrossRef]
- Bjørndal, B.; Alterås, E.K.; Lindquist, C.; Svardal, A.; Skorve, J.; Berge, R.K. Associations between fatty acid oxidation, hepatic mitochondrial function, and plasma acylcarnitine levels in mice. Nutr. Metab. 2018, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Augé, N.; Nègre-Salvayre, A.; Salvayre, R.; Levade, T. Sphingomyelin metabolites in vascular cell signaling and atherogenesis. Prog. Lipid Res. 2000, 39, 207–229. [Google Scholar] [CrossRef] [PubMed]
- Brosche, T.; Platt, D. The biological significance of plasmalogens in defense against oxidative damage. Exp. Gerontol. 1998, 33, 363–369. [Google Scholar] [CrossRef]
- Stella, S.L.; Guadagnin, A.R.; Velasco-Acosta, D.A.; Ferreira, C.R.; Rubessa, M.; Wheeler, M.B.; Luchini, D.; Cardoso, F.C. Rumen-protected methionine supplementation alters lipid profile of preimplantation embryo and endometrial tissue of Holstein cows. Front. Vet. Sci. 2024, 10, 1301986. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Khan, M.Z.; Kou, X.; Chen, Y.; Liang, H.; Ullah, Q.; Khan, N.; Khan, A.; Chai, W.; Wang, C. Enhancing Metabolism and Milk Production Performance in Periparturient Dairy Cattle through Rumen-Protected Methionine and Choline Supplementation. Metabolites 2023, 13, 1080. [Google Scholar] [CrossRef]
- Pate, R.T.; Luchini, D.; Cant, J.P.; Baumgard, L.H.; Cardoso, F.C. Immune and metabolic effects of rumen-protected methionine during a heat stress challenge in lactating Holstein cows. J. Anim. Sci. 2021, 99, skab323. [Google Scholar] [CrossRef]
- Vance, D.E. Phospholipid methylation in mammals: From biochemistry to physiological function. Biochim. Biophys. Acta-Biomembr. 2014, 1838, 1477–1487. [Google Scholar] [CrossRef]
- Potts, S.; Brady, K.; Scholte, C.; Moyes, K.; Sunny, N.; Erdman, R. Rumen-protected choline and methionine during the periparturient period affect choline metabolites, amino acids, and hepatic expression of genes associated with one-carbon and lipid metabolism. J. Dairy Sci. 2023, 106, 4559–4579. [Google Scholar] [CrossRef]
- Herrera, E. Metabolic adaptations in pregnancy and their implications for the availability of substrates to the fetus. Eur. J. Clin. Nutr. 2000, 54, S47–S51. [Google Scholar] [CrossRef]
- Li, C.; Schlabritz-Loutsevitch, N.E.; Hubbard, G.B.; Han, V.; Nygard, K.; Cox, L.A.; McDonald, T.J.; Nathanielsz, P.W. Effects of maternal global nutrient restriction on fetal baboon hepatic insulin-like growth factor system genes and gene products. Endocrinology 2009, 150, 4634–4642. [Google Scholar] [CrossRef]
- Schooneman, M.G.; Vaz, F.M.; Houten, S.M.; Soeters, M.R. Acylcarnitines: Reflecting or inflicting insulin resistance? Diabetes 2013, 62, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.H.; Hoppel, C.L.; Lok, K.H.; Zhao, L.; Wong, S.W.; Minkler, P.E.; Hwang, D.H.; Newman, J.W.; Garvey, W.T. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid β-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J. Nutr. 2009, 139, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Ryckman, K.K.; Donovan, B.M.; Fleener, D.K.; Bedell, B.; Borowski, K.S. Pregnancy-related changes of amino acid and acylcarnitine concentrations: The impact of obesity. Am. J. Perinatol. Rep. 2016, 6, e329–e336. [Google Scholar]
- Sandler, V.; Reisetter, A.C.; Bain, J.R.; Muehlbauer, M.J.; Nodzenski, M.; Stevens, R.D.; Ilkayeva, O.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B. Associations of maternal BMI and insulin resistance with the maternal metabolome and newborn outcomes. Diabetologia 2017, 60, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Pomar, C.A.; Kuda, O.; Kopecky, J.; Rombaldova, M.; Castro, H.; Picó, C.; Sánchez, J.; Palou, A. Alterations in plasma acylcarnitine and amino acid profiles may indicate poor nutrition during the suckling period due to maternal intake of an unbalanced diet and may predict later metabolic dysfunction. FASEB J. 2019, 33, 796–807. [Google Scholar] [CrossRef]
- Virmani, M.A.; Cirulli, M. The role of l-carnitine in mitochondria, prevention of metabolic inflexibility and disease initiation. Int. J. Mol. Sci. 2022, 23, 2717. [Google Scholar] [CrossRef]
- Iessi, E.; Marconi, M.; Manganelli, V.; Sorice, M.; Malorni, W.; Garofalo, T.; Matarrese, P. On the role of sphingolipids in cell survival and death. Int. Rev. Cell Mol. Biol. 2020, 351, 149–195. [Google Scholar]
- Chen, L.; Mir, S.A.; Bendt, A.K.; Chua, E.W.; Narasimhan, K.; Tan, K.M.-L.; Loy, S.L.; Tan, K.H.; Shek, L.P.; Chan, J. Plasma lipidomic profiling reveals metabolic adaptations to pregnancy and signatures of cardiometabolic risk: A preconception and longitudinal cohort study. BMC Med. 2023, 21, 53. [Google Scholar]
- Del Gaudio, I.; Sasset, L.; Di Lorenzo, A.; Wadsack, C. Sphingolipid signature of human feto-placental vasculature in preeclampsia. Int. J. Mol. Sci. 2020, 21, 1019. [Google Scholar] [CrossRef]
- Mizugishi, K.; Li, C.; Olivera, A.; Bielawski, J.; Bielawska, A.; Deng, C.-X.; Proia, R.L. Maternal disturbance in activated sphingolipid metabolism causes pregnancy loss in mice. J. Clin. Investig. 2007, 117, 2993–3006. [Google Scholar] [CrossRef]
- Braverman, N.E.; Moser, A.B. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta-Mol. Basis Dis. 2012, 1822, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.C. Free-radical-induced oxidation of arachidonoyl plasmalogen phospholipids: Antioxidant mechanism and precursor pathway for bioactive eicosanoids. Chem. Res. Toxicol. 2001, 14, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-C.; Pfeiffer, D.R.; Calhoon, E.A.; Madiai, F.; Marcucci, G.; Liu, S.; Jurkowitz, M.S. Purification, identification, and cloning of lysoplasmalogenase, the enzyme that catalyzes hydrolysis of the vinyl ether bond of lysoplasmalogen. J. Biol. Chem. 2011, 286, 24916–24930. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.E.; Park, H.S.; Yoo, H.J.; Baek, I.J.; Yoon, J.E.; Ko, M.S.; Kim, A.R.; Kim, H.S.; Park, H.S.; Lee, S.E. Protective role of endogenous plasmalogens against hepatic steatosis and steatohepatitis in mice. Hepatology 2017, 66, 416–431. [Google Scholar] [CrossRef]
- Karantonis, H.C.; Nomikos, T.; Demopoulos, C.A. Triacylglycerol metabolism. Curr. Drug Targets 2009, 10, 302–319. [Google Scholar] [CrossRef]
- Staels, B.; Fonseca, V.A. Bile acids and metabolic regulation: Mechanisms and clinical responses to bile acid sequestration. Diabetes Care 2009, 32, S237. [Google Scholar] [CrossRef]
- Kars, M.; Yang, L.; Gregor, M.F.; Mohammed, B.S.; Pietka, T.A.; Finck, B.N.; Patterson, B.W.; Horton, J.D.; Mittendorfer, B.; Hotamisligil, G.S. Tauroursodeoxycholic acid may improve liver and muscle but not adipose tissue insulin sensitivity in obese men and women. Diabetes 2010, 59, 1899–1905. [Google Scholar] [CrossRef]
- Moghimipour, E.; Ameri, A.; Handali, S. Absorption-enhancing effects of bile salts. Molecules 2015, 20, 14451–14473. [Google Scholar] [CrossRef]
- Pfeiffer, L.; Wahl, S.; Pilling, L.C.; Reischl, E.; Sandling, J.K.; Kunze, S.; Holdt, L.M.; Kretschmer, A.; Schramm, K.; Adamski, J. DNA methylation of lipid-related genes affects blood lipid levels. Circ. Cardiovasc. Genet. 2015, 8, 334–342. [Google Scholar] [CrossRef]
| Sub-Pathway | Pathway Enrichment Score 1 | ||
|---|---|---|---|
| Gain | OCM | Gain × OCM | |
| Fatty Acid, Dicarboxylate | 2.45 | 1.27 | 1.93 |
| Fatty Acid Metabolism (Acyl Glycine) | 3.97 | 4.29 | 6.53 |
| Primary Bile Acid Metabolism | 2.75 | 4.29 | 5.44 |
| Fatty Acid, Monohydroxy | 1.10 | 1.07 | 0.00 |
| Lysophospholipid | 0.22 | 0.84 | 1.93 |
| Phosphatidylcholine (PC) | 0.35 | 0.68 | 0.34 |
| Fatty Acid Metabolism | 3.31 | 4.29 | 6.53 |
| Glycosyl PE | 2.64 | 3.44 | 5.22 |
| Fatty Acid, Amino | 6.61 | 4.29 | 0.00 |
| Phosphatidylinositol (PI) | 0.66 | 0.00 | 0.00 |
| Sphingolipid Synthesis | 1.65 | 2.15 | 0.00 |
| Fatty Acid Metabolism (Acyl Choline) | 3.31 | 0.00 | 0.00 |
| Ceramides | 0.66 | 0.00 | 1.31 |
| Diacylglycerol | 0.24 | 0.64 | 0.48 |
| Fatty Acid Metabolism (Acyl Carnitine, Dicarboxylate) | 2.20 | 0.00 | 0.00 |
| Plasmalogen | 0.37 | 0.00 | 0.00 |
| Mevalonate Metabolism | 2.20 | 0.00 | 0.00 |
| Sphingomyelins | 0.25 | 0.33 | 0.00 |
| Phosphatidylglycerol (PG) | 1.32 | 5.15 | 2.61 |
| Phospholipid Metabolism | 0.66 | 0.86 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Monounsaturated) | 0.94 | 0.00 | 0.00 |
| Fatty Acid, Branched | 1.32 | 5.15 | 0.00 |
| Ketone Bodies | 6.61 | 0.00 | 0.00 |
| Pregnenolone Steroids | 6.61 | 8.59 | 13.05 |
| Hexosylceramides (HCER) | 3.31 | 0.00 | 0.00 |
| Progestin Steroids | 6.61 | 8.59 | 13.05 |
| Fatty Acid, Dihydroxy | 0.00 | 0.00 | 0.00 |
| Dihydrosphingomyelins | 0.00 | 0.00 | 0.00 |
| Long-Chain Polyunsaturated Fatty Acid (n3 and n6) | 0.00 | 4.07 | 0.00 |
| Endocannabinoid | 0.00 | 1.72 | 0.00 |
| Carnitine Metabolism | 0.00 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Medium-Chain) | 0.00 | 0.00 | 0.00 |
| Eicosanoid | 0.00 | 0.00 | 0.00 |
| Secondary Bile Acid Metabolism | 0.00 | 0.00 | 0.00 |
| Monoacylglycerol | 0.00 | 3.62 | 0.00 |
| Glycerolipid Metabolism | 0.00 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Polyunsaturated) | 0.00 | 0.00 | 3.92 |
| Lysoplasmalogen | 0.00 | 0.00 | 3.73 |
| Long-Chain Saturated Fatty Acid | 0.00 | 4.91 | 0.00 |
| Phosphatidylethanolamine (PE) | 0.00 | 1.91 | 2.18 |
| Fatty Acid, Oxidized | 0.00 | 0.00 | 0.00 |
| Sterol | 0.00 | 0.00 | 0.00 |
| Docosanoid | 0.00 | 4.29 | 0.00 |
| Long-Chain Monounsaturated Fatty Acid | 0.00 | 1.23 | 0.00 |
| Phosphatidylserine (PS) | 0.00 | 0.00 | 0.00 |
| Medium-Chain Fatty Acid | 0.00 | 8.59 | 0.00 |
| Dihydroceramides | 0.00 | 4.29 | 0.00 |
| Lactosylceramides (LCER) | 0.00 | 0.00 | 0.00 |
| Fatty Acid Metabolism (also BCAA Metabolism) | 0.00 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Long-Chain Saturated) | 0.00 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Hydroxy) | 0.00 | 0.00 | 0.00 |
| Sphingosines | 0.00 | 2.86 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Short-Chain) | 0.00 | 0.00 | 0.00 |
| Inositol Metabolism | 0.00 | 0.00 | 0.00 |
| Fatty Acid Synthesis | 0.00 | 0.00 | 0.00 |
| Sub-Pathway | Pathway Enrichment Score 1 | ||
|---|---|---|---|
| Gain | OCM | Gain × OCM | |
| Lysophospholipid | 1.19 | 0.59 | 1.06 |
| Sphingomyelins | 1.92 | 1.04 | 0.62 |
| Phosphatidylcholine (PC) | 1.25 | 0.24 | 0.00 |
| Diacylglycerol | 1.43 | 1.01 | 1.80 |
| Plasmalogen | 1.26 | 0.50 | 0.90 |
| Monoacylglycerol | 1.19 | 1.43 | 3.41 |
| Phosphatidylethanolamine (PE) | 1.13 | 0.50 | 0.00 |
| Phosphatidylinositol (PI) | 1.81 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Long-Chain Saturated) | 2.26 | 0.00 | 0.00 |
| Fatty Acid, Dicarboxylate | 0.59 | 0.34 | 0.60 |
| Long-Chain Polyunsaturated Fatty Acid (n3 and n6) | 0.71 | 0.00 | 1.71 |
| Phospholipid Metabolism | 1.36 | 1.81 | 0.00 |
| Lysoplasmalogen | 1.94 | 1.29 | 2.32 |
| Fatty Acid Metabolism (Acyl Carnitine, Hydroxy) | 1.70 | 1.13 | 2.03 |
| Fatty Acid, Monohydroxy | 0.47 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Polyunsaturated) | 1.13 | 0.91 | 1.62 |
| Fatty Acid Metabolism (Acyl Carnitine, Monounsaturated) | 1.62 | 1.29 | 2.32 |
| Phosphatidylserine (PS) | 1.62 | 2.59 | 0.00 |
| Dihydrosphingomyelins | 1.81 | 0.00 | 0.00 |
| Ceramides | 0.91 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Glycine) | 0.91 | 0.00 | 0.00 |
| Long-Chain Monounsaturated Fatty Acid | 1.29 | 0.00 | 0.00 |
| Endocannabinoid | 0.68 | 0.91 | 8.10 |
| Fatty Acid Metabolism (Acyl Carnitine, Medium-Chain) | 1.70 | 0.00 | 0.00 |
| Eicosanoid | 0.85 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Dicarboxylate) | 2.26 | 0.00 | 0.00 |
| Phosphatidylglycerol (PG) | 1.36 | 0.00 | 0.00 |
| Glycosyl PE | 1.36 | 1.81 | 3.24 |
| Sterol | 1.70 | 0.00 | 0.00 |
| Fatty Acid Metabolism (also BCAA Metabolism) | 0.97 | 1.29 | 0.00 |
| Fatty Acid, Dihydroxy | 0.91 | 0.00 | 0.00 |
| Secondary Bile Acid Metabolism | 0.41 | 1.65 | 2.95 |
| Long-Chain Saturated Fatty Acid | 0.65 | 0.00 | 0.00 |
| Fatty Acid, Branched | 0.91 | 0.00 | 3.24 |
| Hexosylceramides (HCER) | 2.26 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Carnitine, Short-Chain) | 2.26 | 0.00 | 0.00 |
| Primary Bile Acid Metabolism | 0.19 | 1.51 | 0.00 |
| Sphingolipid Synthesis | 0.57 | 2.26 | 0.00 |
| Carnitine Metabolism | 1.13 | 0.00 | 0.00 |
| Mevalonate Metabolism | 0.75 | 0.00 | 0.00 |
| Glycerolipid Metabolism | 0.75 | 0.00 | 0.00 |
| Fatty Acid Metabolism | 0.57 | 2.26 | 4.05 |
| Fatty Acid, Oxidized | 2.26 | 0.00 | 0.00 |
| Docosanoid | 1.13 | 0.00 | 0.00 |
| Medium-Chain Fatty Acid | 2.26 | 0.00 | 0.00 |
| Dihydroceramides | 1.13 | 0.00 | 0.00 |
| Lactosylceramides (LCER) | 2.26 | 0.00 | 0.00 |
| Fatty Acid, Amino | 1.13 | 4.53 | 0.00 |
| Inositol Metabolism | 1.13 | 4.53 | 0.00 |
| Progestin Steroids | 2.26 | 0.00 | 0.00 |
| Fatty Acid Metabolism (Acyl Choline) | 0.00 | 0.00 | 0.00 |
| Sphingosines | 0.00 | 3.02 | 0.00 |
| Ketone Bodies | 0.00 | 0.00 | 0.00 |
| Pregnenolone Steroids | 0.00 | 0.00 | 0.00 |
| Fatty Acid Synthesis | 0.00 | 0.00 | 0.00 |
| Sub-Pathway | Biochemical Name | Two-Way ANOVA Main Effects 1 | Two-Way ANOVA Contrasts 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gain | Supp. | Gain × Supp. | CON + OCM | RES + OCM | RES − OCM | RES + OCM | RES | +OCM | ||
| CON − OCM | RES − OCM | CON − OCM | CON + OCM | CON | −OCM | |||||
| Fatty Acid Metabolism (Acyl Carnitine, Monounsaturated) | cis-4-decenoylcarnitine (C10:1) | 0.04 | 0.21 | 0.57 | 1.22 | 1.17 | 0.8 | 0.77 | 0.79 | 1.2 |
| Fatty Acid Metabolism (Acyl Carnitine, Polyunsaturated) | dihomo-linoleoylcarnitine (C20:2) | 0.29 | 0.74 | 0.04 | 0.79 | 1.43 | 0.88 | 1.59 | 1.24 | 1.11 |
| docosatrienoylcarnitine (C22:3) | 0.44 | 0.69 | 0.04 | 0.65 | 1.42 | 0.85 | 1.87 | 1.36 | 1.04 | |
| Fatty Acid Metabolism (Acyl Carnitine, Dicarboxylate) | pimeloylcarnitine/3-methyladipoylcarnitine (C7-DC) | 0.01 | 0.2 | 0.56 | 1.12 | 1.34 | 0.57 | 0.68 | 0.63 | 1.23 |
| Plasmalogen | 1-palmityl-2-palmitoyl-GPC (O-16:0/16:0) | 0.02 | 0.53 | 0.81 | 1.03 | 1.02 | 1.11 | 1.1 | 1.11 | 1.03 |
| Lysoplasmalogen Monoacylglycerol | 1-palmityl-GPC (O-16:0) | 0.53 | 0.1 | 0.01 | 1.48 | 0.9 | 1.33 | 0.8 | 1.07 | 1.19 |
| 1-stearyl-GPC (O-18:0) | 0.53 | 0.17 | 0.03 | 1.43 | 0.93 | 1.14 | 0.74 | 0.94 | 1.18 | |
| 1-myristoylglycerol (14:0) | 0.17 | 0.05 | 0.78 | 0.67 | 0.51 | 1.58 | 1.2 | 1.39 | 0.59 | |
| 1-pentadecanoylglycerol (15:0) | 0.44 | 0.04 | 0.64 | 0.73 | 0.54 | 1.41 | 1.04 | 1.23 | 0.64 | |
| 1-linoleoylglycerol (18:2) | 0.81 | 0.04 | 0.62 | 0.76 | 0.48 | 1.47 | 0.92 | 1.2 | 0.62 | |
| 1-docosahexaenoylglycerol (22:6) | 0.29 | 0.02 | 0.73 | 0.67 | 0.43 | 1.78 | 1.14 | 1.46 | 0.55 | |
| 2-myristoylglycerol (14:0) | 0.25 | 0.04 | 0.89 | 0.58 | 0.58 | 1.38 | 1.38 | 1.38 | 0.58 | |
| 2-docosahexaenoylglycerol (22:6) | 0.47 | 0.04 | 0.95 | 0.63 | 0.48 | 1.54 | 1.17 | 1.36 | 0.56 | |
| 1-heptadecenoylglycerol (17:1) | 0.21 | 0.02 | 0.4 | 0.49 | 0.54 | 1.3 | 1.43 | 1.37 | 0.52 | |
| 2-heptadecenoylglycerol (17:1) | 0.22 | 0.03 | 0.33 | 0.52 | 0.62 | 1.23 | 1.46 | 1.35 | 0.57 | |
| Diacylglycerol | linoleoyl-docosahexaenoyl-glycerol (18:2/22:6) [2] | 0.77 | 0.03 | 0.61 | 0.81 | 0.63 | 1.04 | 0.81 | 0.93 | 0.72 |
| Hexosylceramides (HCER) | glycosyl-N-stearoyl-sphingosine (d18:1/18:0) | 0.02 | 0.81 | 0.77 | 0.94 | 1.04 | 1.2 | 1.33 | 1.27 | 0.99 |
| Sphingomyelins | sphingomyelin (d18:2/23:1) | 0.28 | 0.03 | 0.64 | 0.77 | 0.86 | 1.05 | 1.17 | 1.11 | 0.82 |
| Sub-Pathway | Biochemical Name | Two-Way ANOVA Main Effects 1 | Two-Way ANOVA Contrasts 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gain | Supp. | Gain × Supp. | CON + OCM | RES + OCM | RES − OCM | RES + OCM | RES | +OCM | ||
| CON − OCM | RES − OCM | CON − OCM | CON + OCM | CON | −OCM | |||||
| Fatty Acid Metabolism (Acyl Carnitine, Short-Chain) | acetylcarnitine (C2) | 0.01 | 0.42 | 0.57 | 1 | 0.84 | 2.02 | 1.71 | 1.87 | 0.92 |
| isocaproylcarnitine | 0.01 | 0.64 | 0.92 | 1.05 | 1.04 | 2 | 1.97 | 1.99 | 1.05 | |
| Fatty Acid Metabolism (Acyl Carnitine, Medium-Chain) | hexanoylcarnitine (C6) | 0.01 | 0.85 | 0.9 | 1.02 | 1 | 2.01 | 1.96 | 1.99 | 1.01 |
| 2-methylhexanoylcarnitine | 0.01 | 0.58 | 0.57 | 0.94 | 1.18 | 1.52 | 1.89 | 1.71 | 1.06 | |
| laurylcarnitine (C12) | 0.01 | 0.2 | 0.69 | 1.37 | 1.14 | 2 | 1.67 | 1.84 | 1.26 | |
| Fatty Acid Metabolism (Acyl Carnitine, Long-Chain Saturated) | myristoylcarnitine (C14) | 0.01 | 0.83 | 0.94 | 0.97 | 0.96 | 1.8 | 1.79 | 1.8 | 0.97 |
| pentadecanoylcarnitine (C15) | 0.01 | 0.9 | 0.79 | 0.91 | 1.09 | 2.34 | 2.79 | 2.57 | 1 | |
| palmitoylcarnitine (C16) | 0.01 | 0.7 | 0.22 | 1.05 | 0.86 | 2.01 | 1.65 | 1.83 | 0.96 | |
| margaroylcarnitine (C17) | 0.01 | 0.62 | 0.77 | 1.07 | 1.06 | 2.7 | 2.68 | 2.69 | 1.07 | |
| stearoylcarnitine (C18) | 0.01 | 0.32 | 0.26 | 1.01 | 0.83 | 2.03 | 1.66 | 1.85 | 0.92 | |
| arachidoylcarnitine (C20) | 0.01 | 0.6 | 0.45 | 0.87 | 1 | 1.78 | 2.06 | 1.92 | 0.94 | |
| behenoylcarnitine (C22) | 0.01 | 0.71 | 0.22 | 0.79 | 1.12 | 1.89 | 2.68 | 2.29 | 0.96 | |
| lignoceroylcarnitine (C24) | 0.01 | 0.67 | 0.82 | 0.96 | 0.87 | 2.83 | 2.56 | 2.7 | 0.92 | |
| Fatty Acid Metabolism (Acyl Carnitine, Monounsaturated) | 5-dodecenoylcarnitine (C12:1) | 0.01 | 0.98 | 0.69 | 1.06 | 0.93 | 3.13 | 2.77 | 2.95 | 1 |
| palmitoleoylcarnitine (C16:1) | 0.01 | 0.54 | 0.35 | 1.12 | 0.97 | 2.29 | 1.98 | 2.14 | 1.05 | |
| oleoylcarnitine (C18:1) | 0.01 | 0.54 | 0.58 | 0.96 | 0.88 | 1.85 | 1.69 | 1.77 | 0.92 | |
| eicosenoylcarnitine (C20:1) | 0.01 | 0.01 | 0.01 | 0.99 | 0.54 | 2.24 | 1.23 | 1.74 | 0.77 | |
| erucoylcarnitine (C22:1) | 0.01 | 0.51 | 0.36 | 0.7 | 1.02 | 2.72 | 3.99 | 3.36 | 0.86 | |
| Fatty Acid Metabolism (Acyl Carnitine, Polyunsaturated) | linoleoylcarnitine (C18:2) | 0.01 | 0.9 | 0.46 | 1.06 | 0.91 | 1.73 | 1.47 | 1.6 | 0.99 |
| linolenoylcarnitine (C18:3) | 0.01 | 0.46 | 0.48 | 0.9 | 0.98 | 1.25 | 1.35 | 1.3 | 0.94 | |
| arachidonoylcarnitine (C20:4) | 0.01 | 0.56 | 0.29 | 1.12 | 0.98 | 1.65 | 1.43 | 1.54 | 1.05 | |
| adrenoylcarnitine (C22:4) | 0.01 | 0.76 | 0.97 | 0.98 | 0.95 | 0.72 | 0.71 | 0.72 | 0.97 | |
| Fatty Acid Metabolism (Acyl Carnitine, Dicarboxylate) | adipoylcarnitine (C6-DC) | 0.01 | 0.43 | 0.59 | 0.88 | 0.94 | 2.35 | 2.52 | 2.44 | 0.91 |
| pimeloylcarnitine/3-methyladipoylcarnitine (C7-DC) | 0.01 | 0.92 | 0.72 | 1.02 | 0.95 | 2.01 | 1.87 | 1.94 | 0.99 | |
| suberoylcarnitine (C8-DC) | 0.01 | 0.4 | 0.48 | 0.91 | 0.97 | 1.75 | 1.88 | 1.82 | 0.94 | |
| Fatty Acid Metabolism (Acyl Carnitine, Hydroxy) | (R)-3-hydroxybutyrylcarnitine | 0.01 | 0.39 | 0.83 | 1.1 | 1.05 | 1.39 | 1.33 | 1.36 | 1.08 |
| (S)-3-hydroxybutyrylcarnitine | 0.01 | 0.32 | 0.47 | 0.95 | 0.78 | 1.62 | 1.33 | 1.48 | 0.87 | |
| 3-hydroxyhexanoylcarnitine (1) | 0.01 | 0.58 | 0.69 | 0.79 | 0.94 | 1.62 | 1.93 | 1.78 | 0.87 | |
| 3-hydroxypalmitoylcarnitine | 0.01 | 0.82 | 0.5 | 1.16 | 0.88 | 2.27 | 1.72 | 2 | 1.02 | |
| 3-hydroxyoleoylcarnitine | 0.01 | 0.86 | 0.52 | 1.06 | 0.92 | 2.24 | 1.94 | 2.09 | 0.99 | |
| Plasmalogen | 1-palmityl-2-palmitoyl-GPC (O-16:0/16:0) | 0.01 | 0.43 | 0.89 | 0.95 | 0.92 | 2.02 | 1.97 | 2 | 0.94 |
| 1-palmityl-2-stearoyl-GPC (O-16:0/18:0) | 0.01 | 0.34 | 1 | 0.93 | 0.93 | 1.64 | 1.64 | 1.64 | 0.93 | |
| 1-palmityl-2-linoleoyl-GPC (O-16:0/18:2) | 0.01 | 0.69 | 0.24 | 1.11 | 0.94 | 1.36 | 1.15 | 1.26 | 1.03 | |
| 1-stearyl-2-arachidonoyl-GPC (O-18:0/20:4) | 0.01 | 0.64 | 0.93 | 0.96 | 0.96 | 1.64 | 1.64 | 1.64 | 0.96 | |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (P-16:0/18:2) | 0.01 | 0.85 | 0.71 | 0.99 | 0.91 | 0.56 | 0.52 | 0.54 | 0.95 | |
| 1-(1-enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0) | 0.01 | 0.66 | 0.83 | 0.98 | 0.96 | 1.51 | 1.47 | 1.49 | 0.97 | |
| 1-(1-enyl-palmitoyl)-2-arachidonoyl-GPE (P-16:0/20:4) | 0.01 | 0.47 | 0.3 | 0.89 | 1.04 | 1.15 | 1.34 | 1.25 | 0.97 | |
| 1-(1-enyl-stearoyl)-2-oleoyl-GPE (P-18:0/18:1) | 0.01 | 0.58 | 0.3 | 1.1 | 0.97 | 1.55 | 1.36 | 1.46 | 1.04 | |
| 1-(1-enyl-stearoyl)-2-arachidonoyl-GPE (P-18:0/20:4) | 0.01 | 0.66 | 0.26 | 1.08 | 0.97 | 1.56 | 1.42 | 1.49 | 1.03 | |
| Lysoplasmalogen | 1-palmityl-GPC (O-16:0) | 0.01 | 0.92 | 0.51 | 0.92 | 1.13 | 2.27 | 2.79 | 2.53 | 1.03 |
| 1-stearyl-GPC (O-18:0) | 0.01 | 0.38 | 0.78 | 0.86 | 0.96 | 2.07 | 2.32 | 2.2 | 0.91 | |
| 1-stearyl-GPE (O-18:0) | 0.01 | 0.43 | 0.61 | 1.02 | 1.15 | 1.72 | 1.93 | 1.83 | 1.09 | |
| 1-(1-enyl-palmitoyl)-GPE (P-16:0) | 0.01 | 0.57 | 0.28 | 1.02 | 0.85 | 1.49 | 1.23 | 1.36 | 0.94 | |
| 1-(1-enyl-oleoyl)-GPE (P-18:1) | 0.01 | 0.48 | 0.22 | 1.05 | 0.8 | 1.68 | 1.29 | 1.49 | 0.93 | |
| Monoacylglycerol | 1-myristoylglycerol (14:0) | 0.01 | 0.22 | 0.17 | 0.98 | 0.54 | 2.35 | 1.3 | 1.83 | 0.76 |
| 1-palmitoleoylglycerol (16:1) | 0.01 | 0.24 | 0.38 | 0.84 | 0.45 | 4.93 | 2.65 | 3.79 | 0.65 | |
| 1-oleoylglycerol (18:1) | 0.01 | 0.44 | 0.59 | 1.15 | 0.59 | 4.9 | 2.53 | 3.72 | 0.87 | |
| 1-linoleoylglycerol (18:2) | 0.05 | 0.8 | 0.01 | 1.45 | 0.53 | 2.63 | 0.97 | 1.8 | 0.99 | |
| 1-docosahexaenoylglycerol (22:6) | 0.1 | 0.04 | 0.09 | 0.92 | 0.51 | 1.79 | 0.99 | 1.39 | 0.72 | |
| 2-myristoylglycerol (14:0) | 0.81 | 0.01 | 0.66 | 0.63 | 0.56 | 1.02 | 0.91 | 0.97 | 0.6 | |
| 2-oleoylglycerol (18:1) | 0.01 | 0.15 | 0.42 | 0.91 | 0.6 | 2.91 | 1.91 | 2.41 | 0.76 | |
| 2-linoleoylglycerol (18:2) | 0.01 | 0.38 | 0.02 | 1.22 | 0.56 | 2.31 | 1.06 | 1.69 | 0.89 | |
| 2-arachidonoylglycerol (20:4) | 0.02 | 0.63 | 0.03 | 1.31 | 0.63 | 2.23 | 1.06 | 1.65 | 0.97 | |
| 2-docosahexaenoylglycerol (22:6) | 0.01 | 0.01 | 0.03 | 0.94 | 0.58 | 1.68 | 1.04 | 1.36 | 0.76 | |
| 1-heptadecenoylglycerol (17:1) | 0.01 | 0.35 | 0.08 | 1.39 | 0.41 | 6.17 | 1.8 | 3.99 | 0.9 | |
| 2-heptadecenoylglycerol (17:1) | 0.01 | 0.38 | 0.34 | 1.02 | 0.56 | 3.19 | 1.74 | 2.47 | 0.79 | |
| Diacylglycerol | diacylglycerol (12:0/18:1, 14:0/16:1, 16:0/14:1) [2] | 0.01 | 0.11 | 0.59 | 0.57 | 0.81 | 4.55 | 6.42 | 5.49 | 0.69 |
| diacylglycerol (14:0/18:1, 16:0/16:1) [2] | 0.01 | 0.69 | 0.66 | 0.86 | 0.86 | 3.53 | 3.53 | 3.53 | 0.86 | |
| diacylglycerol (16:1/18:2 [2], 16:0/18:3 [1]) | 0.01 | 0.95 | 0.77 | 0.98 | 0.95 | 3.26 | 3.15 | 3.21 | 0.97 | |
| palmitoyl-oleoyl-glycerol (16:0/18:1) [1] | 0.01 | 0.78 | 0.78 | 1 | 0.91 | 2.24 | 2.03 | 2.14 | 0.96 | |
| palmitoyl-oleoyl-glycerol (16:0/18:1) [2] | 0.01 | 0.66 | 0.44 | 1 | 0.87 | 3.01 | 2.61 | 2.81 | 0.94 | |
| palmitoyl-linoleoyl-glycerol (16:0/18:2) [2] | 0.01 | 0.28 | 0.29 | 1.02 | 0.83 | 2.13 | 1.73 | 1.93 | 0.93 | |
| palmitoyl-linolenoyl-glycerol (16:0/18:3) [2] | 0.01 | 0.21 | 0.65 | 0.76 | 0.85 | 1.98 | 2.23 | 2.11 | 0.81 | |
| palmitoyl-arachidonoyl-glycerol (16:0/20:4) [2] | 0.01 | 0.4 | 0.32 | 1.02 | 0.88 | 1.89 | 1.63 | 1.76 | 0.95 | |
| palmitoyl-docosahexaenoyl-glycerol (16:0/22:6) [2] | 0.01 | 0.45 | 0.65 | 0.99 | 0.9 | 1.42 | 1.29 | 1.36 | 0.95 | |
| oleoyl-linoleoyl-glycerol (18:1/18:2) [1] | 0.01 | 0.6 | 0.19 | 1.28 | 0.74 | 3.01 | 1.74 | 2.38 | 1.01 | |
| oleoyl-linoleoyl-glycerol (18:1/18:2) [2] | 0.01 | 0.63 | 0.81 | 0.98 | 0.8 | 2.44 | 2.01 | 2.23 | 0.89 | |
| stearoyl-arachidonoyl-glycerol (18:0/20:4) [2] | 0.01 | 0.39 | 0.17 | 1.2 | 0.97 | 1.49 | 1.2 | 1.35 | 1.09 | |
| oleoyl-arachidonoyl-glycerol (18:1/20:4) [2] | 0.01 | 0.92 | 0.31 | 1.12 | 0.94 | 1.46 | 1.23 | 1.35 | 1.03 | |
| linoleoyl-arachidonoyl-glycerol (18:2/20:4) [2] | 0.02 | 0.45 | 0.21 | 1.1 | 0.67 | 2.2 | 1.33 | 1.77 | 0.89 | |
| Hexosylceramides (HCER) | glycosyl-N-palmitoyl-sphingosine (d18:1/16:0) | 0.01 | 0.65 | 0.5 | 1.02 | 0.92 | 1.57 | 1.42 | 1.5 | 0.97 |
| glycosyl-N-stearoyl-sphingosine (d18:1/18:0) | 0.01 | 0.38 | 0.13 | 1.07 | 0.82 | 1.94 | 1.49 | 1.72 | 0.95 | |
| Lactosylceramides (LCER) | lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) | 0.01 | 0.33 | 0.45 | 0.92 | 0.85 | 1.35 | 1.26 | 1.31 | 0.89 |
| Dihydrosphingomyelins | myristoyl dihydrosphingomyelin (d18:0/14:0) | 0.01 | 0.5 | 0.74 | 0.91 | 0.94 | 1.83 | 1.88 | 1.86 | 0.93 |
| palmitoyl dihydrosphingomyelin (d18:0/16:0) | 0.01 | 0.62 | 0.22 | 1.13 | 0.95 | 1.56 | 1.32 | 1.44 | 1.04 | |
| behenoyl dihydrosphingomyelin (d18:0/22:0) | 0.01 | 0.43 | 0.37 | 1.16 | 0.98 | 1.67 | 1.42 | 1.55 | 1.07 | |
| sphingomyelin (d18:0/20:0, d16:0/22:0) | 0.01 | 0.39 | 0.18 | 1.24 | 0.93 | 1.62 | 1.22 | 1.42 | 1.09 | |
| Sphingomyelins | palmitoyl sphingomyelin (d18:1/16:0) | 0.02 | 0.3 | 0.46 | 0.95 | 0.99 | 1.05 | 1.09 | 1.07 | 0.97 |
| behenoyl sphingomyelin (d18:1/22:0) | 0.01 | 0.62 | 0.91 | 0.99 | 0.98 | 1.32 | 1.31 | 1.32 | 0.99 | |
| tricosanoyl sphingomyelin (d18:1/23:0) | 0.01 | 0.83 | 0.99 | 1 | 0.99 | 1.29 | 1.27 | 1.28 | 1 | |
| sphingomyelin (d18:2/18:1) | 0.01 | 0.35 | 0.24 | 1.23 | 0.96 | 2.09 | 1.63 | 1.86 | 1.1 | |
| sphingomyelin (d18:2/23:1) | 0.02 | 0.4 | 0.92 | 0.84 | 0.89 | 1.38 | 1.47 | 1.43 | 0.87 | |
| sphingomyelin (d17:1/14:0, d16:1/15:0) | 0.04 | 0.38 | 0.98 | 0.94 | 0.96 | 0.88 | 0.89 | 0.89 | 0.95 | |
| sphingomyelin (d18:2/14:0, d18:1/14:1) | 0.01 | 0.03 | 0.42 | 0.79 | 0.9 | 0.64 | 0.73 | 0.69 | 0.85 | |
| sphingomyelin (d17:1/16:0, d18:1/15:0, d16:1/17:0) | 0.01 | 0.1 | 0.74 | 0.92 | 0.95 | 1.18 | 1.22 | 1.2 | 0.94 | |
| sphingomyelin (d17:2/16:0, d18:2/15:0) | 0.01 | 0.4 | 0.31 | 0.84 | 1.04 | 0.53 | 0.66 | 0.6 | 0.94 | |
| sphingomyelin (d18:2/16:0, d18:1/16:1) | 0.03 | 0.68 | 0.47 | 0.96 | 1.01 | 0.9 | 0.95 | 0.93 | 0.99 | |
| sphingomyelin (d18:1/17:0, d17:1/18:0, d19:1/16:0) | 0.01 | 0.35 | 0.35 | 0.88 | 0.98 | 0.69 | 0.77 | 0.73 | 0.93 | |
| sphingomyelin (d18:1/19:0, d19:1/18:0) | 0.01 | 0.24 | 0.2 | 1.01 | 0.86 | 1.44 | 1.22 | 1.33 | 0.94 | |
| sphingomyelin (d18:1/20:0, d16:1/22:0) | 0.01 | 0.17 | 0.16 | 1 | 0.9 | 1.34 | 1.21 | 1.28 | 0.95 | |
| sphingomyelin (d18:1/20:1, d18:2/20:0) | 0.03 | 0.87 | 0.79 | 1.03 | 0.99 | 0.78 | 0.76 | 0.77 | 1.01 | |
| sphingomyelin (d18:1/21:0, d17:1/22:0, d16:1/23:0) | 0.01 | 0.04 | 0.77 | 0.93 | 0.91 | 1.41 | 1.39 | 1.4 | 0.92 | |
| sphingomyelin (d18:2/21:0, d16:2/23:0) | 0.01 | 0.26 | 0.35 | 0.96 | 0.82 | 1.63 | 1.39 | 1.51 | 0.89 | |
| sphingomyelin (d18:1/22:1, d18:2/22:0, d16:1/24:1) | 0.01 | 0.15 | 0.22 | 0.99 | 0.83 | 1.42 | 1.2 | 1.31 | 0.91 | |
| sphingomyelin (d18:2/23:0, d18:1/23:1, d17:1/24:1) | 0.01 | 0.29 | 0.72 | 0.93 | 0.95 | 1.25 | 1.28 | 1.27 | 0.94 | |
| sphingomyelin (d18:1/24:1, d18:2/24:0) | 0.01 | 0.19 | 0.59 | 0.96 | 0.92 | 1.31 | 1.25 | 1.28 | 0.94 | |
| sphingomyelin (d18:2/24:1, d18:1/24:2) | 0.01 | 0.95 | 0.38 | 1.06 | 0.95 | 1.29 | 1.15 | 1.22 | 1.01 | |
| sphingomyelin (d18:1/25:0, d19:0/24:1, d20:1/23:0, d19:1/24:0) | 0.01 | 0.78 | 0.69 | 1.01 | 0.97 | 0.84 | 0.81 | 0.83 | 0.99 | |
| Sterol | cholesterol | 0.01 | 0.2 | 0.83 | 0.97 | 0.96 | 1.16 | 1.14 | 1.15 | 0.97 |
| 4-cholesten-3-one | 0.04 | 0.97 | 0.93 | 1 | 0.95 | 1.26 | 1.2 | 1.23 | 0.98 | |
| 7-hydroxycholesterol (alpha or beta) | 0.01 | 0.59 | 0.61 | 1.01 | 1.43 | 1.67 | 2.38 | 2.03 | 1.22 | |
| Secondary Bile Acid Metabolism | taurodeoxycholate | 0.56 | 0.03 | 0.95 | 1.91 | 1.43 | 1.09 | 0.82 | 0.96 | 1.67 |
| glycoursodeoxycholate | 0.01 | 0.15 | 0.02 | 0.35 | 1.52 | 2.15 | 9.29 | 5.72 | 0.94 | |
| tauroursodeoxycholate | 0.01 | 0.23 | 0.3 | 1 | 1.45 | 2.86 | 4.13 | 3.5 | 1.23 | |
| glycocholenate sulfate | 0.2 | 0.01 | 0.79 | 0.67 | 0.73 | 1.09 | 1.18 | 1.14 | 0.7 | |
| taurocholenate sulfate | 0.14 | 0.14 | 0.05 | 1.97 | 0.91 | 1.23 | 0.57 | 0.9 | 1.44 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safain, K.S.; Crouse, M.S.; Syring, J.G.; Entzie, Y.L.; King, L.E.; Ward, A.K.; Reynolds, L.P.; Borowicz, P.P.; Dahlen, C.R.; Swanson, K.C.; et al. Early Gestational Hepatic Lipidomic Profiles Are Modulated by One-Carbon Metabolite Supplementation and Nutrient Restriction in Beef Heifers and Fetuses. Metabolites 2025, 15, 302. https://doi.org/10.3390/metabo15050302
Safain KS, Crouse MS, Syring JG, Entzie YL, King LE, Ward AK, Reynolds LP, Borowicz PP, Dahlen CR, Swanson KC, et al. Early Gestational Hepatic Lipidomic Profiles Are Modulated by One-Carbon Metabolite Supplementation and Nutrient Restriction in Beef Heifers and Fetuses. Metabolites. 2025; 15(5):302. https://doi.org/10.3390/metabo15050302
Chicago/Turabian StyleSafain, Kazi Sarjana, Matthew S. Crouse, Jessica G. Syring, Yssi L. Entzie, Layla E. King, Alison K. Ward, Lawrence P. Reynolds, Pawel P. Borowicz, Carl R. Dahlen, Kendall C. Swanson, and et al. 2025. "Early Gestational Hepatic Lipidomic Profiles Are Modulated by One-Carbon Metabolite Supplementation and Nutrient Restriction in Beef Heifers and Fetuses" Metabolites 15, no. 5: 302. https://doi.org/10.3390/metabo15050302
APA StyleSafain, K. S., Crouse, M. S., Syring, J. G., Entzie, Y. L., King, L. E., Ward, A. K., Reynolds, L. P., Borowicz, P. P., Dahlen, C. R., Swanson, K. C., & Caton, J. S. (2025). Early Gestational Hepatic Lipidomic Profiles Are Modulated by One-Carbon Metabolite Supplementation and Nutrient Restriction in Beef Heifers and Fetuses. Metabolites, 15(5), 302. https://doi.org/10.3390/metabo15050302







